Protein Sensing Device with Multi-Recognition Ability Composed of Self-Organized Glycopeptide Bundle
Abstract
1. Introduction
2. Results and Discussion
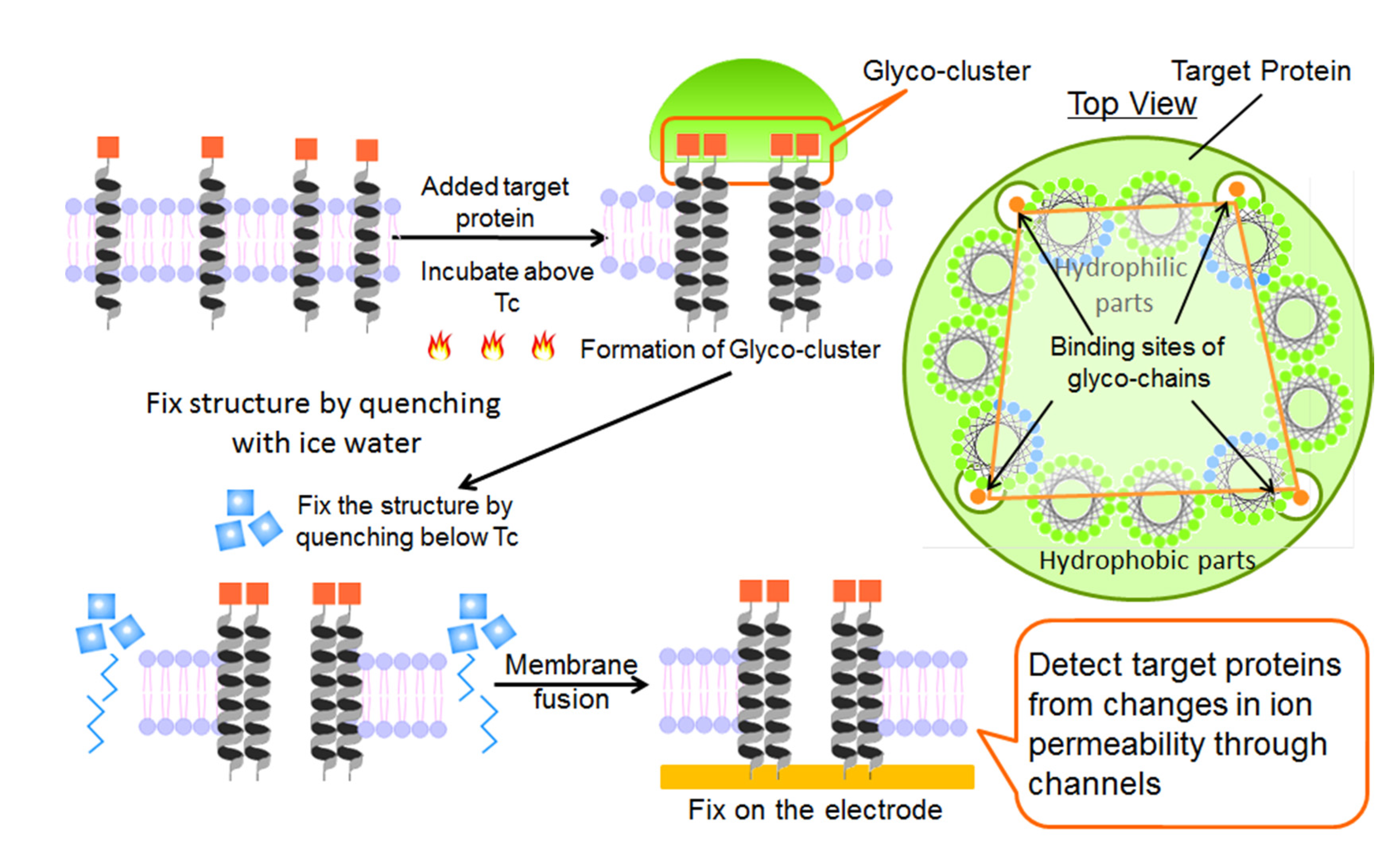
2.1. Fabrication of Protein Sensing Systems Composed of Target Protein-Induced Self-Organized Glycopeptide Bundle
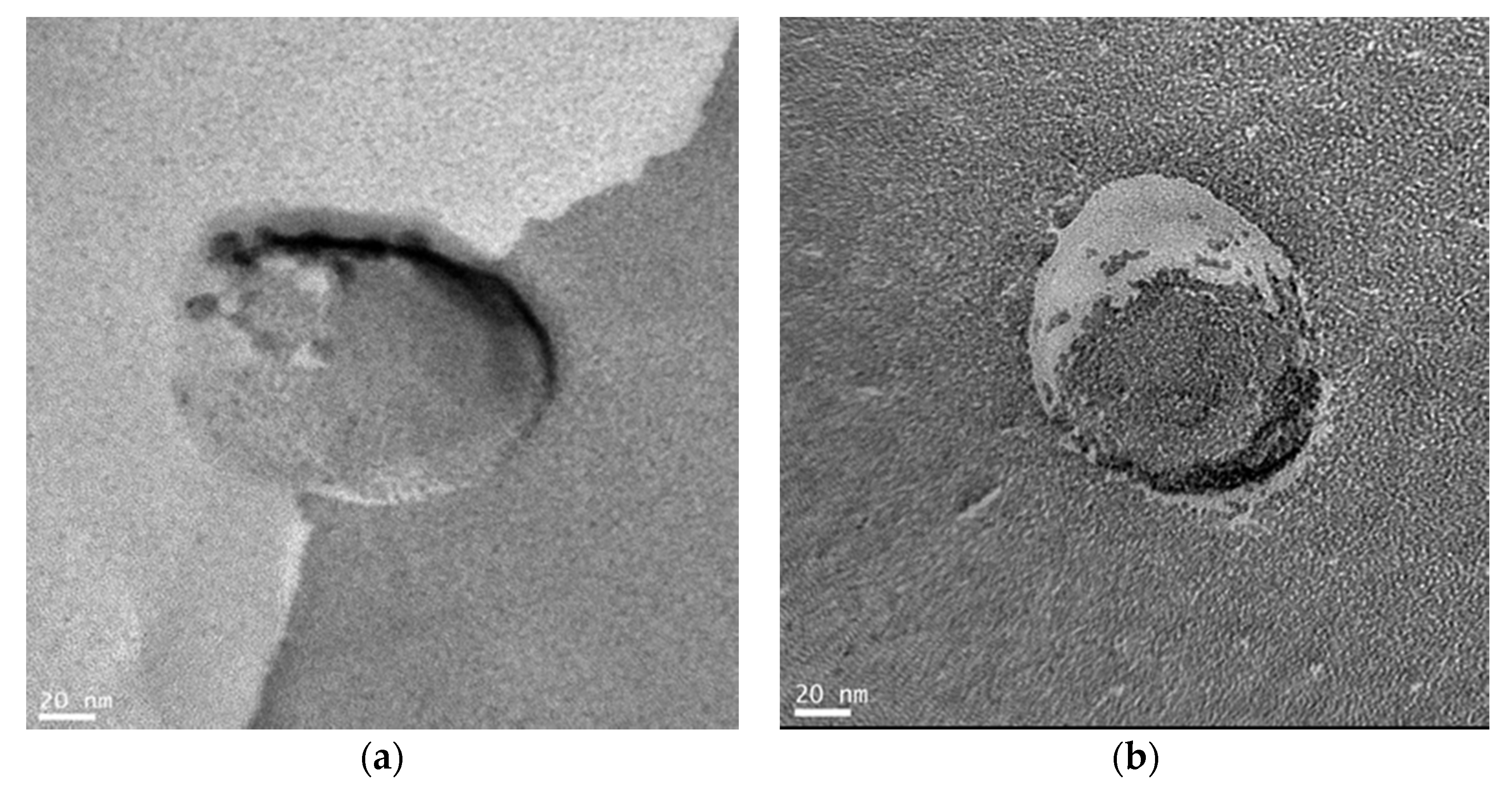
2.2. Structure of Glycopeptide Bundle Induced by Target Protein in the Lipid Bilayer Membrane
2.3. Structure of Bilayer Membrane Containing Protein-Induced Glycopeptide Bundle on the Electrode Surface
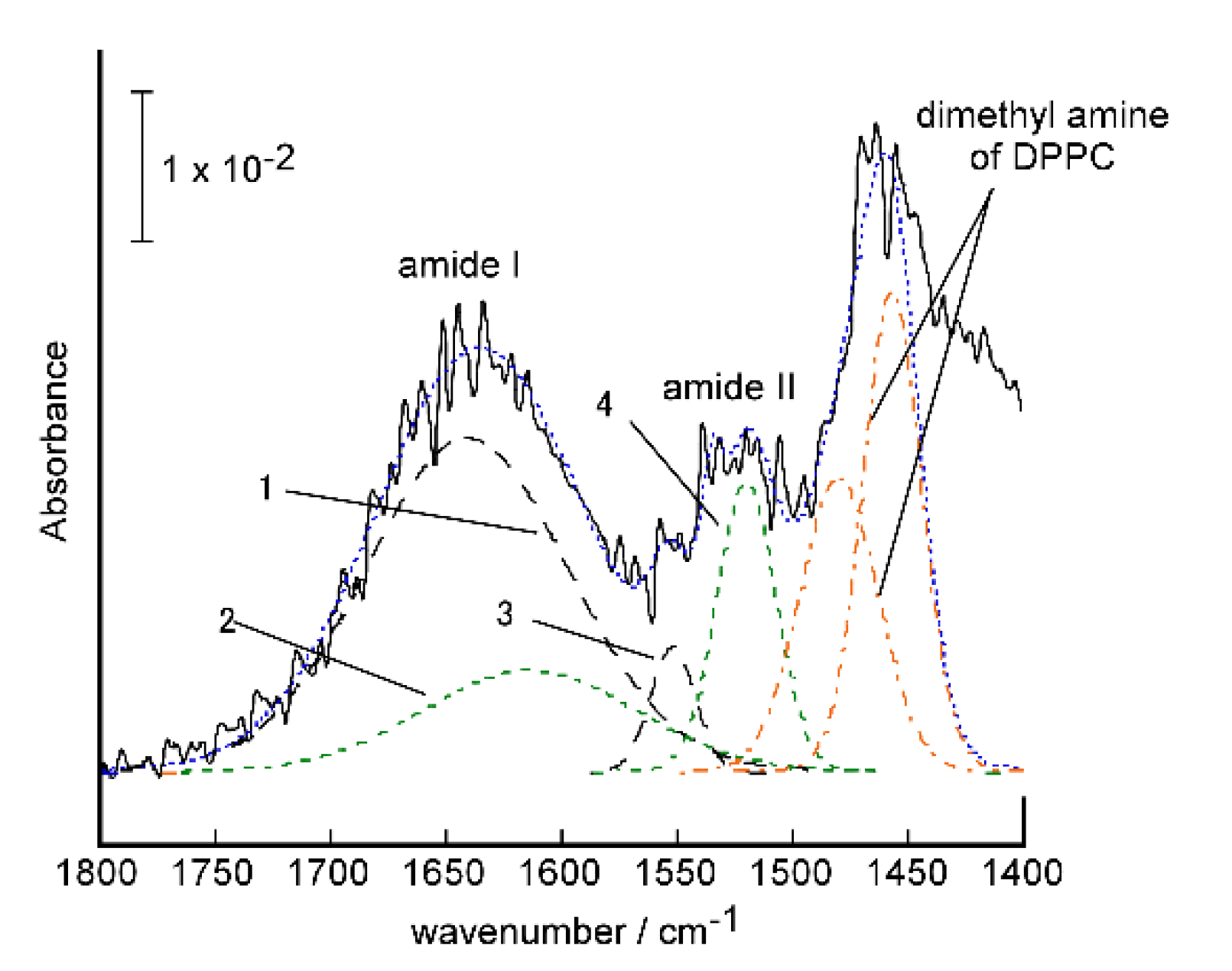
2.3.1. Conformation and Orientation of the Peptides in the Protein-Induced Glycopeptide Bundles Fixed on the Electrodes
2.3.2. Structure of Fixed Bilayer Membranes on the Electrodes
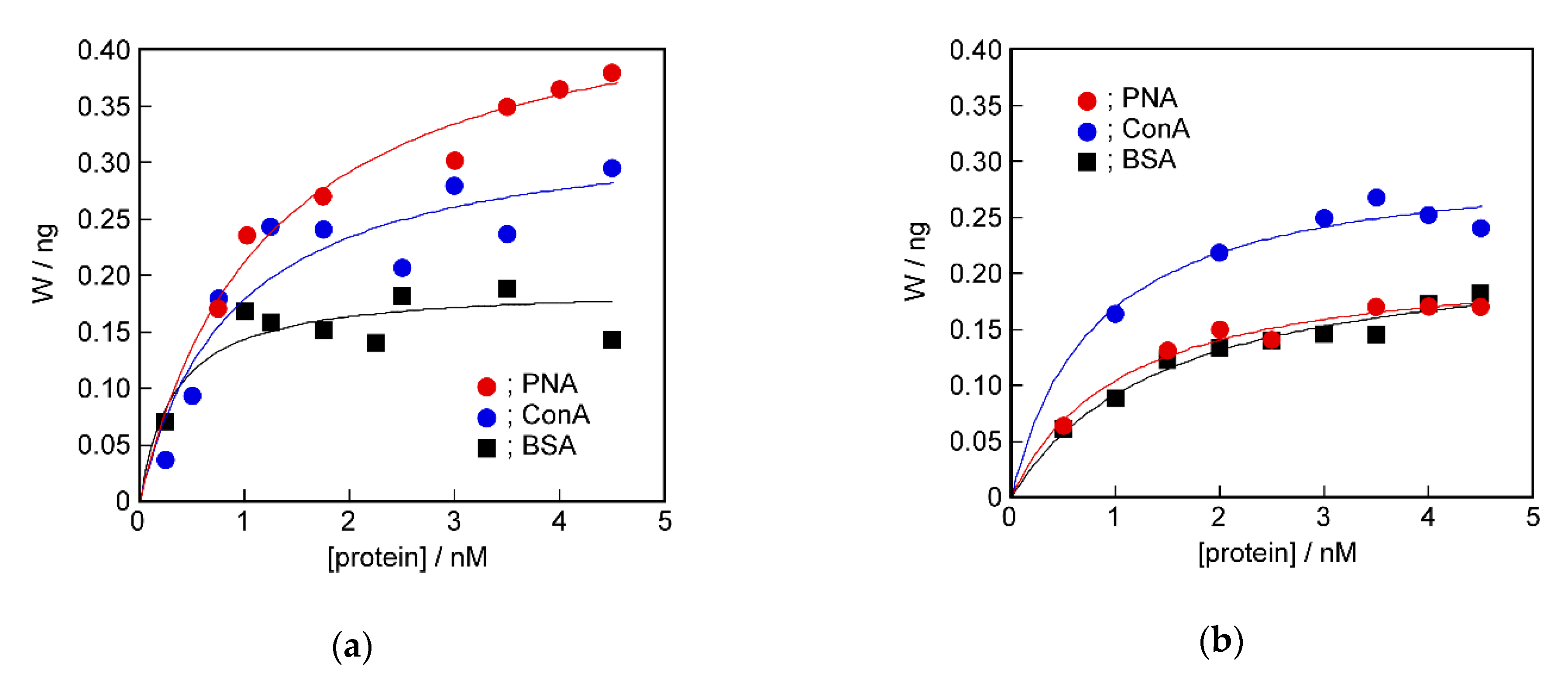
2.4. Recognition Ability of the Target Protein to the Sensing Membrane by QCM Method
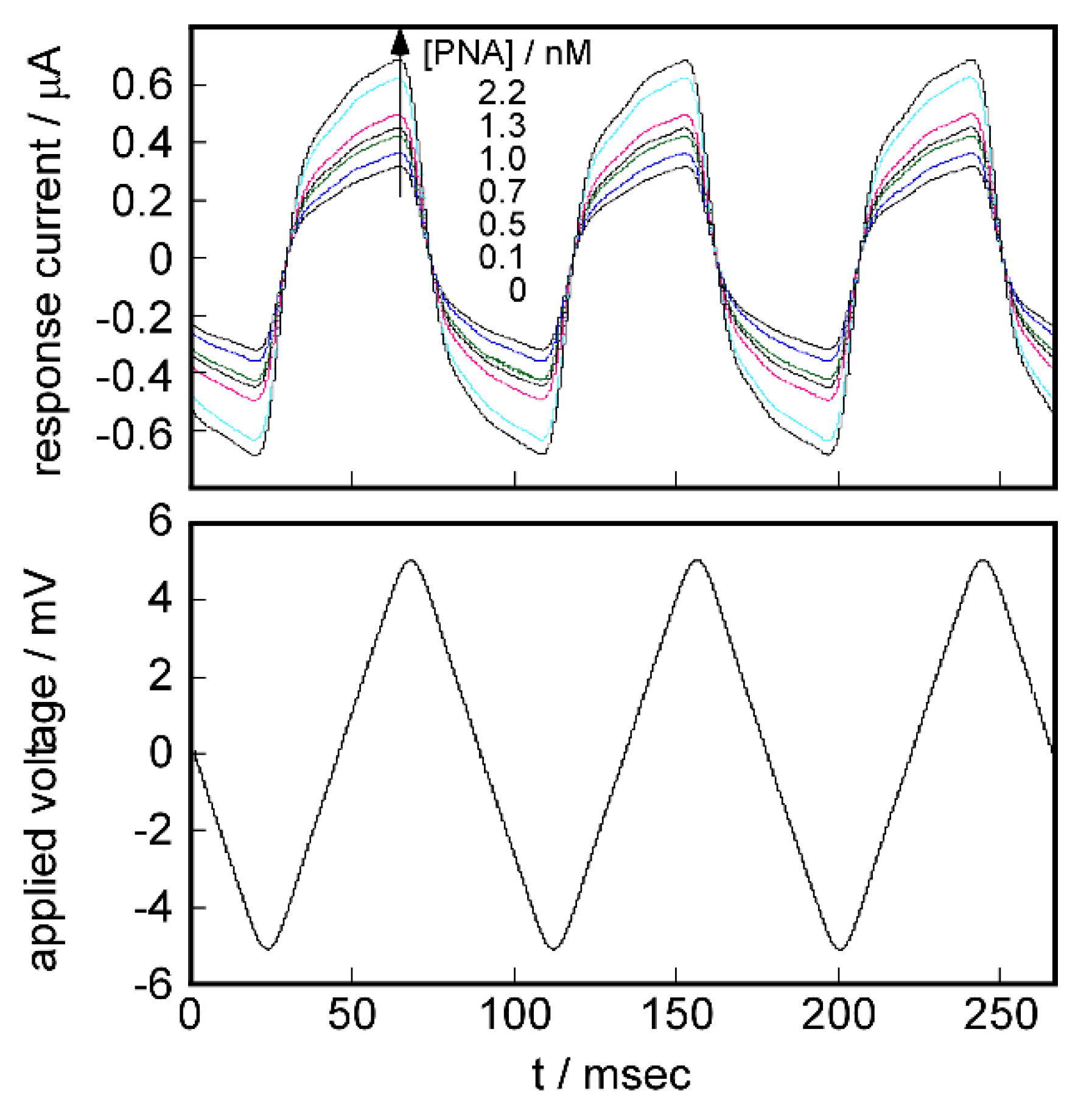
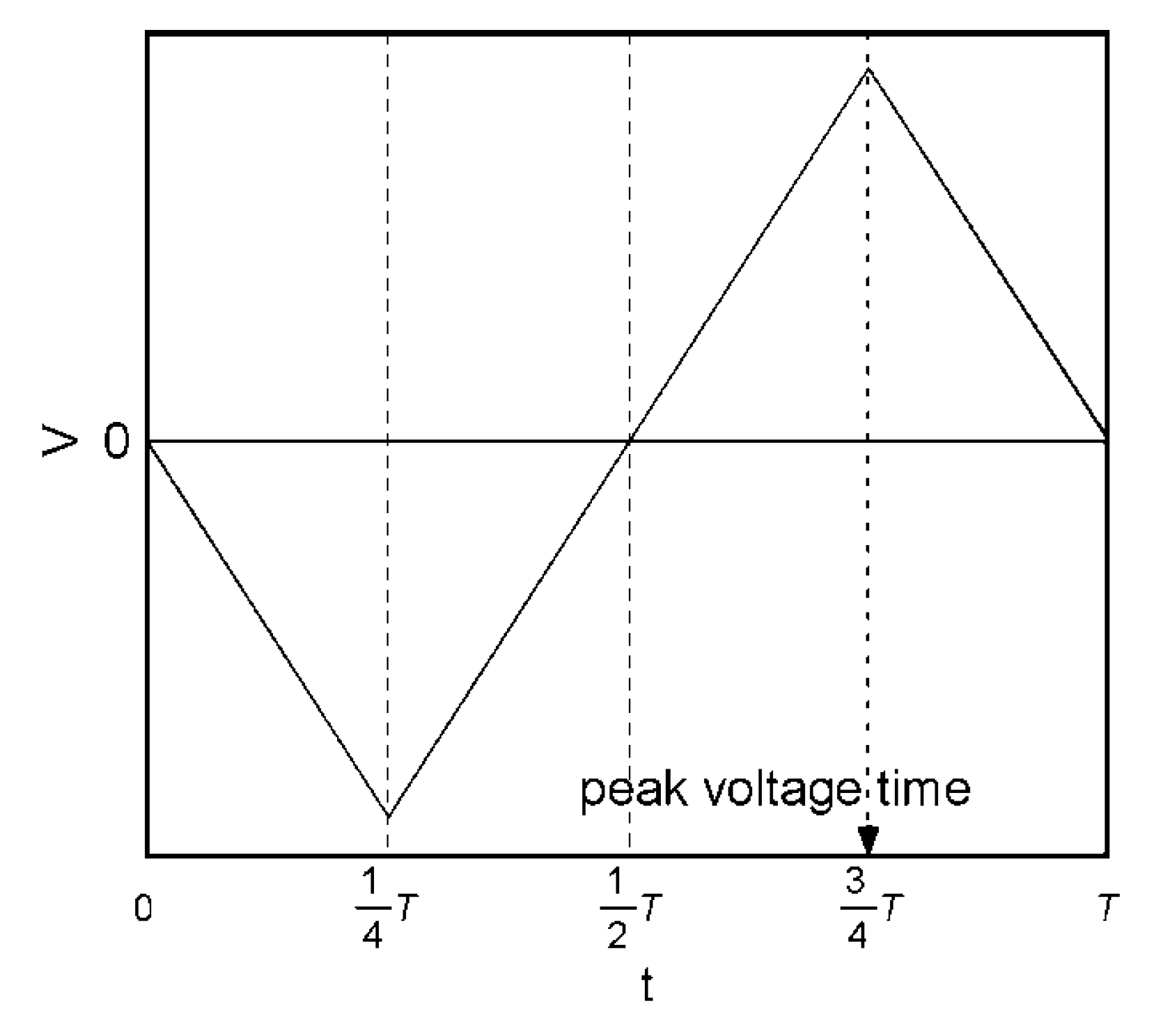
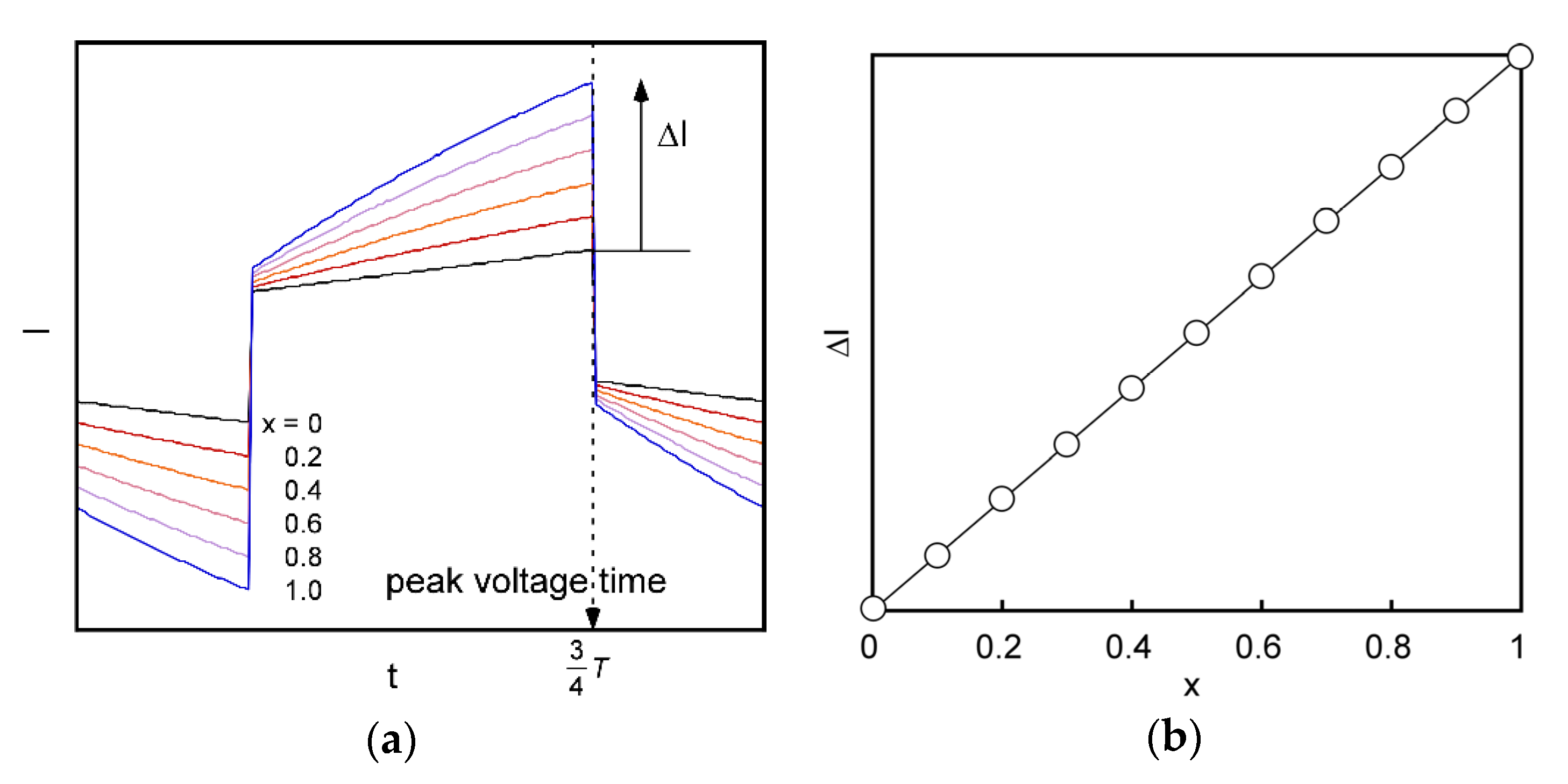
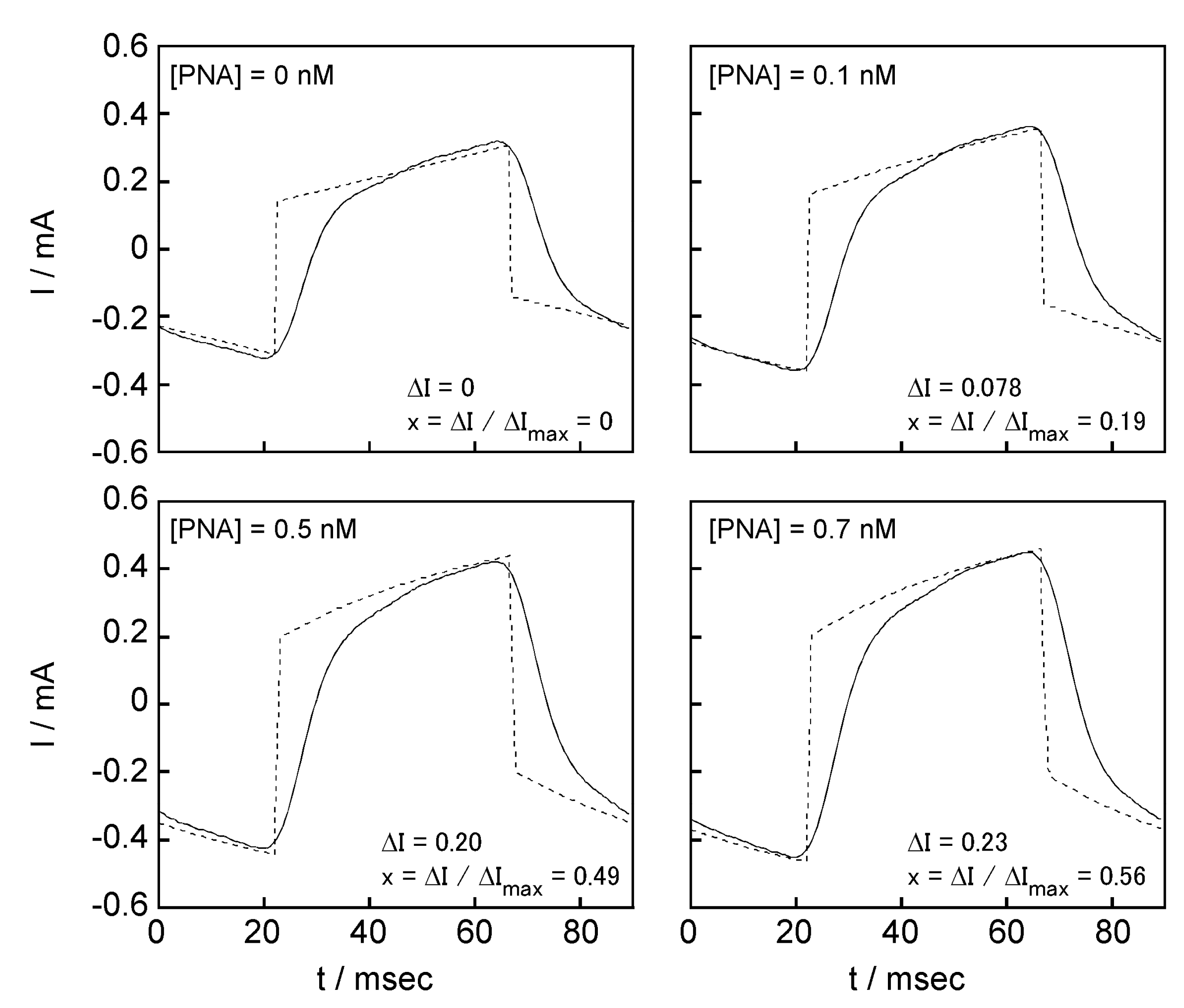
2.5. Signal Transduction Ability of the Target Protein Binding to the Sensing Membrane by Electrochemical Method
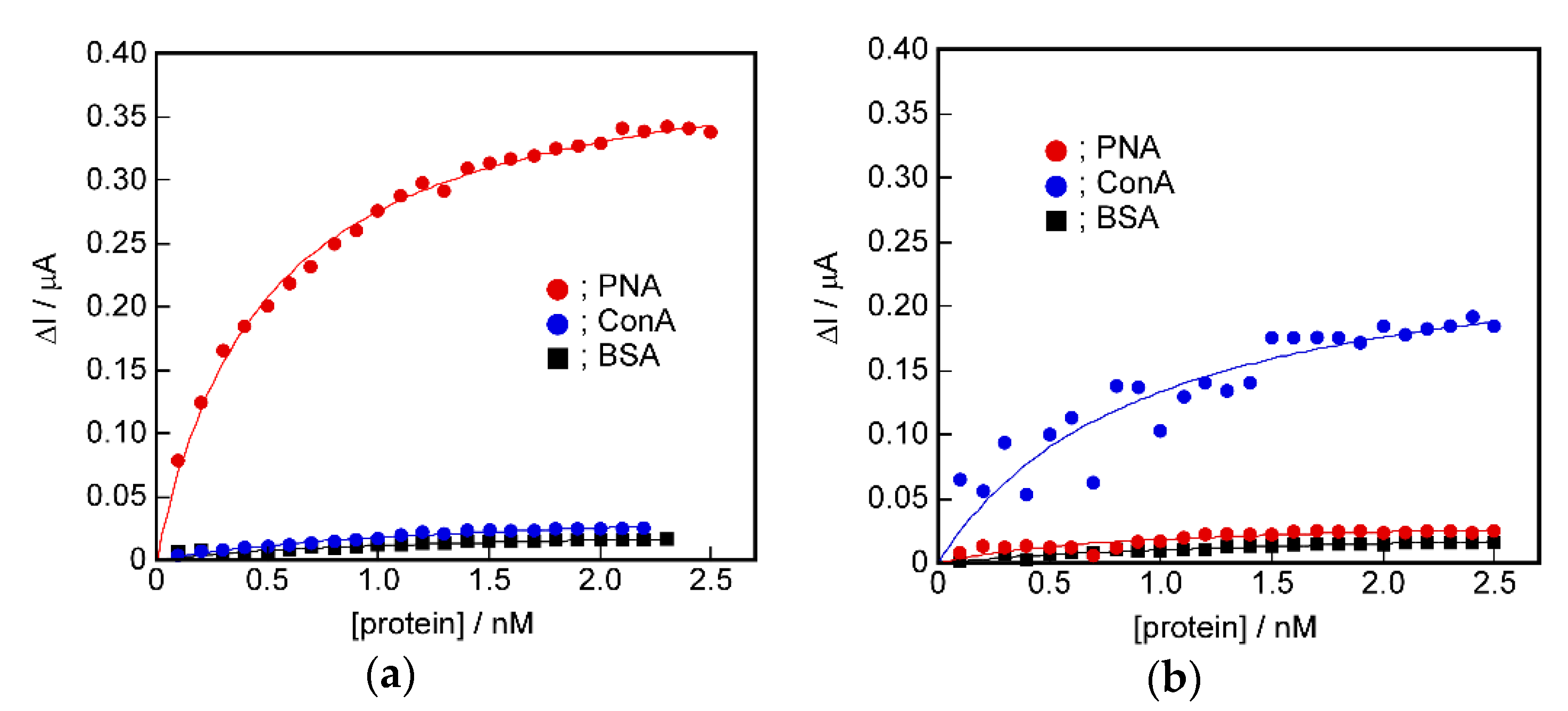
2.6. Difference in Protein Recognition Ability by the QCM and Electrochemical Method
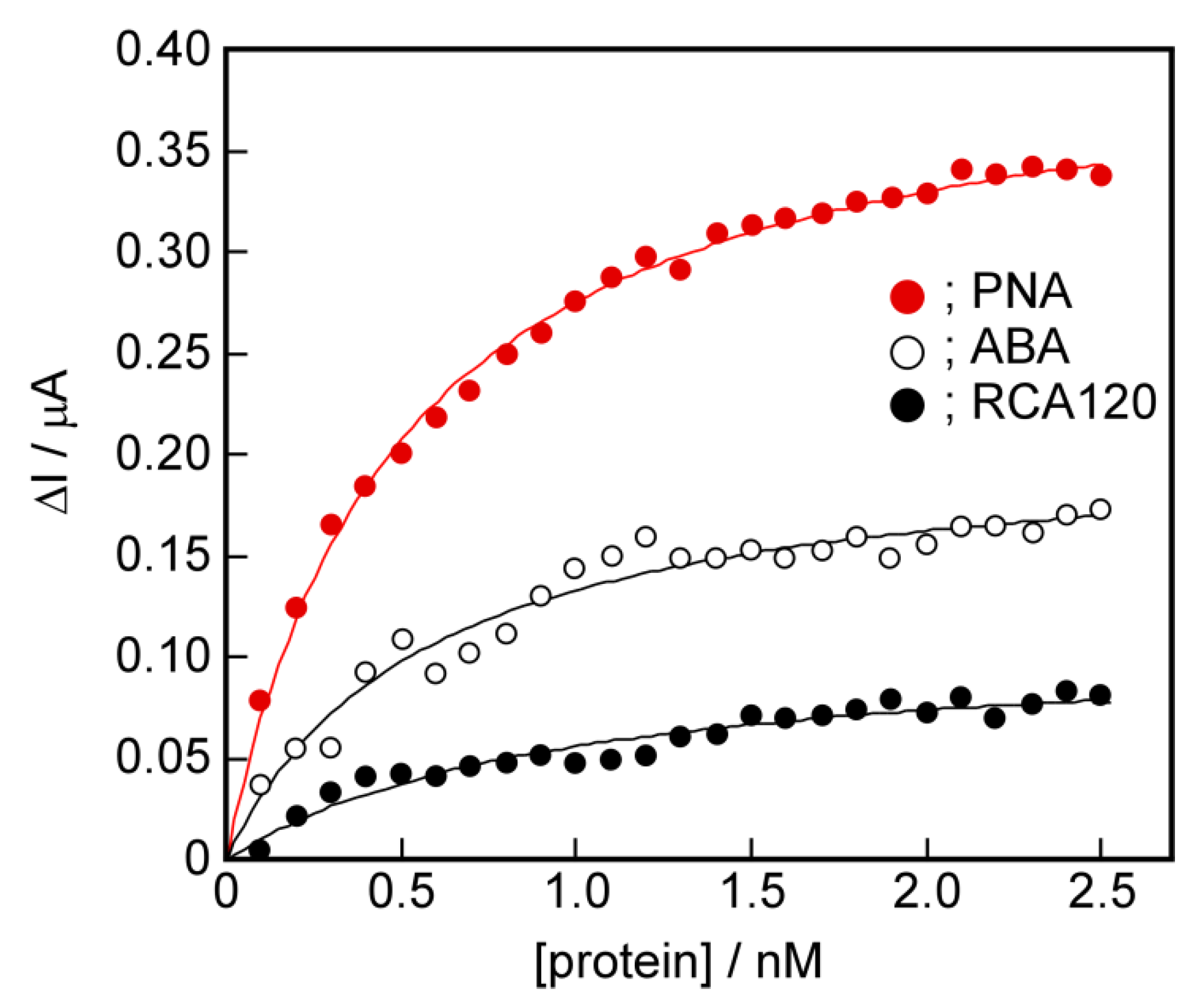
2.7. The Potential for Selective Protein Detection
3. Materials and Methods
3.1. Materials
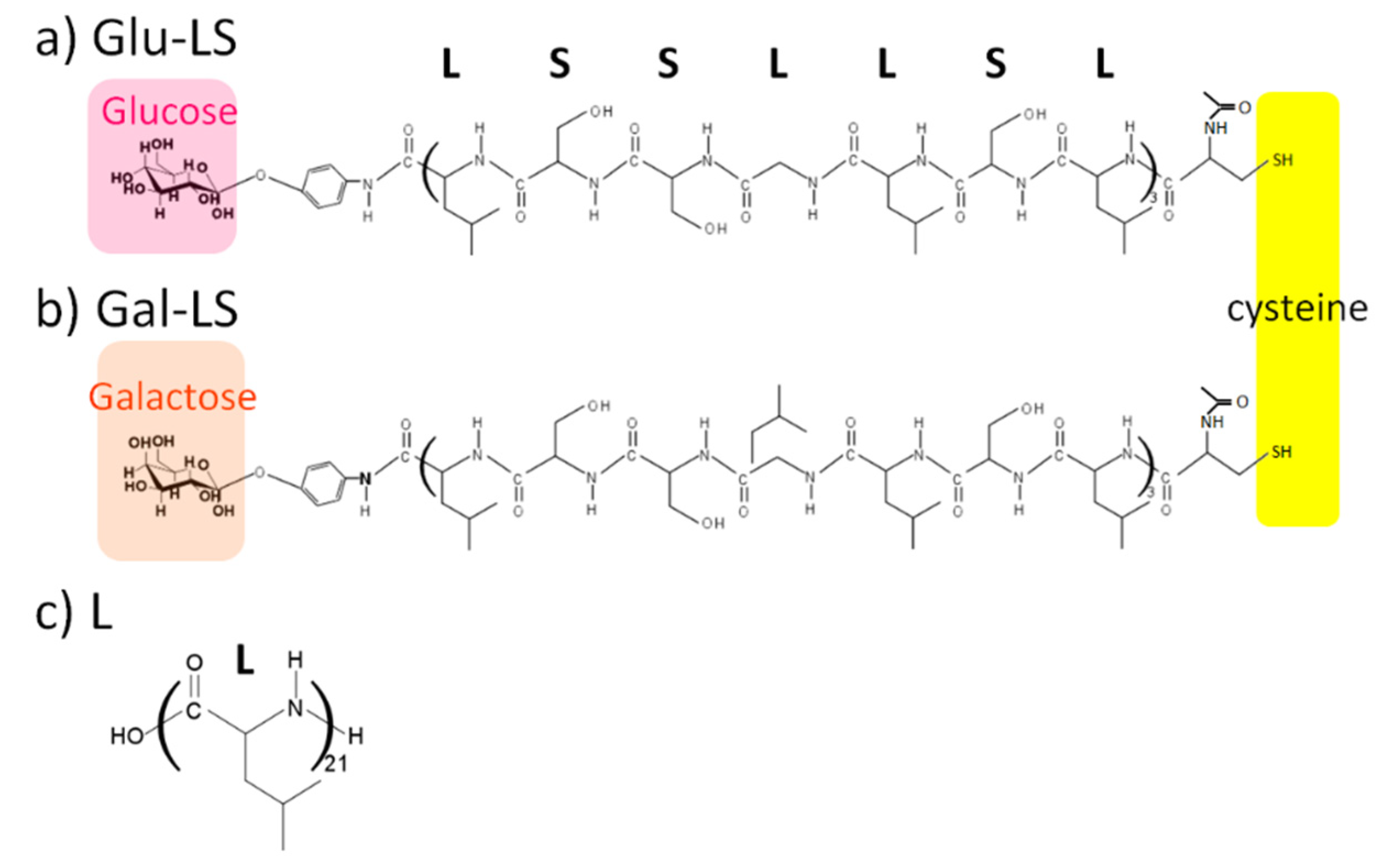
3.1.1. Glycopeptides
3.1.2. Target Proteins
3.1.3. Vesicles
3.1.4. Formation of Glycopeptide Bundle and Fix on the Electrode
3.2. Methods
3.2.1. TEM Observations
3.2.2. FTIR-RAS Measurements
3.2.3. QCM Measurements
3.2.4. Electrochemical Measurements
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A

References
- Daaboul, G.G.; Lopez, C.A.; Chinnala, J.; Goldberg, B.; Connor, J.H.; Ünlu, M.S. Digital sensing and sizing of vesicular stomatitis virus pseudotypes in complex media: A model for Ebola and Marburg detection. ACS Nano 2014, 8, 6047–6055. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Chang, P.; Benton, D.J.; McCauly, J.W.; Iqbal, M.; Cass, A.E.G. Dual recognition element lateral flow assay toward multiplex strain specific influenza virus detection. Anal. Chem. 2017, 89, 6781–6786. [Google Scholar] [CrossRef] [PubMed]
- Brangel, P.; Sobarzo, A.; Parolo, C.; Miller, B.S.; Howes, P.D.; Gelkop, S.; Lutwama, J.J.; Dys, J.M.; Mckendry, R.A.; Lobel, L.; et al. A serological point-of-care test for detection of IgG antibodies against Ebola virus in Human survivors. ACS Nano 2018, 12, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Kim, J.; Tran, V.T.; Lee, D.K.; Ahmed, S.R.; Hong, J.C.; Lee, J.; Park, E.Y.; Lee, J. Magnetic nanozyme-linked immunosorbent assay for ultrasensitive influenza A virus detection. ACS Appl. Mater. Interface 2018, 10, 12534–12543. [Google Scholar] [CrossRef] [PubMed]
- Grossegesse, M.; Hartkopf, F.; Nitsche, A.; Schaade, L.; Doellinger, J.; Muth, T. Prespective on proteomics for virus detection in clinical samples. J. Proteome Res. 2020, 19, 4380–4388. [Google Scholar] [CrossRef]
- Merritt, E.A.; Hol, W.G. AB5 toxins. Curr. Opin. Struct. Biol. 1995, 5, 165–172. [Google Scholar] [CrossRef]
- Matubara, T.; Sumi, M.; Kubota, H.; Taki, T.; Okahata, Y.; Sato, T. Inhibition of influenza virus inflections by sialygalactose-binding peptides selected from a phage library. J. Med. Chem. 2009, 52, 4247–4259. [Google Scholar] [CrossRef]
- Newthouse, E.I.; Xu, D.; Markwick, P.R.L.; Amaro, R.E.; Pao, H.C.; Wu, K.J.; Alam, M.; McCammon, J.A.; Li, W.W. Mechanism of glycan receptor recognition and specific swich for avian, swine, and human adapted influenza virus hemagglutinins: A molecular dyanamics perspective. J. Am. Chem. Soc. 2009, 131, 17430–17442. [Google Scholar] [CrossRef][Green Version]
- Illescas, B.M.; Rojo, J.; Delgado, R.; Martn, N. Multivalent glycosylated nanostructures to inhibit Ebora virus inflection. J. Am. Chem. Soc. 2017, 139, 6018–6025. [Google Scholar] [CrossRef]
- Durrant, J.D.; Kochanek, S.E.; Casalino, L.; Leong, P.U.; Dommer, A.C.; Amaro, R.E. Mesoscale all-atom influenza virus simulations suggesy new substrate binding mechanism. ACS Cent. Sci. 2020, 6, 189–196. [Google Scholar] [CrossRef]
- Hou, Y.; Gochin, M. Artificial ion channel biosensor in human immunodeficiency virus gp41 drug sensing. Anal. Chem. 2008, 80, 5924–5929. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Macras, M.X.; Blake, S.; Jiang, X.; Capone, R.; Eates, D.J.; Mayer, M.; Yang, J. A semi-synthetic ion channel platform for detection of phosphatase and protease activity. ACS Nano 2009, 3, 3569–3580. [Google Scholar]
- Haque, F.; Lunn, J.; Fang, H.; Smithrud, D.; Guo, P. Real-time sensing and discrimination of single chemicals using the channel of Phi29 DNA packaging nanomotor. ACS Nano 2012, 6, 3251–3261. [Google Scholar] [CrossRef]
- Halža, E.; Bro, T.H.; Bilenberg, B.; Koçer, A. Well-defined microapertures for ion channel biosensors. Anal. Chem. 2013, 85, 811–815. [Google Scholar] [CrossRef]
- Muraoka, T.; Endo, T.; Tabata, K.V.; Noji, H.; Nagatoishi, S.; Tsumoto, K.; Li, R.; Kinbara, K. Reversible ion transportation switch by a ligand-gated synthetic supramolecular ion channel. J. Am. Chem. Soc. 2014, 136, 15584–15595. [Google Scholar] [CrossRef]
- Agasid, M.T.; Vomi, T.J.; Saavedra, S.S.; Aspinwall, C.A. Enhanced temporal resolution with ion channel-functionalized sensors using a onductance-based measurement protocol. Anal. Chem. 2017, 89, 1315–1322. [Google Scholar] [CrossRef]
- Chun, K.-Y.; Han, C.-S. Spectral sensor inspired by cone photoreceptors and ion channels without external power. ACS Appl. Mater. Interfaces 2018, 10, 34385–34391. [Google Scholar] [CrossRef]
- Cornell, B.A.; Braach-Marksvytis, V.L.B.; King, L.G.; Osman, D.D.J.; Raguse, B.; Wieczorek, L.; Poce, R.J. A biosensor that use ion-channel switches. Nature 1997, 387, 580–583. [Google Scholar] [CrossRef]
- Terrettaz, S.; Ulrich, W.-P.; Guerrini, R.; Verdini, A.; Vogel, H. Immunosensing by a synthetic ligand-gated ion channel. Angew. Chem. Int. Ed. 2001, 40, 1740–1743. [Google Scholar] [CrossRef]
- Matsuura, K. Synthetic approaches to construct viral capsid-like spherical nanomaterials. Chem. Commun 2018, 54, 8944–8959. [Google Scholar] [CrossRef]
- Inaba, H.; Matsuura, K. Peptide nanomaterials designed from natural supramolecular systems. Chem. Rec. 2019, 19, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Murai, K.; Yamamoto, Y.; Kinoshita, T.; Nagata, K.; Higuchi, M. Self-bonding and the electrochemical properties of silica-coated nanowires composed of cobalt-coordinated peptide bundles. J. Mater. Chem. B 2017, 5, 5539–5548. [Google Scholar] [CrossRef]
- Murai, K.; Inagaki, K.; Hiraoka, C.; Minoshima, S.; Kinoshita, T.; Nagata, K.; Higuchi, M. Mineralization of magnetic nano-tape in selforganized nanospace composed of nucleopeptides and peptides. CrystEngComm. 2019, 21, 3557–3567. [Google Scholar] [CrossRef]
- Higuchi, M.; Koga, T.; Taguchi, K.; Kinoshita, T. Substrate-induced conformation of an artificial receptor with two receptor sites. Langmuir 2002, 18, 813–818. [Google Scholar] [CrossRef]
- Lee, Y.C. Biochemistry of carbohydrate-protein interaction. FASEB J. 1992, 6, 3193–3200. [Google Scholar] [CrossRef]
- DeGrado, W.F.; Wasserman, Z.R.; Lear, J.D. Protein design, a minimalist approach. Science 1989, 243, 622–628. [Google Scholar] [CrossRef]
- Fish, W.W.; Hamlin, L.M.; Miller, R.L. The macromolecular properties of peanut agglutinin. Arch. Biochem. Biophys. 1987, 190, 693–698. [Google Scholar] [CrossRef]
- Severs, N.J. Freeze-fracture electrin microscopy. Nat. Protoc. 2007, 2, 547–576. [Google Scholar] [CrossRef]
- Fujimoto, T.; Miyawaki, A.; Mikoshiba, K. Inositol 1,4,5-trisphosphate receptor-like protein in plasmalemmal caveolae is linked to actin filaments. J. Cell Sci. 1995, 108, 7–15. [Google Scholar]
- Zhang, L.; Longo, M.L.; Stroeve, P. Mobile phospholipid bilayers supported on a polyion/alkylthiol layer pair. Langmuir 2000, 16, 5093–5099. [Google Scholar] [CrossRef]
- Miyazawa, T.; Blout, E.R. The infrared spectra of polypeptides in various conformations: Amide I and II bands. J. Am. Chem. Soc. 1961, 83, 712–719. [Google Scholar] [CrossRef]
- Enriquez, E.P.; Gray, K.H.; Guarisco, V.F.; Linton, R.W.; Mar, K.D.; Samulski, E.T. Behavior of rigid macromolecules in self-assembly at an interface. J. Vac. Sci. Technol. A 1992, 10, 2775–2782. [Google Scholar] [CrossRef]
- Dwyer, J.M.; Johnson, C. The use of concanavalin A to study the immunoregulation of human T cells. Clin. Exp. Immunol. 1981, 46, 237–249. [Google Scholar] [PubMed]
- Rei, H.E. Monomolecular films. Sci. Am. 1961, 204, 152–165. [Google Scholar]
- Shuler, R.L.; Zisman, W.A. A study of films of poly(γ-methyl L-glutamete) adsorbed on water using wase damping and other method. Macromolecules 1972, 5, 487–492. [Google Scholar] [CrossRef]
- Higuchi, M.; Wright, J.P.; Taguchi, K.; Kinoshita, T. Structure and molecular recognition properties of a poly(allylamine) monolayer containing poly(L-alanine) graft chains. Langmuir 2000, 16, 7061–7065. [Google Scholar] [CrossRef]
- Presant, C.A.; Kornfeld, S. Characterization of the cell surface receptor for the agaricus bisporus hemagglutinin. J. Biol. Chem. 1972, 247, 6937–6945. [Google Scholar]
- Shimada, T.; Funatsu, G. Binding of lactose and galactose to native and lodinated Ricin D. Agric. Biol. Chem. 1985, 49, 2125–2130. [Google Scholar]
- Carpino, L.A.; Han, G.Y. 9-Fluorenymethoxycarbonyl amino-protecting group. J. Org. Chem. 1972, 37, 3404–3409. [Google Scholar] [CrossRef]
- Houben, J.L.; Pieroni, O.; Fissi, A.; Ciardelli, F. Optical activity and photochromism of the azo chromophore bound to poly(L-glutamic acid). Biopolymers 1978, 17, 799–804. [Google Scholar] [CrossRef]
- Matsuki, H.; Kaneshina, S. Thermodynamics of bilayer phase transitions of phospholipids. Netsu Sokutei 2006, 33, 74–82. [Google Scholar]
- Sauerbrey, G.H. Verwendung von schwingquarzen zur wägung dünner schichten und zur mikrowägung. Z. Phys. 1959, 155, 206–222. [Google Scholar] [CrossRef]
| ΔF/Hz | W/ng | Coverage/% | |
|---|---|---|---|
| DPPC | 786.0 | 23.6 *1. | 91.9 |
| PNA-induced glycopeptide bundle/DPPC | 874.8 | 26.0 *2. | 104 |
| ConA-induced glycopeptide bundle/DPPC | 859.4 | 25.5 *2. | 102 |
| QCM method | PNA-induced glycopeptide bundle in DPPC bilayer membrane | |||||
| PNA | ConA | BSA | ||||
| K/M−1 | Wmax/ng | K/M−1 | Wmax/ng | K/M−1 | Wmax/ng | |
| 8.2 × 108 | 0.47 | 1.1 × 109 | 0.34 | 3.1 × 109 | 0.19 | |
| ConA-induced glycopeptide bundle in DPPC bilayer, embrane | ||||||
| PNA | ConA | BSA | ||||
| K/M−1 | Wmax/ng | K/M−1 | Wmax/ng | K/M−1 | Wmax/ng | |
| 9.5 × 108 | 0.21 | 1.2 × 109 | 0.31 | 6.7 × 108 | 0.23 | |
| Electrochemical Method | PNA-induced glycopeptide bundle in DPPC bilayer membrane | |||||
| PNA | ConA | BSA | ||||
| K/M−1 | ΔImax/μA | K/M−1 | ΔImax/μA | K/M−1 | ΔImax/μA | |
| 2.1 × 109 | 0.41 | 7.4 × 108 | 0.043 | 9.5 × 108 | 0.023 | |
| ConA-induced glycopeptide bundle in DPPC bilayer membrane | ||||||
| PNA | ConA | BSA | ||||
| K/M−1 | ΔImax/μA | K/M−1 | ΔImax/μA | K/M−1 | ΔImax/μA | |
| 1.1 × 109 | 0.035 | 1.6 × 109 | 0.26 | 5.8 × 108 | 0.028 | |
| QCM method | PNA-induced glycopeptide bundle in DPPC bilayer membrane | |
| PNA to ConA | PNA to BSA | |
| 1.34 | 2.47 | |
| ConA-induced glycopeptide bundle in DPPC bilayer, embrane | ||
| ConA to PNA | ConA to BSA | |
| 1.42 | 1.34 | |
| Electrochemical Method | PNA-induced glycopeptide bundle in DPPC bilayer membrane | |
| PNA to ConA | PNA to BSA | |
| 9.53 | 17.8 | |
| ConA-induced glycopeptide bundle in DPPC bilayer membrane | ||
| ConA to PNA | ConA to BSA | |
| 7.49 | 9.35 | |
| Binding Constant and Saturated Signal Value | |||||
|---|---|---|---|---|---|
| PNA | ABA | RCA120 | |||
| K/M−1 | ΔImax/μA | K/M−1 | ΔImax/μA | K/M−1 | ΔImax/μA |
| 2.1 × 109 | 0.41 | 1.8 × 109 | 0.21 | 1.1 × 109 | 0.11 |
| Selectivity of PNA to other proteins | |||||
| PNA to ABA | PNA to RCA120 | ||||
| 1.95 | 3.76 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arai, M.; Miura, T.; Ito, Y.; Kinoshita, T.; Higuchi, M. Protein Sensing Device with Multi-Recognition Ability Composed of Self-Organized Glycopeptide Bundle. Int. J. Mol. Sci. 2021, 22, 366. https://doi.org/10.3390/ijms22010366
Arai M, Miura T, Ito Y, Kinoshita T, Higuchi M. Protein Sensing Device with Multi-Recognition Ability Composed of Self-Organized Glycopeptide Bundle. International Journal of Molecular Sciences. 2021; 22(1):366. https://doi.org/10.3390/ijms22010366
Chicago/Turabian StyleArai, Mao, Tomohiro Miura, Yuriko Ito, Takatoshi Kinoshita, and Masahiro Higuchi. 2021. "Protein Sensing Device with Multi-Recognition Ability Composed of Self-Organized Glycopeptide Bundle" International Journal of Molecular Sciences 22, no. 1: 366. https://doi.org/10.3390/ijms22010366
APA StyleArai, M., Miura, T., Ito, Y., Kinoshita, T., & Higuchi, M. (2021). Protein Sensing Device with Multi-Recognition Ability Composed of Self-Organized Glycopeptide Bundle. International Journal of Molecular Sciences, 22(1), 366. https://doi.org/10.3390/ijms22010366




