The Dual Role of Autophagy in Cancer Development and a Therapeutic Strategy for Cancer by Targeting Autophagy
Abstract
1. Introduction
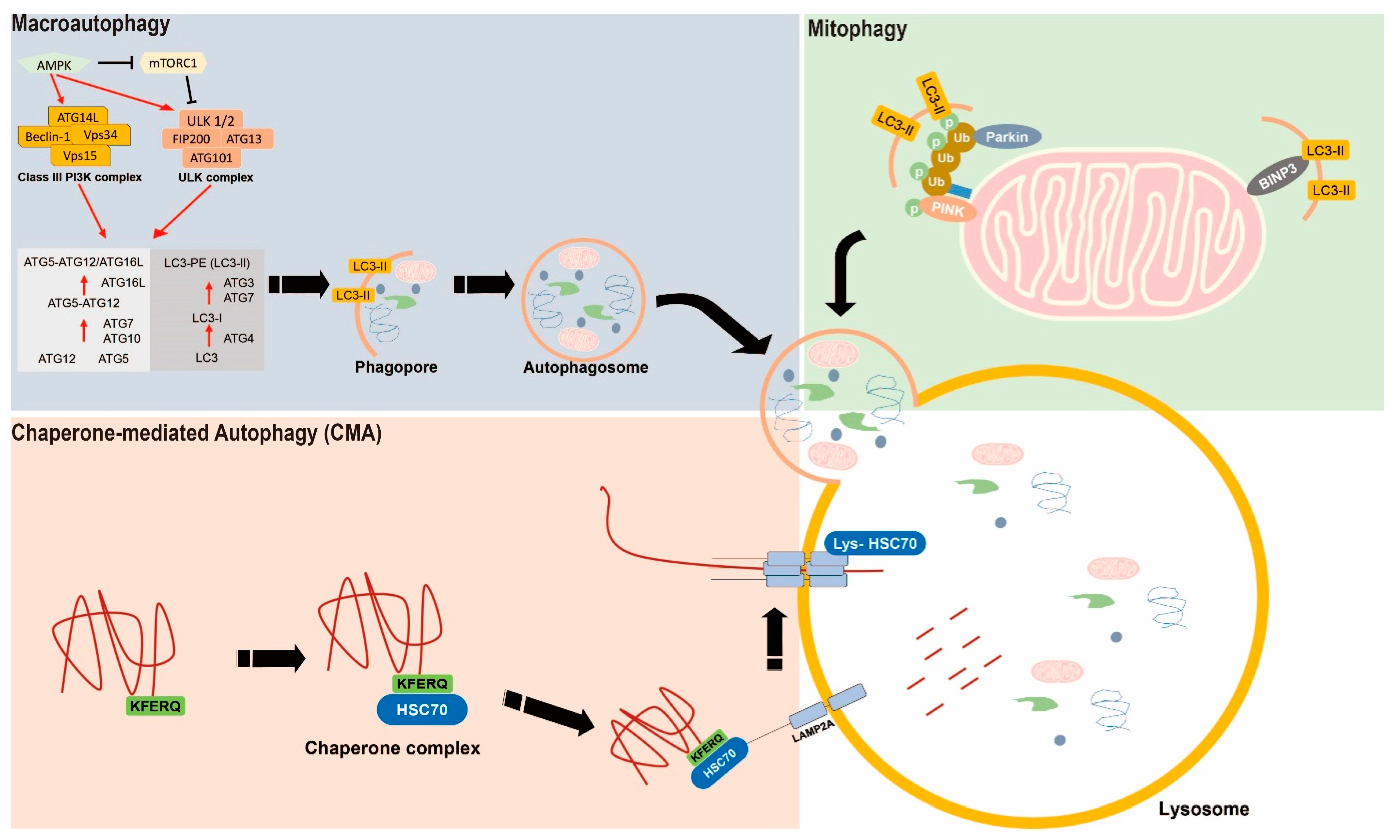
2. The Basic Mechanism of Autophagy
2.1. Macroautophagy
2.2. Mitophagy
2.3. Chaperone-Mediated Autophagy
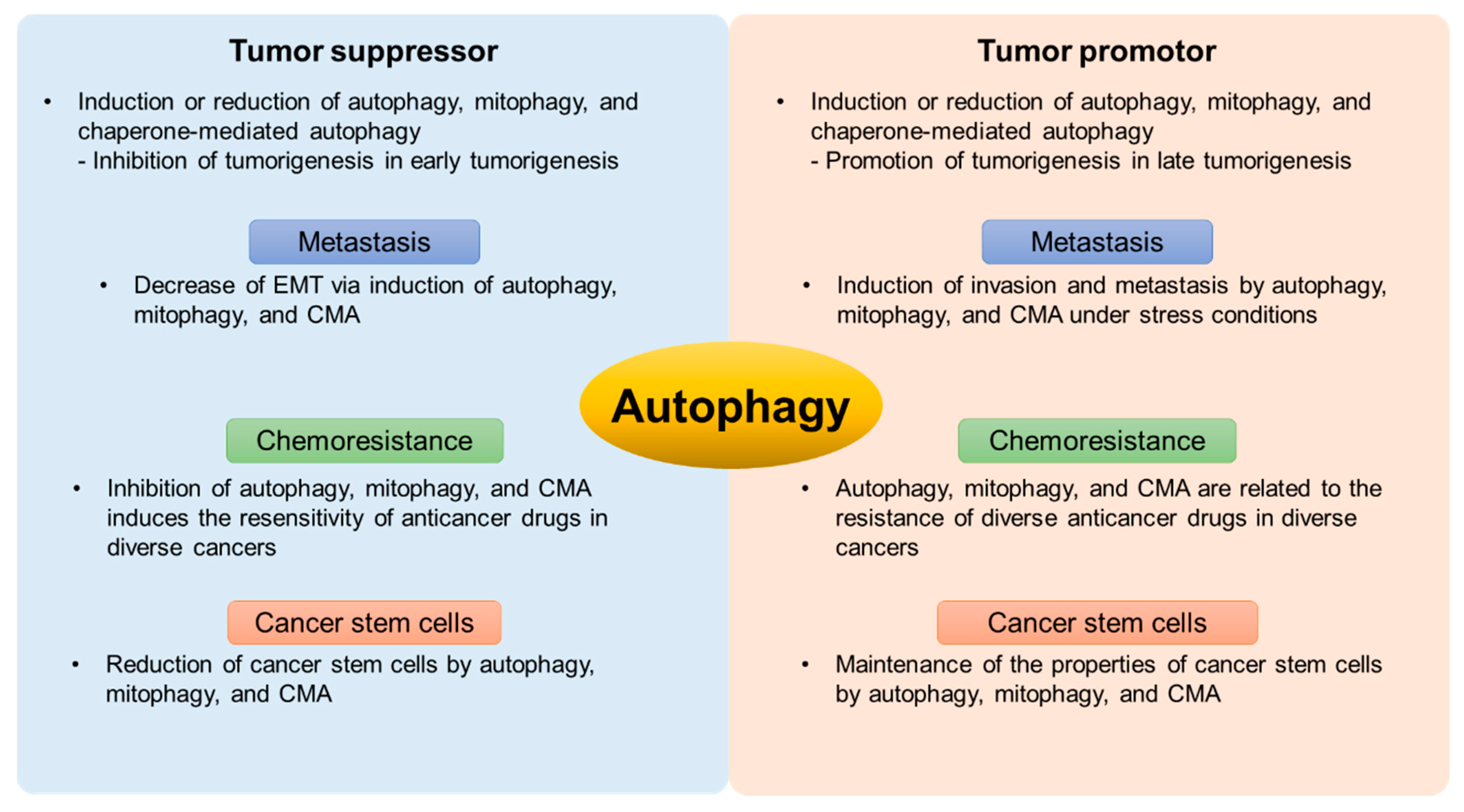
3. The Bipolar Role of Autophagy in Cancer
3.1. The Mechanism of Autophagy in Tumorigenesis
3.2. The Relationship between Autophagy and Metastasis
3.3. The Roles of Autophagy in Chemoresistance
3.4. Autophagy in Cancer Stem Cells
4. Targeting Autophagy as an Anticancer Therapy
4.1. The Effect of Autophagy Inhibitors in Anticancer Therapy
4.2. The Effect of Autophagy Inducers in Anticancer Therapy
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Onorati, A.V.; Dyczynski, M.; Ojha, R.; Amaravadi, R.K. Targeting autophagy in cancer. Cancer 2018, 124, 3307–3318. [Google Scholar] [CrossRef] [PubMed]
- Parzych, K.R.; Klionsky, D.J. An Overview of Autophagy: Morphology, Mechanism, and Regulation. Antioxid. Redox Signal. 2014, 20, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Song, X.; Yang, Y.; Wan, X.; Alvarez, A.A.; Sastry, N.; Feng, H.; Hu, B.; Cheng, S.-Y. Autophagy and Hallmarks of Cancer. Crit. Rev. Oncog. 2018, 23, 247–267. [Google Scholar] [CrossRef] [PubMed]
- Glick, D.; Barth, S.; MacLeod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- Guo, F.; Liu, X.; Cai, H.; Le, W. Autophagy in neurodegenerative diseases: Pathogenesis and therapy. Brain Pathol. 2017, 28, 3–13. [Google Scholar] [CrossRef]
- Mialet-Perez, J.; Vindis, C. Autophagy in health and disease: Focus on the cardiovascular system. Essays Biochem. 2017, 61, 721–732. [Google Scholar] [CrossRef]
- Sarparanta, J.; García-Macia, M.; Singh, R. Autophagy and Mitochondria in Obesity and Type 2 Diabetes. Curr. Diabetes Rev. 2017, 13, 352–369. [Google Scholar] [CrossRef]
- Yun, C.W.; Lee, S.H. The Roles of Autophagy in Cancer. Int. J. Mol. Sci. 2018, 19, 3466. [Google Scholar] [CrossRef]
- Dong, Y.; Wu, Y.; Zhao, G.L.; Ye, Z.Y.; Xing, C.G.; Yang, X.D. Inhibition of autophagy by 3-MA promotes hypox-ia-induced apoptosis in human colorectal cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1047–1054. [Google Scholar] [CrossRef]
- Endo, S.; Nakata, K.; Sagara, A.; Koikawa, K.; Ando, Y.; Kibe, S.; Takesue, S.; Nakayama, H.; Abe, T.; Okumura, T.; et al. Autophagy inhibition enhances antiproliferative effect of salinomycin in pancreatic cancer cells. Pancreatology 2017, 17, 990–996. [Google Scholar] [CrossRef]
- Asl, E.R.; Mansori, B.; Mohammadi, A.; Najafi, S.; Pouya, F.D.; Rami, Y. Interaction between DNA damage response and autophagy in colorectal cancer. Gene 2020, 730, 144323. [Google Scholar] [CrossRef]
- Anderson, C.M.; MacLeod, K.F. Autophagy and cancer cell metabolism. Int. Rev. Cell Mol. Biol. 2019, 347, 145–190. [Google Scholar] [CrossRef]
- Xiong, X.; Lu, B.; Tian, Q.; Zhang, H.; Wu, M.; Guo, H.; Zhang, Q.; Li, X.; Zhou, T.; Wang, Y. Inhibition of autophagy enhances cinobufagin-induced apoptosis in gastric cancer. Oncol. Rep. 2018, 41, 492–500. [Google Scholar] [CrossRef]
- White, E.; Mehnert, J.M.; Chan, C.S. Autophagy, Metabolism, and Cancer. Clin. Cancer Res. 2015, 21, 5037–5046. [Google Scholar] [CrossRef]
- Yorimitsu, T.; Klionsky, D.J. Autophagy: Molecular machinery for self-eating. Cell Death Differ. 2005, 12, 1542–1552. [Google Scholar] [CrossRef]
- Oku, M.; Sakai, Y. Three Distinct Types of Microautophagy Based on Membrane Dynamics and Molecular Machineries. BioEssays 2018, 40, e1800008. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 365–381. [Google Scholar] [CrossRef]
- Shibutani, S.T.; Yoshimori, T. A current perspective of autophagosome biogenesis. Cell Res. 2014, 24, 58–68. [Google Scholar] [CrossRef]
- Gui, D.; Cui, Z.; Zhang, L.; Yu, C.; Yao, D.; Xu, M.; Chen, M.; Wu, P.L.; Liangxing, W.; Wang, L.; et al. Salidroside attenuates hypoxia-induced pulmonary arterial smooth muscle cell proliferation and apoptosis resistance by upregulating autophagy through the AMPK-mTOR-ULK1 pathway. BMC Pulm. Med. 2017, 17, 191. [Google Scholar] [CrossRef]
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef]
- Levine, B.; Klionsky, D.J. Development by Self-Digestion. Dev. Cell 2004, 6, 463–477. [Google Scholar] [CrossRef]
- Matsunaga, K.; Saitoh, T.; Tabata, K.; Omori, H.; Satoh, T.; Kurotori, N.; Maejima, I.; Shirahama-Noda, K.; Ichimura, T.; Isobe, T.; et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009, 11, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Walczak, M.; Martens, S. Dissecting the role of the Atg12–Atg5-Atg16 complex during autophagosome formation. Autophagy 2013, 9, 424–425. [Google Scholar] [CrossRef] [PubMed]
- Dooley, H.C.; Razi, M.; Polson, H.E.; Girardin, S.E.; Wilson, M.I.; Tooze, S.A. WIPI2 Links LC3 Conjugation with PI3P, Autophagosome Formation, and Pathogen Clearance by Recruiting Atg12–5-16L1. Mol. Cell 2014, 55, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, M.B.E.; Keulers, T.G.; Vooijs, M.A.; Rouschop, K.M.A. LC3/GABARAP family proteins: Autophagy-(un)related functions. FASEB J. 2016, 30, 3961–3978. [Google Scholar] [CrossRef]
- Reggiori, F.; Ungermann, C. Autophagosome Maturation and Fusion. J. Mol. Biol. 2017, 429, 486–496. [Google Scholar] [CrossRef]
- Ni, H.-M.; Williams, J.A.; Ding, W.-X. Mitochondrial dynamics and mitochondrial quality control. Redox Biol. 2015, 4, 6–13. [Google Scholar] [CrossRef]
- Pickles, S.; Vigié, P.; Youle, R.J. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 2018, 28, R170–R185. [Google Scholar] [CrossRef]
- Palikaras, K.; Lionaki, E.; Tavernarakis, N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 2018, 20, 1013–1022. [Google Scholar] [CrossRef]
- Jin, S.M.; Youle, R.J. PINK1- and Parkin-mediated mitophagy at a glance. J. Cell Sci. 2012, 125, 795–799. [Google Scholar] [CrossRef]
- Ordureau, A.; Heo, J.-M.; Duda, D.M.; Paulo, J.A.; Olszewski, J.L.; Yanishevski, D.; Rinehart, J.; Schulman, B.A.; Harper, J.W. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc. Natl. Acad. Sci. USA 2015, 112, 6637–6642. [Google Scholar] [CrossRef] [PubMed]
- Durcan, T.M.; Fon, E.A. The three ‘P’s of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev. 2015, 29, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Von Stockum, S.; Marchesan, E.; Ziviani, E. Mitochondrial quality control beyond PINK1/Parkin. Oncotarget 2018, 9, 12550–12551. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.-X.; Yin, X.-M. Mitophagy: Mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012, 393, 547–564. [Google Scholar] [CrossRef]
- Zhang, J.; Ney, P.A. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009, 16, 939–946. [Google Scholar] [CrossRef]
- Yoo, S.-M.; Jung, Y.-K. A Molecular Approach to Mitophagy and Mitochondrial Dynamics. Mol. Cells 2018, 41, 18–26. [Google Scholar]
- Kirchner, P.; Bourdenx, M.; Madrigal-Matute, J.; Tiano, S.; Diaz, A.; Bartholdy, B.A.; Will, B.; Cuervo, A.M. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol. 2019, 17, e3000301. [Google Scholar] [CrossRef]
- Bandyopadhyay, U.; Kaushik, S.; Varticovski, L.; Cuervo, A.M. The Chaperone-Mediated Autophagy Receptor Organizes in Dynamic Protein Complexes at the Lysosomal Membrane. Mol. Cell. Biol. 2008, 28, 5747–5763. [Google Scholar] [CrossRef]
- Bandyopadhyay, U.; Sridhar, S.; Kaushik, S.; Kiffin, R.; Cuervo, A.M. Identification of Regulators of Chaperone-Mediated Autophagy. Mol. Cell 2010, 39, 535–547. [Google Scholar] [CrossRef]
- Cuervo, A.M.; Dice, J.F. Regulation of Lamp2a Levels in the Lysosomal Membrane. Traffic 2000, 1, 570–583. [Google Scholar] [CrossRef]
- Saha, S.; Panigrahi, D.P.; Patil, S.; Bhutia, S.K. Autophagy in health and disease: A comprehensive review. Biomed. Pharmacother. 2018, 104, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Cordani, M.; Butera, G.; Pacchiana, R.; Donadelli, M. Molecular interplay between mutant p53 proteins and autophagy in cancer cells. Biochim. Biophys. Acta (BBA) Bioenerg. 2017, 1867, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.-K.; Zhou, B.; Zhuang, X.-M.; Zhuang, P.-L.; Zhang, D.-M.; Chen, W.-L. Cancer-associated fibroblasts confer cisplatin resistance of tongue cancer via autophagy activation. Biomed. Pharmacother. 2018, 97, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Mainz, L.; Rosenfeldt, M.T. Autophagy and cancer—insights from mouse models. FEBS J. 2017, 285, 792–808. [Google Scholar] [CrossRef] [PubMed]
- Nazio, F.; Bordi, M.; Cianfanelli, V.; Locatelli, F.; Cecconi, F. Autophagy and cancer stem cells: Molecular mechanisms and therapeutic applications. Cell Death Differ. 2019, 26, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Devettere, R.J. Slippery slopes and moral reasoning. J. Clin. Ethic. 1992, 3, 297–301. [Google Scholar]
- Takahashi, Y.; Coppola, D.; Matsushita, N.; Cualing, H.D.; Sun, M.; Sato, Y.; Liang, C.; Jung, J.U.; Cheng, J.Q.; Mulé, J.J.; et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 2007, 9, 1142–1151. [Google Scholar] [CrossRef]
- Toton, E.; Lisiak, N.; Sawicka, P.; Rybczynska, M. Beclin-1 and its role as a target for anticancer therapy. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2014, 65, 459–467. [Google Scholar]
- Tao, H.; Chen, F.; Liu, H.; Hu, Y.; Wang, Y.; Yanling, H. Wnt/β-catenin signaling pathway activation reverses gemcitabine resistance by attenuating Beclin1-mediated autophagy in the MG63 human osteosarcoma cell line. Mol. Med. Rep. 2017, 16, 1701–1706. [Google Scholar] [CrossRef]
- Vega-Rubín-De-Celis, S.; Zou, Z.; Fernández, Á.F.; Ci, B.; Kim, M.; Xiao, G.; Xie, Y.; Levine, B. Increased autophagy blocks HER2-mediated breast tumorigenesis. Proc. Natl. Acad. Sci. USA 2018, 115, 4176–4181. [Google Scholar] [CrossRef]
- Peng, Y.; Miao, H.; Wu, S.; Yang, W.; Zhang, Y.; Xie, G.; Xie, X.; Li, J.; Shi, C.; Ye, L.; et al. ABHD5 interacts with BECN1 to regulate autophagy and tumorigenesis of colon cancer independent of PNPLA2. Autophagy 2016, 12, 2167–2182. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Jeong, E.G.; Lee, S.H.; Yoo, N.J.; Lee, S.H. Somatic mutations of BECN1, an autophagy-related gene, in human cancers. APMIS 2007, 115, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Laddha, S.V.; Ganesan, S.; Chan, C.S.; White, E. Mutational Landscape of the Essential Autophagy Gene BECN1 in Human Cancers. Mol. Cancer Res. 2014, 12, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Yu, J.; Bhagat, G.; Furuya, N.; Hibshoosh, H.; Troxel, A.; Rosen, J.; Eskelinen, E.-L.; Mizushima, N.; Ohsumi, Y.; et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 2003, 112, 1809–1820. [Google Scholar] [CrossRef] [PubMed]
- Takamura, A.; Komatsu, M.; Hara, T.; Sakamoto, A.; Kishi, C.; Waguri, S.; Eishi, Y.; Hino, O.; Tanaka, K.; Mizushima, N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011, 25, 795–800. [Google Scholar] [CrossRef]
- Mathew, R.; Karp, C.M.; Beaudoin, B.; Vuong, N.; Chen, G.; Chen, H.-Y.; Bray, K.; Reddy, A.; Bhanot, G.; Gelinas, C.; et al. Autophagy Suppresses Tumorigenesis through Elimination of p62. Cell 2009, 137, 1062–1075. [Google Scholar] [CrossRef]
- Saito, T.; Ichimura, Y.; Taguchi, K.; Suzuki, T.; Mizushima, T.; Takagi, T.M.K.; Hirose, Y.; Nagahashi, M.; Iso, T.; Fukutomi, T.; et al. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat. Commun. 2016, 7, 12030. [Google Scholar] [CrossRef]
- Moscat, J.; Karin, M.; Diaz-Meco, M.T. p62 in Cancer: Signaling Adaptor Beyond Autophagy. Cell 2016, 167, 606–609. [Google Scholar] [CrossRef]
- Wu, Q.; Xiang, M.; Wang, K.; Chen, Z.; Long, L.; Tao, Y.; Liang, Y.; Yan, Y.; Xiao, Z.; Qiu, S.; et al. Overexpression of p62 Induces Autophagy and Promotes Proliferation, Migration and Invasion of Nasopharyngeal Carcinoma Cells through Promoting ERK Signaling Pathway. Curr. Cancer Drug Targets 2020, 20, 624–637. [Google Scholar] [CrossRef]
- Hwang, S.K.; Jeong, Y.J.; Chang, Y.C. PDCD4 inhibits lung tumorigenesis by the suppressing p62-Nrf2 signaling pathway and upregulating Keap1 expression. Am. J. Cancer Res. 2020, 10, 424–439. [Google Scholar]
- Vara-Perez, M.; Felipe-Abrio, B.; Agostinis, P. Mitophagy in Cancer: A Tale of Adaptation. Cells 2019, 8, 493. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Yi, H.-S.; Kim, H.-W.; Shong, M. Dysregulation of mitophagy in carcinogenesis and tumor progression. Biochim. Biophys. Acta (BBA) Bioenerg. 2017, 1858, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, H.-H.; Cao, Y.-T.; Zhang, L.-L.; Huang, F.; Yi, C. The Role of Mitochondrial Dynamics and Mitophagy in Carcinogenesis, Metastasis and Therapy. Front. Cell Dev. Biol. 2020, 8, 413. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, D.P.; Praharaj, P.P.; Bhol, C.S.; Mahapatra, K.K.; Patra, S.; Behera, B.P.; Mishra, S.R.; Bhutia, S.K. The emerging, multifaceted role of mitophagy in cancer and cancer therapeutics. Semin. Cancer Biol. 2020, 66, 45–58. [Google Scholar] [CrossRef]
- Barazzuol, L.; Giamogante, F.; Brini, M.; Calì, T. PINK1/Parkin Mediated Mitophagy, Ca2+ Signalling, and ER–Mitochondria Contacts in Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 1772. [Google Scholar] [CrossRef]
- Fujiwara, M.; Marusawa, H.; Wang, H.-Q.; Iwai, A.; Ikeuchi, K.; Imai, Y.; Kataoka, A.; Nukina, N.; Takahashi, R.; Chiba, T. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene 2008, 27, 6002–6011. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, C.; Song, J.; Chen, H.; Chen, X.; Ren, L.; Zhou, Z.; Pan, J.; Yang, Z.; Bao, W.; et al. Parkin facilitates proteasome inhibitor-induced apoptosis via suppression of NF-κB activity in hepatocellular carcinoma. Cell Death Dis. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Lu, T.-L.; Huang, G.-J.; Wang, H.-J.; Chen, J.-L.; Hsu, H.-P. Hispolon promotes MDM2 downregulation through chaperone-mediated autophagy. Biochem. Biophys. Res. Commun. 2010, 398, 26–31. [Google Scholar] [CrossRef]
- Bonhoure, A.; Vallentin, A.; Martin, M.; Senff-Ribeiro, A.; Amson, R.; Telerman, A.; Vidal, M. Acetylation of translationally controlled tumor protein promotes its degradation through chaperone-mediated autophagy. Eur. J. Cell Biol. 2017, 96, 83–98. [Google Scholar] [CrossRef]
- Schneider, J.L.; Villarroya, J.; Diaz-Carretero, A.; Patel, B.; Urbanska, A.M.; Thi, M.M.; Villarroya, F.; Santambrogio, L.; Cuervo, A.M. Loss of hepatic chaperone-mediated autophagy accelerates proteostasis failure in aging. Aging Cell 2015, 14, 249–264. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 2015, 17, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Seth, R.K.; Kumar, A.; Kadiiska, M.B.; Michelotti, G.; Diehl, A.M.; Chatterjee, S. Purinergic receptor X7 is a key modulator of metabolic oxidative stress-mediated autophagy and inflammation in experimental nonalcoholic steatohepatitis. Am. J. Physiol. Liver Physiol. 2013, 305, G950–G963. [Google Scholar] [CrossRef]
- Rodriguez-Navarro, J.A.; Kaushik, S.; Koga, H.; Dall’Armi, C.; Shui, G.; Wenk, M.R.; Di Paolo, G.; Cuervo, A.M. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA 2012, 109, E705–E714. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Jogasuria, A.; Yin, H.; Xu, M.-J.; Hu, X.; Wang, J.; Kim, C.; Wu, J.; Lee, K.; Gao, B.; et al. The Detrimental Role Played by Lipocalin-2 in Alcoholic Fatty Liver in Mice. Am. J. Pathol. 2016, 186, 2417–2428. [Google Scholar] [CrossRef]
- Xiongshan, S.; Deng, Y.; Pan, X.; Li, P.; Lai, W.; Luo, H.; Huang, P.; Guan, X.; Deng, Y.; Yan, J.; et al. Downregulation of ATG5-dependent macroautophagy by chaperone-mediated autophagy promotes breast cancer cell metastasis. Sci. Rep. 2017, 7, 4759. [Google Scholar] [CrossRef]
- Dillekås, H.; Rogers, M.S.; Straume, O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019, 8, 5574–5576. [Google Scholar] [CrossRef]
- Saad, A.M.; Gad, M.M.; Al-Husseini, M.J.; AlKhayat, M.A.; Rachid, A.; AlFaar, A.S.; Hamoda, H.M. Suicidal death within a year of a cancer diagnosis: A population-based study. Cancer 2019, 125, 972–979. [Google Scholar] [CrossRef]
- Kenific, C.M.; Thorburn, A.; Debnath, J. Autophagy and metastasis: Another double-edged sword. Curr. Opin. Cell Biol. 2010, 22, 241–245. [Google Scholar] [CrossRef]
- Qiang, L.; Zhao, B.; Ming, M.; Wang, N.; He, T.-C.; Hwang, S.; Thorburn, A.; He, Y.-Y. Regulation of cell proliferation and migration by p62 through stabilization of Twist1. Proc. Natl. Acad. Sci. USA 2014, 111, 9241–9246. [Google Scholar] [CrossRef]
- Lisanti, M.P.; Martinez-Outschoorn, U.E.; Chiavarina, B.; Pavlides, S.; Whitaker-Menezes, D.; Tsirigos, A.; Witkiewicz, A.K.; Lin, Z.; Balliet, R.M.; Howell, A.; et al. Understanding the "lethal" drivers of tumor-stroma co-evolution. Cancer Biol. Ther. 2010, 10, 537–542. [Google Scholar] [CrossRef]
- Mowers, E.E.; Sharifi, M.N.; MacLeod, K.F. Functions of autophagy in the tumor microenvironment and cancer metastasis. FEBS J. 2018, 285, 1751–1766. [Google Scholar] [CrossRef] [PubMed]
- DeNardo, D.G.; Barreto, J.B.; Andreu, P.; Vasquez, L.; Tawfik, D.; Kolhatkar, N.; Coussens, L.M. CD4+ T Cells Regulate Pulmonary Metastasis of Mammary Carcinomas by Enhancing Protumor Properties of Macrophages. Cancer Cell 2009, 16, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Bingle, L.; Brown, N.J.; Lewis, C.E. The role of tumour-associated macrophages in tumour progression: Implications for new anticancer therapies. J. Pathol. 2002, 196, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Li, S.; Li, X.; Wang, W.; Bian, Y.; Wei, S.; Grove, S.; Wang, W.; Vatan, L.; Liu, J.R.; et al. Autophagic adaptation to oxidative stress alters peritoneal residential macrophage survival and ovarian cancer metastasis. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Sharifi, M.N.; Mowers, E.E.; Drake, L.E.; Collier, C.; Chen, H.; Zamora, M.; Mui, S.; MacLeod, K.F. Autophagy Promotes Focal Adhesion Disassembly and Cell Motility of Metastatic Tumor Cells through the Direct Interaction of Paxillin with LC3. Cell Rep. 2016, 15, 1660–1672. [Google Scholar] [CrossRef]
- Guo, W.; Giancotti, F.G. Integrin signalling during tumour progression. Nat. Rev. Mol. Cell Biol. 2004, 5, 816–826. [Google Scholar] [CrossRef]
- Guadamillas, M.C.; Cerezo, A.; Del Pozo, M.Á. Overcoming anoikis—Pathways to anchorage-independent growth in cancer. J. Cell Sci. 2011, 124, 3189–3197. [Google Scholar] [CrossRef]
- Long, X.H.; Zhou, Y.F.; Lan, M.; Huang, S.H.; Liu, Z.L.; Shu, Y. Valosin-containing protein promotes metastasis of osteosarcoma through autophagy induction and anoikis inhibition via the ERK/NF-κβ/beclin-1 signaling pathway. Oncol. Lett. 2019, 18, 3823–3829. [Google Scholar] [CrossRef]
- Schoenherr, C.; Byron, A.; Sandilands, E.; Paliashvili, K.; Baillie, G.S.; Garcia-Munoz, A.; Valacca, C.; Cecconi, F.; Serrels, B.; Frame, M.C. Ambra1 spatially regulates Src activity and Src/FAK-mediated cancer cell invasion via trafficking networks. eLife 2017, 6, 452. [Google Scholar] [CrossRef]
- Li, J.; Yang, B.; Zhou, Q.; Wu, Y.; Shang, D.; Guo, Y.; Song, Z.; Zheng, Q.; Xiong, J. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial–mesenchymal transition. Carcinogenesis 2013, 34, 1343–1351. [Google Scholar] [CrossRef]
- Fan, Q.; Yang, L.; Zhang, X.; Ma, Y.; Li, Y.; Dong, L.; Zong, Z.; Hua, X.; Su, D.; Li, H.; et al. Autophagy promotes metastasis and glycolysis by upregulating MCT1 expression and Wnt/β-catenin signaling pathway activation in hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 2018, 37, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhai, H.; Fesler, A.; Ba, Y.; Wu, S.; Ju, J. Inhibition of colorectal cancer stem cell survival and invasive potential by hsa-miR-140-5p mediated suppression of Smad2 and autophagy. Oncotarget 2015, 6, 19735–19746. [Google Scholar] [CrossRef] [PubMed]
- Zada, S.; Hwang, J.S.; Ahmed, M.; Lai, T.; Pham, T.M.; Kim, D.R. Control of the Epithelial-to-Mesenchymal Transition and Cancer Metastasis by Autophagy-Dependent SNAI1 Degradation. Cells 2019, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Bernardini, J.P.; Lazarou, M.; Dewson, G. Parkin and mitophagy in cancer. Oncogene 2017, 36, 1315–1327. [Google Scholar] [CrossRef]
- Zhang, C.; Lin, M.; Wu, R.; Wang, X.; Yang, B.; Levine, A.J.; Hu, W.; Feng, Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc. Natl. Acad. Sci. USA 2011, 108, 16259–16264. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Gatza, M.L.; Xia, D.; Gao, J.; White, E.; Haffty, B.G.; Hu, W.; Feng, Z.; Zhao, Y.; et al. Parkin targets HIF-1α for ubiquitination and degradation to inhibit breast tumor progression. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef]
- Lee, Y.S.; Jung, Y.Y.; Park, M.H.; Yeo, I.J.; Im, H.S.; Nam, K.T.; Kim, H.D.; Kang, S.K.; Song, J.K.; Kim, Y.R.; et al. Deficiency of parkin suppresses melanoma tumor development and metastasis through inhibition of MFN2 ubiquitination. Cancer Lett. 2018, 433, 156–164. [Google Scholar] [CrossRef]
- Gustafsson, Å.B. Bnip3 as a Dual Regulator of Mitochondrial Turnover and Cell Death in the Myocardium. Pediatr. Cardiol. 2011, 32, 267–274. [Google Scholar] [CrossRef]
- Chourasia, A.H.; Tracy, K.; MacLeod, K.F.; Frankenberger, C.A.; Boland, M.L.; Sharifi, M.N.; Drake, L.E.; Sachleben, J.R.; Asara, J.M.; Locasale, J.W.; et al. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015, 16, 1145–1163. [Google Scholar] [CrossRef]
- Shi, C.; Cai, Y.; Li, Y.; Li, Y.; Hu, N.; Ma, S.; Hu, S.; Zhu, P.; Wang, W.; Zhou, H. Yap promotes hepatocellular carcinoma metastasis and mobilization via governing cofilin/F-actin/lamellipodium axis by regulation of JNK/Bnip3/SERCA/CaMKII pathways. Redox Biol. 2018, 14, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Kon, M.; Kiffin, R.; Koga, H.; Chapochnick, J.; Macian, F.; Varticovski, L.; Cuervo, A.M. Chaperone-Mediated Autophagy Is Required for Tumor Growth. Sci. Transl. Med. 2011, 3, 109ra117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, Y.-Y.; Yao, C.-B.; Li, J.-T.; Zhao, X.-N.; Yang, H.-B.; Zhang, M.; Yin, M.; Chen, J.; Lei, Q.-Y. Acetylation targets HSD17B4 for degradation via the CMA pathway in response to estrone. Autophagy 2017, 13, 538–553. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-J.; Lei, Y.-H.; Yao, N.; Wang, C.-R.; Hu, N.; Ye, W.-C.; Zhang, D.-M.; Chen, Z. Autophagy and multidrug resistance in cancer. Chin. J. Cancer 2017, 36, 52. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, U.K.; Chaudhary, A. Targeting autophagy to overcome drug resistance in cancer therapy. Future Med. Chem. 2015, 7, 1535–1542. [Google Scholar] [CrossRef]
- Wang, F.; Xia, X.; Yang, C.; Shen, J.; Mai, J.; Kim, H.-C.; Kirui, D.; Kang, Y.; Fleming, J.B.; Koay, E.J.; et al. SMAD4Gene Mutation Renders Pancreatic Cancer Resistance to Radiotherapy through Promotion of Autophagy. Clin. Cancer Res. 2018, 24, 3176–3185. [Google Scholar] [CrossRef]
- Taylor, M.A.; Das, B.C.; Ray, S.K. Targeting autophagy for combating chemoresistance and radioresistance in glioblastoma. Apoptosis 2018, 23, 563–575. [Google Scholar] [CrossRef]
- Hao, C.; Liu, G.; Tian, G. Autophagy inhibition of cancer stem cells promotes the efficacy of cisplatin against non-small cell lung carcinoma. Ther. Adv. Respir. Dis. 2019, 13, 1753466619866097. [Google Scholar] [CrossRef]
- Yeo, S.K.; Guan, J.-L. Hierarchical heterogeneity in mammary tumors and its regulation by autophagy. Autophagy 2016, 12, 1960–1961. [Google Scholar] [CrossRef]
- Eapen, V.V.; Waterman, D.P.; Chuartzman, S.G.; Loewith, R.J.; Schuldiner, M.; Denic, V.; Klionsky, D.J.; Haber, J.E.; Bernard, A.; Schiffmann, N.; et al. A pathway of targeted autophagy is induced by DNA damage in budding yeast. Proc. Natl. Acad. Sci. USA 2017, 114, E1158–E1167. [Google Scholar] [CrossRef]
- Weisberg, E.; Nonami, A.; Griffin, J.D. Combination therapy with nilotinib for drug-sensitive and drug-resistant BCR-ABL-positive leukemia and other malignancies. Arch. Toxicol. 2014, 88, 2233–2242. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Yang, L.; Zhang, H.; Wang, Z.; Yu, Y.; Xie, M.; Zhao, M.; Liu, L. S100A8-targeting siRNA enhances arsenic trioxide-induced myeloid leukemia cell death by down-regulating autophagy. Int. J. Mol. Med. 2011, 29, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Wang, W.; Li, Y.; Yang, D.; Li, X.; Shen, C.; Liu, Y.; Ke, X.; Guo, S.; Guo, Z. HSP90AA1-mediated autophagy promotes drug resistance in osteosarcoma. J. Exp. Clin. Cancer Res. 2018, 37, 201. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Chen, W.-M.; Liu, S.; Wang, Z.-H.; Wei, T.-N.; Chen, Z.-Z.; Wu, W.-B. CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk. Res. 2019, 85, 106198. [Google Scholar] [CrossRef] [PubMed]
- Santiago-O’Farrill, J.M.; Weroha, S.J.; Hou, X.; Oberg, A.L.; Bs, E.P.H.; Ms, M.J.M.; Pang, L.; Rask, P.; Amaravadi, R.K.; Becker, S.E.; et al. Poly(adenosine diphosphate ribose) polymerase inhibitors induce autophagy-mediated drug resistance in ovarian cancer cells, xenografts, and patient-derived xenograft models. Cancer 2019, 126, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Choudhury, D.; Das, A.; Das Mukherjee, D.; Dasgupta, M.; Bandopadhyay, S.; Chakrabarti, G. Autophagy inhibition with chloroquine reverts paclitaxel resistance and attenuates metastatic potential in human nonsmall lung adenocarcinoma A549 cells via ROS mediated modulation of β-catenin pathway. Apoptosis 2019, 24, 414–433. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Zhang, C.; Yu, B.; Chen, B.; Liu, Z.; Hou, C.; Wang, F.; Shen, H.; Chen, Z. Autophagic degradation of FOXO3a represses the expression of PUMA to block cell apoptosis in cisplatin-resistant osteosarcoma cells. Am. J. Cancer Res. 2017, 7, 1407–1422. [Google Scholar]
- Chen, S.; Wu, J.; Jiao, K.; Wu, Q.; Ma, J.; Chen, D.; Kang, J.; Zhao, G.; Shi, Y.; Fan, D.; et al. MicroRNA-495-3p inhibits multidrug resistance by modulating autophagy through GRP78/mTOR axis in gastric cancer. Cell Death Dis. 2018, 9, 1070. [Google Scholar] [CrossRef]
- Du, X.; Liu, B.; Luan, X.; Cui, Q.; Li, L. miR-30 decreases multidrug resistance in human gastric cancer cells by modulating cell autophagy. Exp. Ther. Med. 2017, 15, 599–605. [Google Scholar] [CrossRef]
- Scambia, G.; Panici, P.; Baiocchi, G.; Perrone, L.; Greggi, S.; Mancuso, S. CA 15-3 as a tumor marker in gynecological malignancies. Gynecol. Oncol. 1988, 30, 265–273. [Google Scholar] [CrossRef]
- Zhang, H.; Tang, J.; Li, C.; Kong, J.; Wang, J.; Wu, Y.; Xu, E.; Lai, M.-D. MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Lett. 2015, 356, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xiang, N.; Lin, M.; Huang, J.-W.; Zhang, J.; Cheng, B.; Ji, C. miR- 26a Sensitizes Melanoma Cells To Dabrafenib Via Targeting HMGB1-Dependent Autophagy Pathways. Drug Des. Dev. Ther. 2019, 13, 3717–3726. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, K.; Miyata, H.; Makino, T.; Masuike, Y.; Furukawa, H.; Tanaka, K.; Miyazaki, Y.; Takahashi, T.; Kurokawa, Y.; Yamasaki, M.; et al. High Expression of the Mitophagy-Related Protein Pink1 is Associated with a Poor Response to Chemotherapy and a Poor Prognosis for Patients Treated with Neoadjuvant Chemotherapy for Esophageal Squamous Cell Carcinoma. Ann. Surg. Oncol. 2017, 24, 4025–4032. [Google Scholar] [CrossRef] [PubMed]
- Oun, R.; Moussa, Y.E.; Wheate, N.J. Correction: The side effects of platinum-based chemotherapy drugs: A review for chemists. Dalton Trans. 2018, 47, 7848. [Google Scholar] [CrossRef]
- Villa, E.; Proïcs, E.; Rubio-Patiño, C.; Obba, S.; Zunino, B.; Bossowski, J.P.; Rozier, R.M.; Chiche, J.; Mondragón, L.; Riley, J.S.; et al. Parkin-Independent Mitophagy Controls Chemotherapeutic Response in Cancer Cells. Cell Rep. 2017, 20, 2846–2859. [Google Scholar] [CrossRef]
- Yao, N.; Wang, C.; Chen, W.-M.; Chen, Z.; Fu, D.; Ye, W.; Zhang, D.-M.; Hu, N.; Li, Y.; Liu, M.; et al. Inhibition of PINK1/Parkin-dependent mitophagy sensitizes multidrug-resistant cancer cells to B5G1, a new betulinic acid analog. Cell Death Dis. 2019, 10, 232. [Google Scholar] [CrossRef]
- Basit, F.; Van Oppen, L.M.; Schöckel, L.; Bossenbroek, H.M.; Vries, S.E.V.E.-D.; Hermeling, J.C.; Grefte, S.; Kopitz, C.; Heroult, M.; Willems, P.H.; et al. Mitochondrial complex I inhibition triggers a mitophagy-dependent ROS increase leading to necroptosis and ferroptosis in melanoma cells. Cell Death Dis. 2017, 8, e2716. [Google Scholar] [CrossRef]
- Zhou, J.; Li, G.; Zheng, Y.; Shen, H.-M.; Hu, X.; Ming, Q.-L.; Huang, C.; Li, P.; Gao, N. A novel autophagy/mitophagy inhibitor liensinine sensitizes breast cancer cells to chemotherapy through DNM1L-mediated mitochondrial fission. Autophagy 2015, 11, 1259–1279. [Google Scholar] [CrossRef]
- Lobo, N.A.; Shimono, Y.; Qian, D.; Clarke, M.F. The Biology of Cancer Stem Cells. Annu. Rev. Cell Dev. Biol. 2007, 23, 675–699. [Google Scholar] [CrossRef]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef]
- Bellodi, C.; Lidonnici, M.R.; Hamilton, A.; Helgason, G.V.; Soliera, A.R.; Ronchetti, M.; Galavotti, S.; Young, K.W.; Selmi, T.; Yacobi, R.; et al. Targeting autophagy potentiates tyrosine kinase inhibitor–induced cell death in Philadelphia chromosome–positive cells, including primary CML stem cells. J. Clin. Investig. 2009, 119, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Gomez-Manzano, C.; Aoki, H.; Alonso, M.M.; Kondo, S.; McCormick, F.; Xu, J.; Kondo, Y.; Bekele, B.N.; Colman, H.; et al. Examination of the Therapeutic Potential of Delta-24-RGD in Brain Tumor Stem Cells: Role of Autophagic Cell Death. J. Natl. Cancer Inst. 2007, 99, 1410–1414. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, D.; Liu, Y.; Su, Z.; Zhang, L.; Chen, F.; Zhou, Y.; Wu, Y.; Yu, M.; Zhang, Z.; et al. Role of the Hypoxia-inducible factor-1 alpha induced autophagy in the conversion of non-stem pancreatic cancer cells into CD133+ pancreatic cancer stem-like cells. Cancer Cell Int. 2013, 13, 119. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.-L.; Simon, A.K.; Prescott, M.; Menendez, J.A.; Liu, F.; Wang, F.; Wang, C.; Wolvetang, E.J.; Vazquez-Martin, A.; Zhang, J. Autophagy in stem cells. Autophagy 2013, 9, 830–849. [Google Scholar] [CrossRef]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2006, 445, 106–110. [Google Scholar] [CrossRef]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2006, 445, 111–115. [Google Scholar] [CrossRef]
- Gong, C.; Bauvy, C.; Tonelli, G.; Yue, W.; Delomenie, C.; Nicolas, V.; Zhu, Y.; Domergue, V.; Marinesteban, V.; Tharinger, H.; et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene 2012, 32, 2261–2272. [Google Scholar] [CrossRef]
- Chaterjee, M.; Van Golen, K.L. Breast Cancer Stem Cells Survive Periods of Farnesyl-Transferase Inhibitor-Induced Dormancy by Undergoing Autophagy. Bone Marrow Res. 2011, 2011, 3629381. [Google Scholar] [CrossRef]
- Hermann, P.C.; Huber, S.L.; Herrler, T.; Aicher, A.; Ellwart, J.W.; Guba, M.; Bruns, C.J.; Heeschen, C. Distinct Populations of Cancer Stem Cells Determine Tumor Growth and Metastatic Activity in Human Pancreatic Cancer. Cell Stem Cell 2007, 1, 313–323. [Google Scholar] [CrossRef]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.S.; Clarke, M.F.; Simeone, D.M. Identification of Pancreatic Cancer Stem Cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef]
- Momburg, F.; Roelse, J.; Howard, J.C.; Butcher, G.W.; Hämmerling, G.J.; Neefjes, J.J. Selectivity of MHC-encoded peptide transporters from human, mouse and rat. Nat. Cell Biol. 1994, 367, 648–651. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Balch, C.; Chan, M.W.; Lai, H.-C.; Matei, D.; Schilder, J.M.; Yan, P.S.; Huang, T.H.-M.; Nephew, K.P. Identification and Characterization of Ovarian Cancer-Initiating Cells from Primary Human Tumors. Cancer Res. 2008, 68, 4311–4320. [Google Scholar] [CrossRef] [PubMed]
- Buccarelli, M.; Marconi, M.; Pacioni, S.; De Pasqualis, I.; D’Alessandris, Q.G.; Martini, M.; Ascione, B.; Malorni, W.; LaRocca, L.M.; Pallini, R.; et al. Inhibition of autophagy increases susceptibility of glioblastoma stem cells to temozolomide by igniting ferroptosis. Cell Death Dis. 2018, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kantara, C.; O’Connell, M.; Sarkar, S.; Moya, S.; Ullrich, R.; Singh, P. Curcumin promotes autophagic survival of a subset of colon cancer stem cells, which are ablated by DCLK1-siRNA. Cancer Res. 2014, 74, 2487–2498. [Google Scholar] [CrossRef]
- Han, Y.; Fan, S.; Qin, T.; Yang, J.; Sun, Y.; Lu, Y.; Mao, J.; Li, L. Role of autophagy in breast cancer and breast cancer stem cells (Review). Int. J. Oncol. 2018, 52, 1057–1070. [Google Scholar] [CrossRef]
- Gong, C.; Song, E.; Codogno, P.; Mehrpour, M. The roles of BECN1 and autophagy in cancer are context dependent. Autophagy 2012, 8, 1853–1855. [Google Scholar] [CrossRef]
- Wolf, J.; Dewi, D.L.; Fredebohm, J.; Müller-Decker, K.; Flechtenmacher, C.; Hoheisel, J.; Boettcher, M. A mammosphere formation RNAi screen reveals that ATG4A promotes a breast cancer stem-like phenotype. Breast Cancer Res. 2013, 15, R109. [Google Scholar] [CrossRef]
- Sanchez, C.G.; Penfornis, P.; Oskowitz, A.Z.; Boonjindasup, A.G.; Cai, D.Z.; Dhule, S.S.; Rowan, B.G.; Kelekar, A.; Krause, D.S.; Pochampally, R.R. Activation of autophagy in mesenchymal stem cells provides tumor stromal support. Carcinogenesis 2011, 32, 964–972. [Google Scholar] [CrossRef]
- Rahman, A.; Saha, S.K.; Rahman, S.; Uddin, J.; Uddin, S.; Pang, M.-G.; Rhim, H.; Cho, S.-G. Molecular Insights Into Therapeutic Potential of Autophagy Modulation by Natural Products for Cancer Stem Cells. Front. Cell Dev. Biol. 2020, 8, 283. [Google Scholar] [CrossRef]
- Altman, B.J.; Jacobs, S.R.; Mason, E.F.; Michalek, R.D.; MacIntyre, A.N.; Coloff, J.L.; Ilkayeva, O.; Jia, W.; He, Y.-W.; Rathmell, J.C. Autophagy is essential to suppress cell stress and to allow BCR-Abl-mediated leukemogenesis. Oncogene 2010, 30, 1855–1867. [Google Scholar] [CrossRef]
- Karvela, M.; Baquero, P.; Kuntz, E.M.; Mukhopadhyay, A.; Mitchell, R.; Allan, E.K.; Chan, E.; Kranc, K.R.; Calabretta, B.; Salomoni, P.; et al. ATG7 regulates energy metabolism, differentiation and survival of Philadelphia-chromosome-positive cells. Autophagy 2016, 12, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Rothe, K.; Lin, H.; Lin, K.B.L.; Leung, A.; Wang, H.M.; Malekesmaeili, M.; Brinkman, R.; Forrest, D.L.; Gorski, S.M.; Jiang, X. The core autophagy protein ATG4B is a potential biomarker and therapeutic target in CML stem/progenitor cells. Blood 2014, 123, 3622–3634. [Google Scholar] [CrossRef] [PubMed]
- Ojha, R.; Jha, V.; Singh, S.K.; Bhattacharyya, S. Autophagy inhibition suppresses the tumorigenic potential of cancer stem cell enriched side population in bladder cancer. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2014, 1842, 2073–2086. [Google Scholar] [CrossRef] [PubMed]
- Kenific, C.M.; Debnath, J. Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol. 2014, 25, 37–45. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, L.; Zhou, H.; Wang, W.; Luo, Y.; Yang, H.; Yi, H. Inhibition of autophagy promotes cisplatin-induced apoptotic cell death through Atg5 and Beclin 1 in A549 human lung cancer cells. Mol. Med. Rep. 2018, 17, 6859–6865. [Google Scholar] [CrossRef]
- Quan, Y.; Lei, H.; Wahafu, W.; Liu, Y.; Ping, H.; Zhang, X. Inhibition of autophagy enhances the anticancer effect of enzalutamide on bladder cancer. Biomed. Pharmacother. 2019, 120, 109490. [Google Scholar] [CrossRef]
- Sheng, B.; Song, Y.; Zhang, J.; Li, R.; Wang, Z.; Zhu, X. Atorvastatin suppresses the progression of cervical cancer via regulation of autophagy. Am J Transl Res 2020, 12, 5252–5268. [Google Scholar]
- Shin, D.; Kim, E.H.; Lee, J.; Roh, J.-L. RITA plus 3-MA overcomes chemoresistance of head and neck cancer cells via dual inhibition of autophagy and antioxidant systems. Redox Biol. 2017, 13, 219–227. [Google Scholar] [CrossRef]
- Cheng, X.; Feng, H.; Wu, H.; Jin, Z.; Shen, X.; Kuang, J.; Huo, Z.; Chen, X.; Gao, H.; Ye, F.; et al. Targeting autophagy enhances apatinib-induced apoptosis via endoplasmic reticulum stress for human colorectal cancer. Cancer Lett. 2018, 431, 105–114. [Google Scholar] [CrossRef]
- Yuan, N.; Song, L.; Cai, J.; Wang, J.; Zhang, Y.; Mao, X.; Zhao, W.; Hu, S.; Chen, S.; Zhang, S.; et al. Bafilomycin A1 targets both autophagy and apoptosis pathways in pediatric B-cell acute lymphoblastic leukemia. Haematologica 2015, 100, 345–356. [Google Scholar] [CrossRef]
- Lin, J.-F.; Lin, Y.-C.; Tsai, T.-F.; Chen, H.-E.; Chou, K.-Y.; Hwang, T.I.-S. Cisplatin induces protective autophagy through activation of BECN1 in human bladder cancer cells. Drug Des. Dev. Ther. 2017, 11, 1517–1533. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-Q.; Xie, W.; Pan, D.; Chen, H.; Zhang, L. Inhibition of autophagy by bafilomycin A1 promotes chemosensitivity of gastric cancer cells. Tumor Biol. 2016, 37, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Lin, J.-F.; Wen, S.-I.; Yang, S.-C.; Tsai, T.-F.; Chen, H.-E.; Chou, K.-Y.; Hwang, T.I.-S. Chloroquine and hydroxychloroquine inhibit bladder cancer cell growth by targeting basal autophagy and enhancing apoptosis. Kaohsiung J. Med Sci. 2017, 33, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.-F.; Kim, C.-F.; Chow, H.-Y.; Chong, H.-C.; Tam, S.-Y.; Leung, Y.-C.; Lo, W.-H. Recombinant Bacillus caldovelox Arginase Mutant (BCA-M) Induces Apoptosis, Autophagy, Cell Cycle Arrest and Growth Inhibition in Human Cervical Cancer Cells. Int. J. Mol. Sci. 2020, 21, 7445. [Google Scholar] [CrossRef]
- Scott, E.C.; Maziarz, R.T.; Cascio, M.J.; Podolak, J.; Gordon, M.; Botelho, J.; Stadtmauer, E.; Amaravadi, R.; Vogl, D.T.; Spurgeon, S.E.; et al. Double autophagy stimulation using chemotherapy and mTOR inhibition combined with hydroxychloroquine for autophagy modulation in patients with relapsed or refractory multiple myeloma. Haematologica 2017, 102, e261–e265. [Google Scholar] [CrossRef]
- Patel, S.; Hurez, V.; Nawrocki, S.T.; Goros, M.; Michalek, J.; Sarantopoulos, J.; Curiel, T.; Mahalingam, D. Vorinostat and hydroxychloroquine improve immunity and inhibit autophagy in metastatic colorectal cancer. Oncotarget 2016, 7, 59087–59097. [Google Scholar] [CrossRef]
- Boone, B.A.; Bahary, N.; Espina, V.; Loughran, P.; Lotze, M.T.; Zeh, H.J.; Zureikat, A.H.; Moser, A.J.; Normolle, D.P.; Wu, W.-C.; et al. Safety and Biologic Response of Pre-operative Autophagy Inhibition in Combination with Gemcitabine in Patients with Pancreatic Adenocarcinoma. Ann. Surg. Oncol. 2015, 22, 4402–4410. [Google Scholar] [CrossRef]
- Egan, D.F.; Chun, M.G.; Vamos, M.; Zou, H.; Rong, J.; Miller, C.J.; Lou, H.J.; Raveendra-Panickar, D.; Yang, C.-C.; Sheffler, D.J.; et al. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol. Cell 2015, 59, 285–297. [Google Scholar] [CrossRef]
- Martin, K.R.; Celano, S.L.; Solitro, A.R.; Gunaydin, H.; Scott, M.; O’Hagan, R.C.; Shumway, S.D.; Fuller, P.; MacKeigan, J.P. A Potent and Selective ULK1 Inhibitor Suppresses Autophagy and Sensitizes Cancer Cells to Nutrient Stress. iScience 2018, 8, 74–84. [Google Scholar] [CrossRef]
- Pasquier, B. SAR405, a PIK3C3/Vps34 inhibitor that prevents autophagy and synergizes with MTOR inhibition in tumor cells. Autophagy 2015, 11, 725–726. [Google Scholar] [CrossRef]
- Dyczynski, M.; Yu, Y.; Otrocka, M.; Parpal, S.; Braga, T.; Henley, A.B.; Zazzi, H.; Lerner, M.; Wennerberg, K.; Viklund, J.; et al. Targeting autophagy by small molecule inhibitors of vacuolar protein sorting 34 (Vps34) improves the sensitivity of breast cancer cells to Sunitinib. Cancer Lett. 2018, 435, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Luanpitpong, S.; Chanvorachote, P.; Nimmannit, U.; Leonard, S.S.; Stehlik, C.; Wang, L.; Rojanasakul, Y. Mitochondrial superoxide mediates doxorubicin-induced keratinocyte apoptosis through oxidative modification of ERK and Bcl-2 ubiquitination. Biochem. Pharmacol. 2012, 83, 1643–1654. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Luo, L.; Guo, C.-Y.; Goto, S.; Urata, Y.; Shao, J.-H.; Li, T.-S. Doxorubicin-induced mitophagy contributes to drug resistance in cancer stem cells from HCT8 human colorectal cancer cells. Cancer Lett. 2017, 388, 34–42. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Gu, K. Tanshinone IIA regulates colorectal cancer apoptosis via attenuation of Parkin-mediated mitophagy by suppressing AMPK/Skp2 pathways. Mol. Med. Rep. 2018, 18, 1692–1703. [Google Scholar] [CrossRef]
- Si, L.; Fu, J.; Liu, W.; Hayashi, T.; Mizuno, K.; Hattori, S.; Fujisaki, H.; Onodera, S.; Ikejima, T. Silibinin-induced mitochondria fission leads to mitophagy, which attenuates silibinin-induced apoptosis in MCF-7 and MDA-MB-231 cells. Arch. Biochem. Biophys. 2020, 685, 108284. [Google Scholar] [CrossRef]
- Kocaturk, N.M.; Akkoc, Y.; Kig, C.; Bayraktar, O.; Gozuacik, D.; Kutlu, O. Autophagy as a molecular target for cancer treatment. Eur. J. Pharm. Sci. 2019, 134, 116–137. [Google Scholar] [CrossRef]
- Singh, S.S.; Vats, S.; Chia, A.Y.-Q.; Tan, T.Z.; Deng, S.; Ong, M.S.; Arfuso, F.; Yap, C.T.; Goh, B.C.; Sethi, G.; et al. Dual role of autophagy in hallmarks of cancer. Oncogene 2018, 37, 1142–1158. [Google Scholar] [CrossRef]
- Russo, M.; Milito, A.; Spagnuolo, C.; Carbone, V.; Rosén, A.; Minasi, P.; Lauria, F.; Russo, G.L. CK2 and PI3K are direct molecular targets of quercetin in chronic lymphocytic leukaemia. Oncotarget 2017, 8, 42571–42587. [Google Scholar] [CrossRef]
- Karpel-Massler, G.; Ishida, C.T.; Zhang, Y.; Halatsch, M.-E.; Westhoff, M.-A.; Siegelin, M.D. Targeting intrinsic apoptosis and other forms of cell death by BH3-mimetics in glioblastoma. Expert Opin. Drug Discov. 2017, 12, 1031–1040. [Google Scholar] [CrossRef]
- Takahashi, A.; Kimura, F.; Yamanaka, A.; Takebayashi, A.; Kita, N.; Takahashi, K.; Murakami, T. Metformin impairs growth of endometrial cancer cells via cell cycle arrest and concomitant autophagy and apoptosis. Cancer Cell Int. 2014, 14, 53. [Google Scholar] [CrossRef]
- Nazim, U.M.; Moon, J.-H.; Lee, J.-H.; Lee, Y.-J.; Seol, J.-W.; Eo, S.-K.; Lee, J.-H.; Park, S.-Y. Activation of autophagy flux by metformin downregulates cellular FLICE-like inhibitory protein and enhances TRAIL- induced apoptosis. Oncotarget 2016, 7, 23468–23481. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hao, Y.; Li, Y.; Zheng, Y.; Dai, J.; Zhong, F.; Wei, W.; Fang, Z. Salinomycin induces autophagic cell death in salinomycin-sensitive melanoma cells through inhibition of autophagic flux. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Ding, N.; Ge, J.; Wang, Y.; Wang, L.; Wu, N.; Wei, Q.; Xu, S.; Liu, X.; Zhou, G. Esomeprazole overcomes paclitaxel-resistance and enhances anticancer effects of paclitaxel by inducing autophagy in A549/Taxol cells. Cell Biol. Int. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ozates, N.P.; Soğutlu, F.; Lerminoglu, F.; Demir, B.; Gunduz, C.; Shademan, B.; Avci, C.B. Effects of rapamycin and AZD3463 combination on apoptosis, autophagy, and cell cycle for resistance control in breast cancer. Life Sci. 2020, 10, 118643. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Luo, Y.; Li, S.; Hong, M.; Wang, Q.; Chi, X.; Yang, C. ISL Induces Apoptosis and Autophagy in Hepatocellular Carcinoma via Downregulation of PI3K/AKT/mTOR Pathway in vivo and in vitro. Drug Des. Dev. Ther. 2020, 14, 4363–4376. [Google Scholar] [CrossRef] [PubMed]
- Boutouja, F.; Stiehm, C.M.; Platta, H.W. mTOR: A Cellular Regulator Interface in Health and Disease. Cells 2019, 8, 18. [Google Scholar] [CrossRef]
- Blagosklonny, M.V. Rapalogs in cancer prevention. Cancer Biol. Ther. 2012, 13, 1349–1354. [Google Scholar] [CrossRef]
- Wang, H.; Li, D.; Li, X.; Ou, X.; Liu, S.; Zhang, Y.; Ding, J.; Xie, B. Mammalian target of rapamycin inhibitor RAD001 sensitizes endometrial cancer cells to paclitaxel-induced apoptosis via the induction of autophagy. Oncol. Lett. 2016, 12, 5029–5035. [Google Scholar] [CrossRef]
- Dai, Z.; Gao, J.; Ma, X.; Kang, H.-F.; Wang, B.; Lu, W.-F.; Lin, S.; Wang, X.-J.; Wu, W.-Y. Antitumor Effects of Rapamycin in Pancreatic Cancer Cells by Inducing Apoptosis and Autophagy. Int. J. Mol. Sci. 2012, 14, 273–285. [Google Scholar] [CrossRef]
| Compound | Combination Treatment | Cancer Type | Experimental Model | Function | Reference |
|---|---|---|---|---|---|
| ATG5 siRNA Beclin-1 siRNA | Cisplatin | Lung cancer (A549) | In vitro | Inhibition of autophagy Restore the sensitivity of cisplatin Enhancement of cisplatin-mediated apoptosis Upregulation of caspase activity Reduction of cell viability | [155] |
| 3-MA | Colorectal cancer (HCT116) | In vitro | Promotion of hypoxia-mediated apoptosis | [9] | |
| Enzalutamide | Chloroquine 3-MA Bafilomycin A1 | Bladder cancer (J82, T24, and UMUC3) | In vitro /In vivo | Restores the sensitivity against ENZ Reduction of autophagy and tumor growth Induction of apoptosis | [156] |
| Atorvastatin | 3-MA Bafilomycin A1 | Cervical cancer (SiHa and Caski) | In vitro /In vivo | Enhancement of ATO-mediated apoptosis Reduction of autophagy | [157] |
| Reactivation of p53 and induction of tumor cell apoptosis (RITA) | 3-MA | Head and neck cancer (AMC-HN2-10) | In vitro /In vivo | Promotion of therapeutic effects of cisplatin resistance or RITA-resistant cancer Inhibition of autophagy Induction of apoptosis | [158] |
| Bafilomycin A1 | Pediatric B-cell acute lymphoblastic leukemia (RS4;11, NB4, HL-60, K562 and BV173) | In vitro /In vivo | Therapeutic effect at low concentrations Inhibition of autophagy Targeting mitochondria Induction of apoptosis | [160] | |
| Cisplatin | Bafilomycin A1 chloroquine | Bladder cancer (5637 and T25) | In vitro | Enhancement of the therapeutic effect to cisplatin Inhibition of autophagy | [161] |
| Bafilomycin A1 | 5-FU | Gastric cancer (SGC-7901) | In vitro | Inhibition of cell viability, colony formation, invasion, and migration Enhancement of apoptosis Suppression of autophagy | [162] |
| Chloroquine and hydroxychloroquine | Bladder cancer (RT4, 5637, T24, PC3, and MCF-7) | In vitro | Inhibition of autophagy Induction of apoptosis | [163] | |
| Recombinant Bacillus caldovelox arginase mutant | Chloroquine | Cervical cancer (Hela, ME-180, C-33A and SiHa) | In vitro | Reduction of tumor growth Increased apoptosis and cell cycle arrest Reduction of autophagy | [164] |
| SBI-0206965 | mTOR inhibitors | Prostate cancer Lung cancer glioblastoma (HEK-293T, U87MG, PC3 and A549) | In vitro | Inhibition of autophagy Reduction of cell survival Promotion of cell death | [169] |
| SAR405 | Everolimus | Renal cancer | In vitro | Inhibition of autophagy Suppression of catalytic activity of PI3KC3 Reduction of cell proliferation | [170] |
| SB02024 | Sunitinib Erlotinib | Breast cancer (HOS and MDA-MB-231) | In vitro /In vivo | Inhibition of autophagy Improvement of sensitivity to Sunitinib and Erlotinib | [171] |
| Doxorubicin | BNIP3L | Colorectal cancer (HCT8) | In vitro | Inhibition of mitophagy Restoration of the sensitivity of doxorubicin | [173] |
| Tanshinone IIA | 3-MA | Colorectal cancer (SW837 and SW480) | In vitro | Reduction of mitophagy Promotion of mitochondrial apoptosis Decrease of AMPK and Parkin | [174] |
| Mitochondrial division inhibitor 1 | Silibinin | Breast cancer (MCF7 and MDA-MB-231) | In vitro | Inhibition of DRP1 and Dynamin I Decrease of mitophagy Enhancement of silibinin-induced apoptosis | [175] |
| Compound | Combination Treatment | Cancer Type | Experimental Model | Function | Reference |
|---|---|---|---|---|---|
| Quercetin | ABT-737 ABT-263 | Leukemic cell lines B-cells (HG3) | In vitro | Inhibition of the PI3K/AKT pathway Induction of autophagy Restoration of the sensitivity to ABT-737 | [178] |
| ABT-737 ABT-263 ABT-199 | Glioblastoma cells | In vitro | Induction of autophagic cell death Interruption of the interaction with Beclin-1 and Bcl2 | [179] | |
| Metformin | 3-MA Chloroquine | Endometrial cancer cells (Ishikawa cells) | In vitro | Inhibition of cell viability and proliferation Increased cell cycle arrest and apoptosis Enhancement of autophagy | [180] |
| TRAIL-resistant lung cancer (A549, Calu-3 and HCC-15) | In vitro | Promotion of autophagic flux Accumulation of LC3-II Reduction of p62 | [181] | ||
| Salinomycin | Melanoma cells (M7, M8, M21, M29, SK-MEL-1, SK-MEL-12 and A375) | In vitro /In vivo | Induction of cell death Accumulation of abnormal mitochondria Increased ER stress | [182] | |
| Esomeprazole | Paclitaxel | Non-small cell lung cancer (A549) | In vitro | Restoration of the sensitivity to paclitaxel Inhibition of V-ATPase and cell proliferation Enhancement of autophagy | [183] |
| AZD3463 | Rapamycin | Breast cancer (MCF7) | In vitro | AZD3463: ALK/IGF1R inhibitor Promotion of apoptosis and autophagy Reduction of cell proliferation | [184] |
| Isoliquiritigenin | Hepatocellular carcinoma (MHCC97-H, LO2 and SMMC7721) | In vitro /In vivo | Inhibition of cell growth Enhancement of apoptosis and autophagy Modulation of the PI3K/AKT/mTOR pathway | [185] | |
| RAD-001 | Paclitaxel | Endometrial cancer cells (Ishikawa and HEC-1A) | In vitro | Induction of sensitivity to paclitaxel Promotion of apoptosis and autophagy Downregulation of AKT/mTOR Accumulation of LC3 | [188] |
| Rapamycin | Pancreatic carcinoma (PC-2) | In vitro | Activation of Beclin-1 Induction of autophagic vacuoles Inhibition of proliferation and induction of apoptosis | [189] | |
| Everolimus | Hydroxychloroquine | Lymphangioleiomyomatosis | Phase I /Complete | Investigation of the effect on the regulation of autophagy in lymphangioleiomyomatosis | NCT01687179 |
| Rapamune | Hydroxychloroquine | Advanced cancer | Phase I /Active | Investigation of the effect on the regulation of autophagy in advanced cancer | NCT01266057 |
| NCT Number | Title | Status | Cancer Type | Drugs | Phase |
|---|---|---|---|---|---|
| NCT03037437 | Sorafenib Induced Autophagy Using Hydroxychloroquine in Hepatocellular Cancer | Recruiting | Hepatocellular cancer | Sorafenib Hydroxychloroquine | Phase II |
| NCT01649947 | Modulation of Autophagy in Patients With Advanced/Recurrent Non-small Cell Lung Cancer | Complete | Non-small cell lung cancer | Paclitaxel Carboplatin Hydroxychloroquine Bevacizumab | Phase II |
| NCT04214418 | Study of Combination Therapy With the MEK Inhibitor, Cobimetinib, Immune Checkpoint Blockade, Atezolizumab, and the AUTOphagy Inhibitor, Hydroxychloroquine in KRAS-mutated Advanced Malignancies | Recruiting | Gastrointestinal cancer | Cobimetinib Hydroxychloroquine Atezolizumab | Phase I/II |
| NCT04333914 | Prospective Study in Patients With Advanced or Metastatic Cancer and SARS-CoV-2 Infection | Recruiting | Advanced or Metastatic Hematological or Solid Tumor | Autophagy inhibitor (GNS651) Avdoralimab Monalizumab | Phase II |
| NCT01206530 | FOLFOX/Bevacizumab/Hydroxychloroquine (HCQ) in Colorectal Cancer | Complete | Rectal and colon cancer | Hydroxychloroquine Oxaliplatin Leucovorin | Phase I/II |
| NCT03774472 | Hydroxychloroquine, Palbociclib, and Letrozole Before Surgery in Treating Participants With Estrogen Receptor Positive, HER2 Negative Breast Cancer | Recruiting | Breast cancer | Hydroxychloroquine Letrozole Palbociclib | Phase I/II |
| NCT02316340 | Vorinostat Plus Hydroxychloroquine Versus Regorafenib in Colorectal Cancer | Complete | Colorectal cancer | Voriostat Hydroxychloroquine Regorafenib | Phase II |
| NCT04132505 | Binimetinib and Hydroxychloroquine in Treating Patients With KRAS Mutant Metastatic Pancreatic Cancer | Recruiting | Pancreatic cancer | Binimetinib Hydroxychloroquine | Phase I |
| NCT04524702 | Paricalcitol and Hydroxychloroquine in Combination With Gemcitabine and Nab-Paclitaxel for the Treatment of Advanced or Metastatic Pancreatic Cancer | Recruiting | Pancreatic cancer | Cemcitabine Hydroxychloroquine Nab-paclitaxel Paricaitol | Phase II |
| NCT03377179 | A Study of ABC294640 (Yeliva ®) Alone and in Combination With Hydroxychloroquine Sulfate in Treatment of Patients With Advanced Cholangiocarcinoma | Recruiting | Cholangiocarcinoma | ABC294640 Hydroxychloroquine | Phase II |
| NCT04163107 | Combined Carfilzomib and Hydroxychloroquine in Patients With Relapsed/Refractory Multiple Myeloma | Recruiting | Multiple Myeloma | Hydroxychloroquine Carfizomib Dexamethasone | Phase I |
| NCT03598595 | Gemcitabine, Docetaxel, and Hydroxychloroquine in Treating Participants With Recurrent or Refractory Osteosarcoma | Recruiting | Osteosarcoma | Docetaxel Gemcitabine Hydroxychloroquine | Phase I/II |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yun, C.W.; Jeon, J.; Go, G.; Lee, J.H.; Lee, S.H. The Dual Role of Autophagy in Cancer Development and a Therapeutic Strategy for Cancer by Targeting Autophagy. Int. J. Mol. Sci. 2021, 22, 179. https://doi.org/10.3390/ijms22010179
Yun CW, Jeon J, Go G, Lee JH, Lee SH. The Dual Role of Autophagy in Cancer Development and a Therapeutic Strategy for Cancer by Targeting Autophagy. International Journal of Molecular Sciences. 2021; 22(1):179. https://doi.org/10.3390/ijms22010179
Chicago/Turabian StyleYun, Chul Won, Juhee Jeon, Gyeongyun Go, Jun Hee Lee, and Sang Hun Lee. 2021. "The Dual Role of Autophagy in Cancer Development and a Therapeutic Strategy for Cancer by Targeting Autophagy" International Journal of Molecular Sciences 22, no. 1: 179. https://doi.org/10.3390/ijms22010179
APA StyleYun, C. W., Jeon, J., Go, G., Lee, J. H., & Lee, S. H. (2021). The Dual Role of Autophagy in Cancer Development and a Therapeutic Strategy for Cancer by Targeting Autophagy. International Journal of Molecular Sciences, 22(1), 179. https://doi.org/10.3390/ijms22010179





