Exploring the Triple Interaction between the Host Genome, the Epigenome, and the Gut Microbiome in Type 1 Diabetes
Abstract
1. Introduction
2. Genetic Predisposition to T1D
3. Gut Microbiome, Immunity, and T1D
3.1. Mode of Delivery and Risk of T1D
3.2. Environmental Factors, Gut Bacteriome, and T1D
3.3. Gut Virome, Immunity, and T1D
3.4. Gut Mycobiome and T1D
3.5. Role of the Gut Microbiome in Animal Models with T1D
4. Microbial Metabolites, Probiotics, and T1D
5. Role of Genetic Predisposition on the Gut Microbiome Composition in Individuals with T1D
6. Role of Gut Microbiome in Gene Expression and Epigenetic Regulations of T1D
6.1. Non-Coding RNA Binding
6.2. Histones Modifications
6.3. DNA Methylation
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| T1D | Type 1 diabetes |
| MHC-II | Major histocompatibility class II |
| SCFA | Short-chain fatty acid |
| Treg | T regulatory cells |
| TCR | T cell antigen receptor |
| IGRP-reactive CD8+ T cells | Islet-specific glucose-6-phosphatase catalytic subunit–related protein (IGRP) reactive CD8 T cell |
| LPS | Lipopolysaccharide |
| IAA | Insulin autoantibody |
| GADA | Glutamic acid decarboxylase antibody |
| FOXP3 | Forkhead box P3 |
| HDACi | Histone deactylase inhibitor |
| HMGB1 | High-mobility group protein 1 |
| NF-κB p65 | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| mTOR complex | Mammalian target of rapamycin complex |
| HATs | Histone acetyltransferases |
| PSA | Polysaccharide A |
| TEDDY | The Environmental Determinants of Diabetes in the Young |
| Rorc | RAR-related orphan receptor C gene |
| Stat3 transcription factor | Signal transducer and activator of transcription 3 |
| TGF-β gene | Transforming growth factor β gene |
| TGF-β | Transforming growth factor β |
| ATRA | Alltrans retinoic acid |
| INS | Insulin gene 3 |
| PTPN-22 | Protein tyrosine phosphatase non-receptor type 22 gene |
| CTLA4 | Cytotoxic T-lymphocyte-associated protein 4 gene |
| IL2RA | Interleukin 2 receptor alpha |
| IFIH1 | Interferon-induced with helicase C domain 1 |
| CDHR5 | Cadherin-related family member 5 |
| CDH1 | Cadherin-1 |
| FCGBP | IgGFc-binding protein |
| CEACAM5 | Carcinoembryonic antigen-related cell adhesion molecule 5 |
| MUC2 | Mucin-2 |
| MGAM | Maltase-glucoamylase |
| NAALADL1 | N-acetylated alpha-linked acidic dipeptidase like 1 |
| PXK | PX domain-containing protein kinase-like protein |
| PDHB | Pyruvate Dehydrogenase E1 Subunit Beta |
| PPIL2 | Peptidylprolyl Isomerase Like 2 |
References
- Siljander, H.; Honkanen, J.; Knip, M. Microbiome and type 1 diabetes. EBioMedicine 2019, 46, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Rewers, M.; Ludvigsson, J. Environmental risk factors for type 1 diabetes. Lancet 2016, 387, 2340–2348. [Google Scholar] [CrossRef]
- Kemppainen, K.M.; Ardissone, A.N.; Davis-Richardson, A.G.; Fagen, J.R.; Gano, K.A.; León-Novelo, L.G.; Vehik, K.; Casella, G.; Simell, O.; Ziegler, A.G.; et al. Early childhood gut microbiomes show strong geographic differences among subjects at high risk for type 1 diabetes. Diabetes Care 2015, 38, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Knip, M.; Simell, O. Environmental triggers of type 1 diabetes. Cold Spring Harb. Perspect. Med. 2012, 2, a007690. [Google Scholar] [CrossRef] [PubMed]
- Blohmé, G.; Nyström, L.; Arnqvist, H.; Lithner, F.; Littorin, B.; Olsson, P.O.; Scherstén, B.; Wibell, L.; Ostman, J. Male predominance of type 1 (insulin-dependent) diabetes mellitus in young adults: Results from a 5-year prospective nationwide study of the 15–34-year age group in Sweden. Diabetologia 1992, 35, 56–62. [Google Scholar] [CrossRef]
- Ostman, J.; Lönnberg, G.; Arnqvist, H.J.; Blohmé, G.; Bolinder, J.; Ekbom, S.A.; Eriksson, J.W.; Gudbjörnsdottir, S.; Sundkvist, G.; Nyström, L. Gender differences and temporal variation in the incidence of type 1 diabetes: Results of 8012 cases in the nationwide Diabetes Incidence Study in Sweden 1983–2002. J. Intern. Med. 2008, 263, 386–394. [Google Scholar] [CrossRef]
- Fazeli Farsani, S.; Souverein, P.C.; van der Vorst, M.M.; Knibbe, C.A.; Herings, R.M.; de Boer, A.; Mantel-Teeuwisse, A.K. Increasing trends in the incidence and prevalence rates of type 1 diabetes among children and adolescents in the Netherlands. Pediatric Diabetes 2016, 17, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Majeed, N.A.; Shiruhana, S.A.; Maniam, J.; Eigenmann, C.A.; Siyan, A.; Ogle, G.D. Incidence, prevalence and mortality of diabetes in children and adolescents aged under 20 years in the Republic of Maldives. J. Paediatr. Child Health 2020, 56, 746–750. [Google Scholar] [CrossRef] [PubMed]
- International Diabetes Federation. IDF Diabetes Atlas, 9th ed.; International Diabetes Federation: Brussels, Belgium, 2019; Available online: https://www.diabetesatlas.org (accessed on 5 December 2019).
- Divers, J.; Mayer-Davis, E.J.; Lawrence, J.M.; Isom, S.; Dabelea, D.; Dolan, L.; Imperatore, G.; Marcovina, S.; Pettitt, D.J.; Pihoker, C.; et al. Trends in incidence of type 1 and type 2 diabetes among youths—Selected counties and Indian reservations, United States, 2002–2015. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Li, Y.; Fang, J.; Liu, G.; Yin, J.; Li, T.; Yin, Y. Gut Microbiota and Type 1 Diabetes. Int. J. Mol. Sci. 2018, 19, 995–1006. [Google Scholar] [CrossRef]
- Zheng, P.; Li, Z.; Zhou, Z. Gut microbiome in type 1 diabetes: A comprehensive review. Diabetes Metab. Res. Rev. 2018, 34, e3043. [Google Scholar] [CrossRef] [PubMed]
- Sohail, M.U.; Althani, A.; Anwar, H.; Rizzi, R.; Marei, H.E. Role of the Gastrointestinal Tract Microbiome in the Pathophysiology of Diabetes Mellitus. J. Diabetes Res. 2017, 2017, 9631435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Lu, Q. Genetic and epigenetic influences on the loss of tolerance in autoimmunity. Cell. Mol. Immunol. 2018, 15, 575–585. [Google Scholar] [CrossRef]
- Cerna, M. Epigenetic Regulation in Etiology of Type 1 Diabetes Mellitus. Int. J. Mol. Sci. 2019, 21, 36. [Google Scholar] [CrossRef] [PubMed]
- Krischer, J.P.; Liu, X.; Vehik, K.; Akolkar, B.; Hagopian, W.A.; Rewers, M.J.; She, J.X.; Toppari, J.; Ziegler, A.G.; Lernmark, Å. Predicting islet cell autoimmunity and type 1 diabetes: An 8-year TEDDY study progress report. Diabetes Care 2019, 42, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Mullaney, J.A.; Stephens, J.E.; Costello, M.E.; Fong, C.; Geeling, B.E.; Gavin, P.G.; Wright, C.M.; Spector, T.D.; Brown, M.A.; Hamilton-Williams, E.E. Type 1 diabetes susceptibility alleles are associated with distinct alterations in the gut microbiota. Microbiome 2018, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jia, H. Metagenome-wide association studies: Fine-mining the microbiome. Nat. Rev. Microbiol. 2016, 14, 508–522. [Google Scholar] [CrossRef]
- Lee, E.-S.; Song, E.-J.; Nam, Y.-D. Dysbiosis of gut microbiome and its impact on epigenetic regulation. J. Clin. Epigenetics 2017, 3, 1–7. [Google Scholar]
- Bach, J.F.; Chatenoud, L. The hygiene hypothesis: An explanation for the increased frequency of insulin-dependent diabetes. Cold Spring Harb. Perspect. Med. 2012, 2, a007799. [Google Scholar] [CrossRef]
- Sharp, S.A.; Weedon, M.N.; Hagopian, W.A.; Oram, R.A. Clinical and research uses of genetic risk scores in type 1 diabetes. Curr. Opin. Genet. Dev. 2018, 50, 96–102. [Google Scholar] [CrossRef]
- Sharma, A.; Liu, X.; Hadley, D.; Hagopian, W.; Chen, W.M.; Onengut-Gumuscu, S.; Torn, C.; Steck, A.K.; Frohnert, B.I.; Rewers, M.; et al. Identification of non-HLA genes associated with development of islet autoimmunity and type 1 diabetes in the prospective TEDDY cohort. J. Autoimmun. 2018, 89, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Bonifacio, E.; Beyerlein, A.; Hippich, M.; Winkler, C.; Vehik, K.; Weedon, M.N.; Laimighofer, M.; Hattersley, A.T.; Krumsiek, J.; Frohnert, B.I.; et al. Genetic scores to stratify risk of developing multiple islet autoantibodies and type 1 diabetes: A prospective study in children. PLoS Med. 2018, 15, e1002548. [Google Scholar] [CrossRef] [PubMed]
- Krischer, J.P.; Lynch, K.F.; Lernmark, A.; Hagopian, W.A.; Rewers, M.J.; She, J.X.; Toppari, J.; Ziegler, A.G.; Akolkar, B.; Group, T.S. Genetic and environmental interactions modify the risk of diabetes-related autoimmunity by 6 years of age: The TEDDY study. Diabetes Care 2017, 40, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Cousminer, D.L.; Ahlqvist, E.; Mishra, R.; Andersen, M.K.; Chesi, A.; Hawa, M.I.; Davis, A.; Hodge, K.M.; Bradfield, J.P.; Zhou, K.; et al. First genome-wide association study of latent autoimmune diabetes in adults reveals novel insights linking immune and metabolic diabetes. Diabetes Care 2018, 41, 2396–2403. [Google Scholar] [CrossRef] [PubMed]
- Pociot, F. Type 1 diabetes genome-wide association studies: Not to be lost in translation. Clin. Transl. Immunol. 2017, 6, e162. [Google Scholar] [CrossRef] [PubMed]
- Steck, A.K.; Rewers, M.J. Genetics of type 1 diabetes. Clin. Chem. 2011, 57, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Cucca, F.; Lampis, R.; Congia, M.; Angius, E.; Nutland, S.; Bain, S.C.; Barnett, A.H.; Todd, J.A. A correlation between the relative predisposition of MHC class II alleles to type 1 diabetes and the structure of their proteins. Hum. Mol. Genet. 2001, 10, 2025–2037. [Google Scholar] [CrossRef]
- Redondo, M.J.; Steck, A.K.; Pugliese, A. Genetics of type 1 diabetes. Pediatric Diabetes 2018, 19, 346–353. [Google Scholar] [CrossRef]
- Hagopian, W.A.; Erlich, H.; Lernmark, A.; Rewers, M.; Ziegler, A.G.; Simell, O.; Akolkar, B.; Vogt, R., Jr.; Blair, A.; Ilonen, J.; et al. The environmental determinants of diabetes in the young (TEDDY): Genetic criteria and international diabetes risk screening of 421,000 infants. Pediatric Diabetes 2011, 12, 733–743. [Google Scholar] [CrossRef]
- Turtinen, M.; Härkönen, T.; Parkkola, A.; Ilonen, J.; Knip, M. Characteristics of familial type 1 diabetes: Effects of the relationship to the affected family member on phenotype and genotype at diagnosis. Diabetologia 2019, 62, 2025–2039. [Google Scholar] [CrossRef]
- Harjutsalo, V.; Reunanen, A.; Tuomilehto, J. Differential transmission of type 1 diabetes from diabetic fathers and mothers to their offspring. Diabetes 2006, 55, 1517–1524. [Google Scholar] [CrossRef] [PubMed]
- Todd, J.A.; Walker, N.M.; Cooper, J.D.; Smyth, D.J.; Downes, K.; Plagnol, V.; Bailey, R.; Nejentsev, S.; Field, S.F.; Payne, F.; et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 2007, 39, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Bottini, N.; Musumeci, L.; Alonso, A.; Rahmouni, S.; Nika, K.; Rostamkhani, M.; MacMurray, J.; Meloni, G.F.; Lucarelli, P.; Pellecchia, M.; et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat. Genet. 2004, 36, 337–338. [Google Scholar] [CrossRef]
- Bell, G.I.; Horita, S.; Karam, J.H. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes 1984, 33, 176–183. [Google Scholar] [CrossRef]
- Winkler, C.; Lauber, C.; Adler, K.; Grallert, H.; Illig, T.; Ziegler, A.G.; Bonifacio, E. An interferon-induced helicase (IFIH1) gene polymorphism associates with different rates of progression from autoimmunity to type 1 diabetes. Diabetes 2011, 60, 685–690. [Google Scholar] [CrossRef]
- Hyttinen, V.; Kaprio, J.; Kinnunen, L.; Koskenvuo, M.; Tuomilehto, J. Genetic liability of type 1 diabetes and the onset age among 22,650 young Finnish twin pairs: A nationwide follow-up study. Diabetes 2003, 52, 1052–1055. [Google Scholar] [CrossRef]
- Skov, J.; Eriksson, D.; Kuja-Halkola, R.; Höijer, J.; Gudbjörnsdottir, S.; Svensson, A.-M.; E Magnusson, P.K.; Ludvigsson, J.F.; Kämpe, O.; Bensing, S. Co-aggregation and heritability of organ-specific autoimmunity: A population-based twin study. Eur. J. Endocrinol. 2020, 182, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Nistico, L.; Iafusco, D.; Galderisi, A.; Fagnani, C.; Cotichini, R.; Toccaceli, V.; Stazi, M.A. Emerging effects of early environmental factors over genetic background for type 1 diabetes susceptibility: Evidence from a Nationwide Italian Twin Study. J. Clin. Endocrinol. Metab. 2012, 97, E1483–E1491. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, A.; Kuzuya, T. Diabetic twins in Japan. Diabetes Res. Clin. Pract. 1994, 24, S63–S67. [Google Scholar] [CrossRef]
- Kaprio, J.; Tuomilehto, J.; Koskenvuo, M.; Romanov, K.; Reunanen, A.; Eriksson, J.; Stengård, J.; Kesäniemi, Y.A. Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia 1992, 35, 1060–1067. [Google Scholar] [CrossRef]
- Russell, J.T.; Roesch, L.F.W.; Ordberg, M.; Ilonen, J.; Atkinson, M.A.; Schatz, D.A.; Triplett, E.W.; Ludvigsson, J. Genetic risk for autoimmunity is associated with distinct changes in the human gut microbiome. Nat. Commun. 2019, 10, 3621. [Google Scholar] [CrossRef] [PubMed]
- Stiemsma, L.T.; Michels, K.B. The role of the microbiome in the developmental origins of health and disease. Pediatrics 2018, 141, e20172437. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Scepanovic, P.; Hodel, F.; Mondot, S.; Partula, V.; Byrd, A.; Hammer, C.; Alanio, C.; Bergstedt, J.; Patin, E.; Touvier, M.; et al. A comprehensive assessment of demographic, environmental, and host genetic associations with gut microbiome diversity in healthy individuals. Microbiome 2019, 7, 130. [Google Scholar] [CrossRef] [PubMed]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef]
- Kumar, M.; Singh, P.; Murugesan, S.; Vetizou, M.; McCulloch, J.; Badger, J.H.; Trinchieri, G.; Al Khodor, S. Microbiome as an immunological modifier. Methods Mol. Biol. 2020, 2055, 595–638. [Google Scholar]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef]
- Kumar, M.; Mathur, T.; Joshi, V.; Upadhyay, D.J.; Inoue, S.I.; Masuda, N. Effect of DS-2969b, a novel GyrB inhibitor, on rat and monkey intestinal microbiota. Anaerobe 2018, 51, 120–123. [Google Scholar] [CrossRef]
- de la Cuesta-Zuluaga, J.; Mueller, N.T.; Alvarez-Quintero, R.; Velasquez-Mejia, E.P.; Sierra, J.A.; Corrales-Agudelo, V.; Carmona, J.A.; Abad, J.M.; Escobar, J.S. Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients 2018, 11, 51. [Google Scholar] [CrossRef]
- Salguero, M.V.; Al-Obaide, M.A.I.; Singh, R.; Siepmann, T.; Vasylyeva, T.L. Dysbiosis of Gram-negative gut microbiota and the associated serum lipopolysaccharide exacerbates inflammation in type 2 diabetic patients with chronic kidney disease. Exp. Ther. Med. 2019, 18, 3461–3469. [Google Scholar] [CrossRef]
- Maya-Lucas, O.; Murugesan, S.; Nirmalkar, K.; Alcaraz, L.D.; Hoyo-Vadillo, C.; Pizano-Zarate, M.L.; Garcia-Mena, J. The gut microbiome of Mexican children affected by obesity. Anaerobe 2019, 55, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Saffouri, G.B.; Shields-Cutler, R.R.; Chen, J.; Yang, Y.; Lekatz, H.R.; Hale, V.L.; Cho, J.M.; Battaglioli, E.J.; Bhattarai, Y.; Thompson, K.J.; et al. Small intestinal microbial dysbiosis underlies symptoms associated with functional gastrointestinal disorders. Nat. Commun. 2019, 10, 2012. [Google Scholar] [CrossRef] [PubMed]
- Murri, M.; Leiva, I.; Gomez-Zumaquero, J.M.; Tinahones, F.J.; Cardona, F.; Soriguer, F.; Queipo-Ortuno, M.I. Gut microbiota in children with type 1 diabetes differs from that in healthy children: A case-control study. BMC Med. 2013, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Leiva-Gea, I.; Sanchez-Alcoholado, L.; Martin-Tejedor, B.; Castellano-Castillo, D.; Moreno-Indias, I.; Urda-Cardona, A.; Tinahones, F.J.; Fernandez-Garcia, J.C.; Queipo-Ortuno, M.I. Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: A case-control study. Diabetes Care 2018, 41, 2385–2395. [Google Scholar] [CrossRef] [PubMed]
- de Goffau, M.C.; Luopajarvi, K.; Knip, M.; Ilonen, J.; Ruohtula, T.; Harkonen, T.; Orivuori, L.; Hakala, S.; Welling, G.W.; Harmsen, H.J.; et al. Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes 2013, 62, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, B.S.; Rodrigues, N.; Gonzaga, M.I.; Paiolo, J.C.C.; Stefanutto, N.; Omori, W.P.; Pinheiro, D.G.; Brisotti, J.L.; Matheucci, E., Jr.; Mariano, V.S.; et al. Intestinal dysbiosis in autoimmune diabetes is correlated with poor glycemic control and increased Interleukin-6: A pilot study. Front. Immunol. 2018, 9, 1689. [Google Scholar] [CrossRef] [PubMed]
- Vatanen, T.; Franzosa, E.A.; Schwager, R.; Tripathi, S.; Arthur, T.D.; Vehik, K.; Lernmark, Å.; Hagopian, W.A.; Rewers, M.J.; She, J.X.; et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018, 562, 589–594. [Google Scholar] [CrossRef]
- Zhao, G.; Vatanen, T.; Droit, L.; Park, A.; Kostic, A.D.; Poon, T.W.; Vlamakis, H.; Siljander, H.; Härkönen, T.; Hämäläinen, A.M.; et al. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc. Natl. Acad. Sci. USA 2017, 114, e6166–e6175. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Jakobsson, H.E.; Abrahamsson, T.R.; Jenmalm, M.C.; Harris, K.; Quince, C.; Jernberg, C.; Bjorksten, B.; Engstrand, L.; Andersson, A.F. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 2014, 63, 559–566. [Google Scholar] [CrossRef]
- Neu, J.; Rushing, J. Cesarean versus vaginal delivery: Long-term infant outcomes and the hygiene hypothesis. Clin. Perinatol. 2011, 38, 321–331. [Google Scholar] [CrossRef]
- Khashan, A.S.; Kenny, L.C.; Lundholm, C.; Kearney, P.M.; Gong, T.; Almqvist, C. Mode of obstetrical delivery and type 1 diabetes: A sibling design study. Pediatrics 2014, 134, e806–e813. [Google Scholar] [CrossRef] [PubMed]
- Tanoey, J.; Gulati, A.; Patterson, C.; Becher, H. Risk of type 1 diabetes in the offspring born through elective or non-elective caesarean section in comparison to vaginal delivery: A meta-analysis of observational studies. Curr. Diabetes Rep. 2019, 19, 124. [Google Scholar] [CrossRef] [PubMed]
- Begum, M.; Pilkington, R.; Chittleborough, C.; Lynch, J.; Penno, M.; Smithers, L. Caesarean section and risk of type 1 diabetes: Whole-of-population study. Diabet. Med. 2019, 36, 1686–1693. [Google Scholar] [CrossRef] [PubMed]
- Cardwell, C.R.; Stene, L.C.; Joner, G.; Cinek, O.; Svensson, J.; Goldacre, M.J.; Parslow, R.C.; Pozzilli, P.; Brigis, G.; Stoyanov, D.; et al. Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: A meta-analysis of observational studies. Diabetologia 2008, 51, 726–735. [Google Scholar] [CrossRef]
- Samuelsson, U.; Lindell, N.; Bladh, M.; Akesson, K.; Carlsson, A.; Josefsson, A. Caesarean section per se does not increase the risk of offspring developing type 1 diabetes: A Swedish population-based study. Diabetologia 2015, 58, 2517–2524. [Google Scholar] [CrossRef]
- Bayer, A.L.; Fraker, C.A. The folate cycle as a cause of natural killer cell dysfunction and viral etiology in type 1 diabetes. Front. Endocrinol. 2017, 8, 315. [Google Scholar] [CrossRef]
- Leonard, M.M.; Karathia, H.; Pujolassos, M.; Troisi, J.; Valitutti, F.; Subramanian, P.; Camhi, S.; Kenyon, V.; Colucci, A.; Serena, G.; et al. Multi-omics analysis reveals the influence of genetic and environmental risk factors on developing gut microbiota in infants at risk of celiac disease. Microbiome 2020, 8, 130. [Google Scholar] [CrossRef]
- Clausen, T.D.; Bergholt, T.; Bouaziz, O.; Arpi, M.; Eriksson, F.; Rasmussen, S.; Keiding, N.; Lokkegaard, E.C. Broad-spectrum antibiotic treatment and subsequent childhood type 1 diabetes: A nationwide Danish cohort study. PLoS ONE 2016, 11, e0161654. [Google Scholar] [CrossRef]
- Insel, R.A.; Dunne, J.L.; Atkinson, M.A.; Chiang, J.L.; Dabelea, D.; Gottlieb, P.A.; Greenbaum, C.J.; Herold, K.C.; Krischer, J.P.; Lernmark, A.; et al. Staging presymptomatic type 1 diabetes: A scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care 2015, 38, 1964–1974. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Davis-Richardson, A.G.; Triplett, E.W. A model for the role of gut bacteria in the development of autoimmunity for type 1 diabetes. Diabetologia 2015, 58, 1386–1393. [Google Scholar] [CrossRef] [PubMed]
- Ts, D.; Sf, P.; Aa, J.; Md, A.; Dr, M. Lactobacillus and Bifidobacterium Promote Antibacterial and Antiviral Immune Response in Human Macrophages. J. Probiotics Health 2018, 6, 1–7. [Google Scholar] [CrossRef]
- de Goffau, M.C.; Fuentes, S.; van den Bogert, B.; Honkanen, H.; de Vos, W.M.; Welling, G.W.; Hyoty, H.; Harmsen, H.J. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia 2014, 57, 1569–1577. [Google Scholar] [CrossRef] [PubMed]
- van den Bogert, B.; Meijerink, M.; Zoetendal, E.G.; Wells, J.M.; Kleerebezem, M. Immunomodulatory properties of Streptococcus and Veillonella isolates from the human small intestine microbiota. PLoS ONE 2014, 9, e114277. [Google Scholar] [CrossRef] [PubMed]
- Harbison, J.E.; Roth-Schulze, A.J.; Giles, L.C.; Tran, C.D.; Ngui, K.M.; Penno, M.A.; Thomson, R.L.; Wentworth, J.M.; Colman, P.G.; Craig, M.E.; et al. Gut microbiome dysbiosis and increased intestinal permeability in children with islet autoimmunity and type 1 diabetes: A prospective cohort study. Pediatric Diabetes 2019, 20, 574–583. [Google Scholar] [CrossRef]
- Abdellatif, A.M.; Jensen Smith, H.; Harms, R.Z.; Sarvetnick, N.E. Human Islet Response to Selected Type 1 Diabetes-Associated Bacteria: A Transcriptome-Based Study. Front. Immunol. 2019, 10, 2623. [Google Scholar] [CrossRef]
- Tuovinen, E.; Keto, J.; Nikkila, J.; Matto, J.; Lahteenmaki, K. Cytokine response of human mononuclear cells induced by intestinal Clostridium species. Anaerobe 2013, 19, 70–76. [Google Scholar] [CrossRef]
- Gackowska, L.; Michalkiewicz, J.; Krotkiewski, M.; Helmin-Basa, A.; Kubiszewska, I.; Dzierzanowska, D. Combined effect of different lactic acid bacteria strains on the mode of cytokines pattern expression in human peripheral blood mononuclear cells. J. Physiol. Pharmacol. 2006, 57, 13–21. [Google Scholar]
- Haller, D.; Holt, L.; Kim, S.C.; Schwabe, R.F.; Sartor, R.B.; Jobin, C. Transforming growth factor-beta 1 inhibits non-pathogenic Gram negative bacteria-induced NF-kappa B recruitment to the interleukin-6 gene promoter in intestinal epithelial cells through modulation of histone acetylation. J. Biol. Chem. 2003, 278, 23851–23860. [Google Scholar] [CrossRef]
- Altindis, E.; Vomund, A.N.; Chow, I.T.; Damasio, M.; Kwok, W.; Unanue, E.R.; Kahn, C.R. Identification of cross reactive insulin immunogenic epitopes from commensal gut microbes. Diabetes 2018, 67. [Google Scholar] [CrossRef]
- Lakhdari, O.; Tap, J.; Beguet-Crespel, F.; Le Roux, K.; de Wouters, T.; Cultrone, A.; Nepelska, M.; Lefevre, F.; Dore, J.; Blottiere, H.M. Identification of NF-kappaB modulation capabilities within human intestinal commensal bacteria. J. Biomed. Biotechnol. 2011, 2011, 282356. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.; Brown, S.M.; Hao, Y.; Tetz, V. Type 1 diabetes: An association between autoimmunity, the dynamics of gut amyloid-producing E. coli and their phages. Sci. Rep. 2019, 9, 9685. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Hervas, S.; Sanchez-Garcia, V.; Herrero-Cervera, A.; Vinue, A.; Real, J.T.; Ascaso, J.F.; Burks, D.J.; Gonzalez-Navarro, H. Type 1 diabetic mellitus patients with increased atherosclerosis risk display decreased CDKN2A/2B/2BAS gene expression in leukocytes. J. Transl. Med. 2019, 17, 222. [Google Scholar] [CrossRef] [PubMed]
- Cinek, O.; Kramna, L.; Mazankova, K.; Odeh, R.; Alassaf, A.; Ibekwe, M.U.; Ahmadov, G.; Elmahi, B.M.E.; Mekki, H.; Lebl, J.; et al. The bacteriome at the onset of type 1 diabetes: A study from four geographically distant African and Asian countries. Diabetes Res. Clin. Pract. 2018, 144, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.S.; Wang, J.; Yannie, P.J.; Ghosh, S. Intestinal Barrier Dysfunction, LPS Translocation, and Disease Development. J. Endocrinol. Soc. 2020, 4, bvz039. [Google Scholar] [CrossRef]
- Kong, X.N.; Yan, H.X.; Chen, L.; Dong, L.W.; Yang, W.; Liu, Q.; Yu, L.X.; Huang, D.D.; Liu, S.Q.; Liu, H.; et al. LPS-induced down-regulation of signal regulatory protein {alpha} contributes to innate immune activation in macrophages. J. Exp. Med. 2007, 204, 2719–2731. [Google Scholar] [CrossRef]
- Huang, Y.; Li, S.C.; Hu, J.; Ruan, H.B.; Guo, H.M.; Zhang, H.H.; Wang, X.; Pei, Y.F.; Pan, Y.; Fang, C. Gut microbiota profiling in Han Chinese with type 1 diabetes. Diabetes Res. Clin. Pract. 2018, 141, 256–263. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Zhang, M.; Zhou, Q.; Dorfman, R.G.; Huang, X.; Fan, T.; Zhang, H.; Zhang, J.; Yu, C. Butyrate inhibits interleukin-17 and generates Tregs to ameliorate colorectal colitis in rats. BMC Gastroenterol. 2016, 16, 84. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Larasati, R.A.; Harbuwono, D.S.; Rahajeng, E.; Pradipta, S.; Nuraeni, H.S.; Susilowati, A.; Wibowo, H. The role of butyrate on monocyte migration and inflammation response in patient with type 2 diabetes mellitus. Biomedicines 2019, 7, 74. [Google Scholar] [CrossRef] [PubMed]
- Endesfelder, D.; Engel, M.; Davis-Richardson, A.G.; Ardissone, A.N.; Achenbach, P.; Hummel, S.; Winkler, C.; Atkinson, M.; Schatz, D.; Triplett, E.; et al. Towards a functional hypothesis relating anti-islet cell autoimmunity to the dietary impact on microbial communities and butyrate production. Microbiome 2016, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Tai, N.; Wong, F.S.; Wen, L. The role of gut microbiota in the development of type 1, type 2 diabetes mellitus and obesity. Rev. Endocr. Metab. Disord. 2015, 16, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Davis-Richardson, A.G.; Ardissone, A.N.; Dias, R.; Simell, V.; Leonard, M.T.; Kemppainen, K.M.; Drew, J.C.; Schatz, D.; Atkinson, M.A.; Kolaczkowski, B.; et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front. Microbiol. 2014, 5, 678. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A common factor in human diseases. Biomed. Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef]
- Alkanani, A.K.; Hara, N.; Gottlieb, P.A.; Ir, D.; Robertson, C.E.; Wagner, B.D.; Frank, D.N.; Zipris, D. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes 2015, 64, 3510–3520. [Google Scholar] [CrossRef]
- Vatanen, T.; Kostic, A.D.; d’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Arthur, T.D.; Hamalainen, A.M.; et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 2016, 165, 842–853. [Google Scholar] [CrossRef]
- Berezow, A.B.; Ernst, R.K.; Coats, S.R.; Braham, P.H.; Karimi-Naser, L.M.; Darveau, R.P. The structurally similar, penta-acylated lipopolysaccharides of Porphyromonas gingivalis and Bacteroides elicit strikingly different innate immune responses. Microb. Pathog. 2009, 47, 68–77. [Google Scholar] [CrossRef] [PubMed]
- d’Hennezel, E.; Abubucker, S.; Murphy, L.O.; Cullen, T.W. Total Lipopolysaccharide from the Human Gut Microbiome Silences Toll-Like Receptor Signaling. Msystems 2017, 2. [Google Scholar] [CrossRef]
- Coats, S.R.; Pham, T.T.; Bainbridge, B.W.; Reife, R.A.; Darveau, R.P. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J. Immunol. 2005, 175, 4490–4498. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, W.D.; Wang, Y.D. The relationship between gut microbiota and inflammatory diseases: The role of macrophages. Front. Microbiol. 2020, 11, 1065. [Google Scholar] [CrossRef] [PubMed]
- Parisi, L.; Gini, E.; Baci, D.; Tremolati, M.; Fanuli, M.; Bassani, B.; Farronato, G.; Bruno, A.; Mortara, L. Macrophage polarization in chronic inflammatory diseases: Killers or builders? J. Immunol. Res. 2018, 2018, 8917804. [Google Scholar] [CrossRef] [PubMed]
- Arif, S.; Tree, T.I.; Astill, T.P.; Tremble, J.M.; Bishop, A.J.; Dayan, C.M.; Roep, B.O.; Peakman, M. Autoreactive T cell responses show proinflammatory polarization in diabetes but a regulatory phenotype in health. J. Clin. Investig. 2004, 113, 451–463. [Google Scholar] [CrossRef]
- Toshchakov, V.; Jones, B.W.; Perera, P.Y.; Thomas, K.; Cody, M.J.; Zhang, S.; Williams, B.R.; Major, J.; Hamilton, T.A.; Fenton, M.J.; et al. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat. Immunol. 2002, 3, 392–398. [Google Scholar] [CrossRef]
- Eun, S.Y.; Seo, J.; Park, S.W.; Lee, J.H.; Chang, K.C.; Kim, H.J. LPS potentiates nucleotide-induced inflammatory gene expression in macrophages via the upregulation of P2Y2 receptor. Int. Immunopharmacol. 2014, 18, 270–276. [Google Scholar] [CrossRef]
- Marro, B.S.; Legrain, S.; Ware, B.C.; Oldstone, M.B. Macrophage IFN-I signaling promotes autoreactive T cell infiltration into islets in type 1 diabetes model. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Huang, Q.; Chow, I.T.; Brady, C.; Raisingani, A.; Li, D.; Ostrov, D.A.; Atkinson, M.A.; Kwok, W.W.; Kahn, C.R.; Altindis, E. Parabacteroides distasonis insulin B:9-23 epitope mimic stimulates insulin specific T-cells and enhances Type 1 Diabetes in NOD mice. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wang, Y.; Sosinowski, T.; Novikov, A.; Crawford, F.; White, J.; Jin, N.; Liu, Z.; Zou, J.; Neau, D.; Davidson, H.W.; et al. How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes. Sci. Immunol. 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, M.; Beilke, J.N.; Jasinski, J.M.; Kobayashi, M.; Miao, D.; Li, M.; Coulombe, M.G.; Liu, E.; Elliott, J.F.; Gill, R.G.; et al. Priming and effector dependence on insulin B:9-23 peptide in NOD islet autoimmunity. J. Clin. Investig. 2007, 117, 1835–1843. [Google Scholar] [CrossRef] [PubMed]
- Gavin, P.G.; Mullaney, J.A.; Loo, D.; Cao, K.L.; Gottlieb, P.A.; Hill, M.M.; Zipris, D.; Hamilton-Williams, E.E. Intestinal Metaproteomics Reveals Host-Microbiota Interactions in Subjects at Risk for Type 1 Diabetes. Diabetes Care 2018, 41, 2178–2186. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, X.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. The role of MUC2 mucin in intestinal homeostasis and the impact of dietary components on MUC2 expression. Int. J. Biol. Macromol. 2020, 164, 884–891. [Google Scholar] [CrossRef]
- Kim, Y.S.; Ho, S.B. Intestinal goblet cells and mucins in health and disease: Recent insights and progress. Curr. Gastroenterol. Rep. 2010, 12, 319–330. [Google Scholar] [CrossRef]
- Shao, L.; Allez, M.; Park, M.S.; Mayer, L. Immunomodulatory roles of the carcinoembryonic antigen family of glycoproteins. Ann. N.Y. Acad. Sci. 2006, 1072, 194–209. [Google Scholar] [CrossRef]
- Honkanen, H.; Oikarinen, S.; Nurminen, N.; Laitinen, O.H.; Huhtala, H.; Lehtonen, J.; Ruokoranta, T.; Hankaniemi, M.M.; Lecouturier, V.; Almond, J.W.; et al. Detection of enteroviruses in stools precedes islet autoimmunity by several months: Possible evidence for slowly operating mechanisms in virus-induced autoimmunity. Diabetologia 2017, 60, 424–431. [Google Scholar] [CrossRef]
- Sioofy-Khojine, A.B.; Lehtonen, J.; Nurminen, N.; Laitinen, O.H.; Oikarinen, S.; Huhtala, H.; Pakkanen, O.; Ruokoranta, T.; Hankaniemi, M.M.; Toppari, J.; et al. Coxsackievirus B1 infections are associated with the initiation of insulin-driven autoimmunity that progresses to type 1 diabetes. Diabetologia 2018, 61, 1193–1202. [Google Scholar] [CrossRef]
- Kim, K.W.; Horton, J.L.; Pang, C.N.I.; Jain, K.; Leung, P.; Isaacs, S.R.; Bull, R.A.; Luciani, F.; Wilkins, M.R.; Catteau, J.; et al. Higher abundance of enterovirus A species in the gut of children with islet autoimmunity. Sci. Rep. 2019, 9, 1749. [Google Scholar] [CrossRef]
- Lin, H.C.; Wang, C.H.; Tsai, F.J.; Hwang, K.P.; Chen, W.; Lin, C.C.; Li, T.C. Enterovirus infection is associated with an increased risk of childhood type 1 diabetes in Taiwan: A nationwide population-based cohort study. Diabetologia 2015, 58, 79–86. [Google Scholar] [CrossRef]
- Yeung, W.C.; Al-Shabeeb, A.; Pang, C.N.; Wilkins, M.R.; Catteau, J.; Howard, N.J.; Rawlinson, W.D.; Craig, M.E. Children with islet autoimmunity and enterovirus infection demonstrate a distinct cytokine profile. Diabetes 2012, 61, 1500–1508. [Google Scholar] [CrossRef]
- Oikarinen, M.; Tauriainen, S.; Oikarinen, S.; Honkanen, T.; Collin, P.; Rantala, I.; Maki, M.; Kaukinen, K.; Hyoty, H. Type 1 diabetes is associated with enterovirus infection in gut mucosa. Diabetes 2012, 61, 687–691. [Google Scholar] [CrossRef] [PubMed]
- Stene, L.C.; Oikarinen, S.; Hyoty, H.; Barriga, K.J.; Norris, J.M.; Klingensmith, G.; Hutton, J.C.; Erlich, H.A.; Eisenbarth, G.S.; Rewers, M. Enterovirus infection and progression from islet autoimmunity to type 1 diabetes: The Diabetes and Autoimmunity Study in the Young (DAISY). Diabetes 2010, 59, 3174–3180. [Google Scholar] [CrossRef]
- Jaidane, H.; Sauter, P.; Sane, F.; Goffard, A.; Gharbi, J.; Hober, D. Enteroviruses and type 1 diabetes: Towards a better understanding of the relationship. Rev. Med. Virol. 2010, 20, 265–280. [Google Scholar] [CrossRef]
- Cinek, O.; Kramna, L.; Lin, J.; Oikarinen, S.; Kolarova, K.; Ilonen, J.; Simell, O.; Veijola, R.; Autio, R.; Hyoty, H. Imbalance of bacteriome profiles within the Finnish Diabetes Prediction and Prevention study: Parallel use of 16S profiling and virome sequencing in stool samples from children with islet autoimmunity and matched controls. Pediatric Diabetes 2017, 18, 588–598. [Google Scholar] [CrossRef]
- Santiago-Rodriguez, T.M.; Hollister, E.B. Human virome and disease: High-throughput sequencing for virus discovery, identification of phage-bacteria dysbiosis and development of therapeutic approaches with emphasis on the human gut. Viruses 2019, 11, 656. [Google Scholar] [CrossRef]
- Neil, J.A.; Cadwell, K. The intestinal virome and immunity. J. Immunol. 2018, 201, 1615–1624. [Google Scholar] [CrossRef]
- Lietzen, N.; Hirvonen, K.; Honkimaa, A.; Buchacher, T.; Laiho, J.E.; Oikarinen, S.; Mazur, M.A.; Flodstrom-Tullberg, M.; Dufour, E.; Sioofy-Khojine, A.B.; et al. Coxsackievirus B persistence modifies the proteome and the secretome of pancreatic ductal cells. iScience 2019, 19, 340–357. [Google Scholar] [CrossRef]
- Nekoua, M.P.; Bertin, A.; Sane, F.; Alidjinou, E.K.; Lobert, D.; Trauet, J.; Hober, C.; Engelmann, I.; Moutairou, K.; Yessoufou, A.; et al. Pancreatic beta cells persistently infected with coxsackievirus B4 are targets of NK cell-mediated cytolytic activity. Cell. Mol. Life Sci. 2020, 77, 179–194. [Google Scholar] [CrossRef]
- Ylipaasto, P.; Kutlu, B.; Rasilainen, S.; Rasschaert, J.; Salmela, K.; Teerijoki, H.; Korsgren, O.; Lahesmaa, R.; Hovi, T.; Eizirik, D.L.; et al. Global profiling of coxsackievirus- and cytokine-induced gene expression in human pancreatic islets. Diabetologia 2005, 48, 1510–1522. [Google Scholar] [CrossRef] [PubMed]
- Vehik, K.; Lynch, K.F.; Wong, M.C.; Tian, X.; Ross, M.C.; Gibbs, R.A.; Ajami, N.J.; Petrosino, J.F.; Rewers, M.; Toppari, J.; et al. Prospective virome analyses in young children at increased genetic risk for type 1 diabetes. Nat. Med. 2019, 25, 1865–1872. [Google Scholar] [CrossRef] [PubMed]
- Lietzen, N.; An, L.T.T.; Jaakkola, M.K.; Kallionpaa, H.; Oikarinen, S.; Mykkanen, J.; Knip, M.; Veijola, R.; Ilonen, J.; Toppari, J.; et al. Enterovirus-associated changes in blood transcriptomic profiles of children with genetic susceptibility to type 1 diabetes. Diabetologia 2018, 61, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Lien, E.; Zipris, D. The role of Toll-like receptor pathways in the mechanism of type 1 diabetes. Curr. Mol. Med. 2009, 9, 52–68. [Google Scholar] [CrossRef]
- Harkonen, T.; Lankinen, H.; Davydova, B.; Hovi, T.; Roivainen, M. Enterovirus infection can induce immune responses that cross-react with beta-cell autoantigen tyrosine phosphatase IA-2/IAR. J. Med. Virol. 2002, 66, 340–350. [Google Scholar] [CrossRef]
- Honeyman, M.C.; Stone, N.L.; Falk, B.A.; Nepom, G.; Harrison, L.C. Evidence for molecular mimicry between human T cell epitopes in rotavirus and pancreatic islet autoantigens. J. Immunol. 2010, 184, 2204–2210. [Google Scholar] [CrossRef]
- Huang, Q.; Kahn, C.R.; Altindis, E. Viral Hormones: Expanding Dimensions in Endocrinology. Endocrinology 2019, 160, 2165–2179. [Google Scholar] [CrossRef]
- Altindis, E.; Cai, W.; Sakaguchi, M.; Zhang, F.; GuoXiao, W.; Liu, F.; De Meyts, P.; Gelfanov, V.; Pan, H.; DiMarchi, R.; et al. Viral insulin-like peptides activate human insulin and IGF-1 receptor signaling: A paradigm shift for host-microbe interactions. Proc. Natl. Acad. Sci. USA 2018, 115, 2461–2466. [Google Scholar] [CrossRef]
- Perrett, K.P.; Jachno, K.; Nolan, T.M.; Harrison, L.C. Association of rotavirus vaccination with the incidence of type 1 diabetes in children. JAMA Pediatrics 2019, 173, 280–282. [Google Scholar] [CrossRef]
- Rogers, M.A.M.; Basu, T.; Kim, C. Lower Incidence Rate of Type 1 Diabetes after Receipt of the Rotavirus Vaccine in the United States, 2001–2017. Sci. Rep. 2019, 9, 7727. [Google Scholar] [CrossRef]
- Wook Kim, K.; Allen, D.W.; Briese, T.; Couper, J.J.; Barry, S.C.; Colman, P.G.; Cotterill, A.M.; Davis, E.A.; Giles, L.C.; Harrison, L.C.; et al. Distinct gut virome profile of pregnant women with type 1 diabetes in the ENDIA study. Open Forum Infect. Dis. 2019, 6, ofz025. [Google Scholar] [CrossRef] [PubMed]
- Viskari, H.; Knip, M.; Tauriainen, S.; Huhtala, H.; Veijola, R.; Ilonen, J.; Simell, O.; Surcel, H.M.; Hyoty, H. Maternal enterovirus infection as a risk factor for type 1 diabetes in the exposed offspring. Diabetes Care 2012, 35, 1328–1332. [Google Scholar] [CrossRef] [PubMed]
- Lind, A.; Lynch, K.F.; Lundgren, M.; Lernmark, A.; Almgren, P.; Ramelius, A.; Puustinen, L.; Hyoty, H.; Lundstig, A. First trimester enterovirus IgM and beta cell autoantibodies in mothers to children affected by type 1 diabetes autoimmunity before 7 years of age. J. Reprod. Immunol. 2018, 127, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Elfving, M.; Svensson, J.; Oikarinen, S.; Jonsson, B.; Olofsson, P.; Sundkvist, G.; Lindberg, B.; Lernmark, A.; Hyoty, H.; Ivarsson, S.A. Maternal enterovirus infection during pregnancy as a risk factor in offspring diagnosed with type 1 diabetes between 15 and 30 years of age. Exp. Diabetes Res. 2008, 2008, 271958. [Google Scholar] [CrossRef] [PubMed]
- Kowalewska, B.; Zorena, K.; Szmigiero-Kawko, M.; Waz, P.; Mysliwiec, M. Higher diversity in fungal species discriminates children with type 1 diabetes mellitus from healthy control. Patient Prefer. Adherence 2016, 10, 591–599. [Google Scholar] [PubMed]
- Gosiewski, T.; Salamon, D.; Szopa, M.; Sroka, A.; Malecki, M.T.; Bulanda, M. Quantitative evaluation of fungi of the genus Candida in the feces of adult patients with type 1 and 2 diabetes—A pilot study. Gut Pathog. 2014, 6, 43. [Google Scholar] [CrossRef]
- Martin, F.-P.J.; Sprenger, N.; Montoliu, I.; Rezzi, S.; Kochhar, S.; Nicholson, J.K. Dietary modulation of gut functional ecology studied by fecal metabonomics. J. Proteome Res. 2010, 9, 5284–5295. [Google Scholar] [CrossRef]
- Martin, F.P.; Sprenger, N.; Montoliu, I.; Rezzi, S.; Kochhar, S.; Nicholson, J.K. Metabolome progression during early gut microbial colonization of gnotobiotic mice. Sci. Rep. 2015, 5, 11589. [Google Scholar]
- Boerner, B.P.; Sarvetnick, N.E. Type 1 diabetes: Role of intestinal microbiome in humans and mice. Ann. N.Y. Acad. Sci. 2011, 1243, 103–118. [Google Scholar] [CrossRef]
- Alam, C.; Bittoun, E.; Bhagwat, D.; Valkonen, S.; Saari, A.; Jaakkola, U.; Eerola, E.; Huovinen, P.; Hanninen, A. Effects of a germ-free environment on gut immune regulation and diabetes progression in non-obese diabetic (NOD) mice. Diabetologia 2011, 54, 1398–1406. [Google Scholar] [CrossRef]
- Hu, Y.; Peng, J.; Li, F.; Wong, F.S.; Wen, L. Evaluation of different mucosal microbiota leads to gut microbiota-based prediction of type 1 diabetes in NOD mice. Sci. Rep. 2018, 8, 15451. [Google Scholar] [CrossRef] [PubMed]
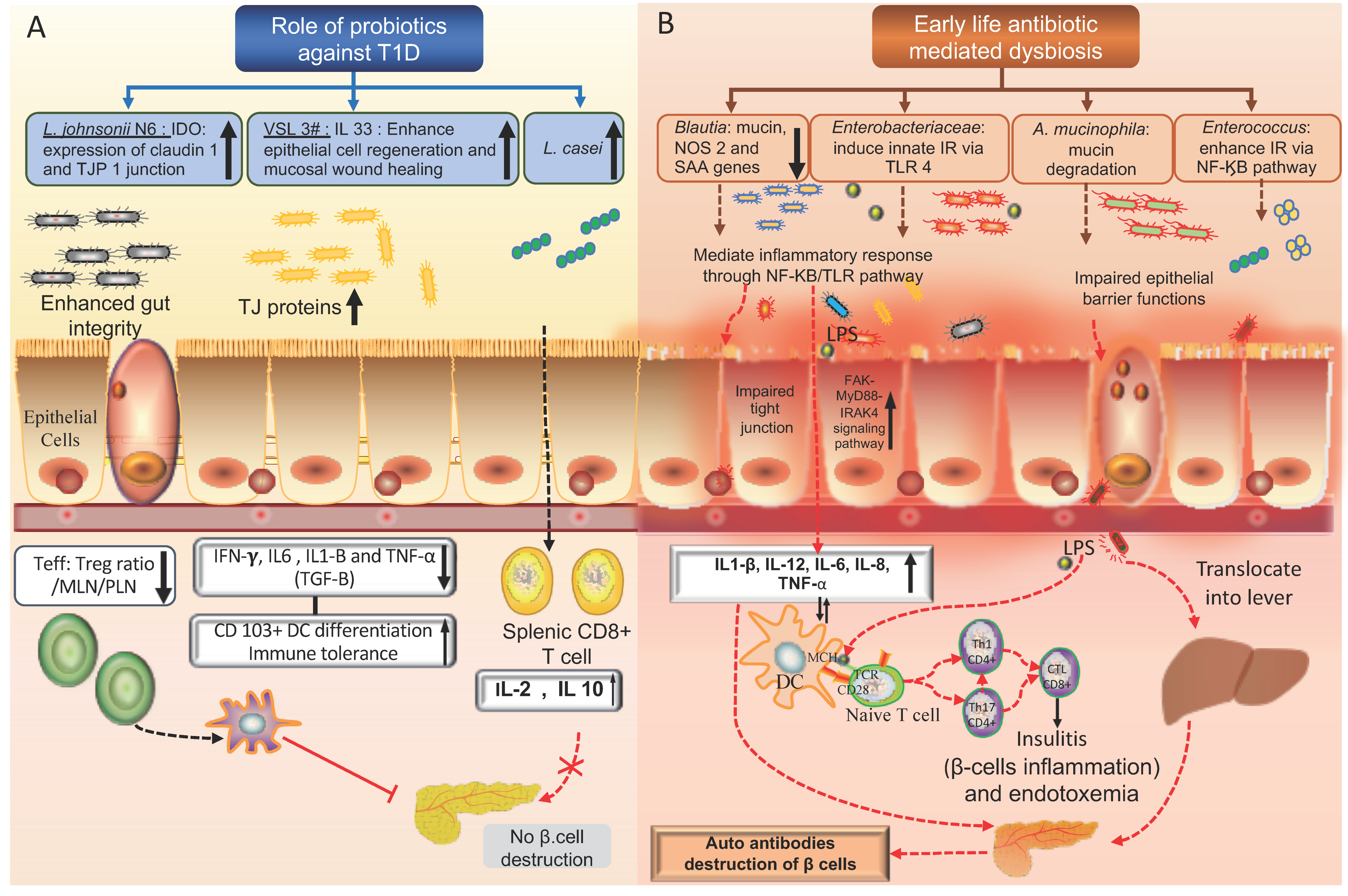
- Brown, K.; Godovannyi, A.; Ma, C.; Zhang, Y.; Ahmadi-Vand, Z.; Dai, C.; Gorzelak, M.A.; Chan, Y.; Chan, J.M.; Lochner, A.; et al. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J. 2016, 10, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Li, Y.; Wang, J.; Li, P.; Duan, Y.; Dai, H.; An, Y.; Cheng, L.; Wang, T.; Wang, C.; et al. Investigation of gut microbiome changes in type 1 diabetic mellitus rats based on high-throughput sequencing. Biomed. Pharmacother. 2020, 124, 109873. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.R.; Francozo, M.C.; de Oliveira, G.G.; Ignacio, A.; Castoldi, A.; Zamboni, D.S.; Ramos, S.G.; Camara, N.O.; de Zoete, M.R.; Palm, N.W.; et al. Gut microbiota translocation to the pancreatic lymph nodes triggers NOD2 activation and contributes to T1D onset. J. Exp. Med. 2016, 213, 1223–1239. [Google Scholar] [CrossRef]
- Livanos, A.E.; Greiner, T.U.; Vangay, P.; Pathmasiri, W.; Stewart, D.; McRitchie, S.; Li, H.; Chung, J.; Sohn, J.; Kim, S.; et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat. Microbiol. 2016, 1, 16140. [Google Scholar] [CrossRef]
- Hansen, C.H.; Krych, L.; Nielsen, D.S.; Vogensen, F.K.; Hansen, L.H.; Sorensen, S.J.; Buschard, K.; Hansen, A.K. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 2012, 55, 2285–2294. [Google Scholar] [CrossRef]
- Dolpady, J.; Sorini, C.; Di Pietro, C.; Cosorich, I.; Ferrarese, R.; Saita, D.; Clementi, M.; Canducci, F.; Falcone, M. Oral Probiotic VSL#3 Prevents Autoimmune Diabetes by Modulating Microbiota and Promoting Indoleamine 2,3-Dioxygenase-Enriched Tolerogenic Intestinal Environment. J. Diabetes Res. 2016, 2016, 7569431. [Google Scholar]
- Kataoka, S.; Satoh, J.; Fujiya, H.; Toyota, T.; Suzuki, R.; Itoh, K.; Kumagai, K. Immunologic aspects of the nonobese diabetic (NOD) mouse. Abnormalities of cellular immunity. Diabetes 1983, 32, 247–253. [Google Scholar] [CrossRef]
- Chen, D.; Thayer, T.C.; Wen, L.; Wong, F.S. Mouse Models of Autoimmune Diabetes: The Nonobese Diabetic (NOD) Mouse. Methods Mol. Biol. 2020, 2128, 87–92. [Google Scholar]
- Burrows, M.P.; Volchkov, P.; Kobayashi, K.S.; Chervonsky, A.V. Microbiota regulates type 1 diabetes through Toll-like receptors. Proc. Natl. Acad. Sci. USA 2015, 112, 9973–9977. [Google Scholar] [CrossRef]
- Prasad, R.; Duan, Y.; Floyd, J.L.; Grant, M.B. 48-OR: Gut dysbiosis promotes diabetic retinopathy (DR) through TLR-2 activation by peptidoglycan (PGN) in angiotensin converting enzyme 2 (ACE2) deficient type 1 diabetic (T1D) mice. Diabetes 2019, 68. [Google Scholar] [CrossRef]
- Sorini, C.; Cosorich, I.; Lo Conte, M.; De Giorgi, L.; Facciotti, F.; Luciano, R.; Rocchi, M.; Ferrarese, R.; Sanvito, F.; Canducci, F.; et al. Loss of gut barrier integrity triggers activation of islet-reactive T cells and autoimmune diabetes. Proc. Natl. Acad. Sci. USA 2019, 116, 15140–15149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.S.; Li, J.; Krautkramer, K.A.; Badri, M.; Battaglia, T.; Borbet, T.C.; Koh, H.; Ng, S.; Sibley, R.A.; Li, Y.; et al. Antibiotic-induced acceleration of type 1 diabetes alters maturation of innate intestinal immunity. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Kriegel, M.A.; Sefik, E.; Hill, J.A.; Wu, H.J.; Benoist, C.; Mathis, D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. USA 2011, 108, 11548–11553. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Monin, L.; Castillo, P.; Elsegeiny, W.; Horne, W.; Eddens, T.; Vikram, A.; Good, M.; Schoenborn, A.A.; Bibby, K.; et al. Intestinal Interleukin-17 Receptor Signaling Mediates Reciprocal Control of the Gut Microbiota and Autoimmune Inflammation. Immunity 2016, 44, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Tato, C.M.; Joyce-Shaikh, B.; Gulen, M.F.; Cayatte, C.; Chen, Y.; Blumenschein, W.M.; Judo, M.; Ayanoglu, G.; McClanahan, T.K.; et al. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity 2015, 43, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Nighot, M.; Al-Sadi, R.; Alhmoud, T.; Nighot, P.; Ma, T.Y. Lipopolysaccharide regulation of intestinal tight junction permeability is mediated by TLR4 signal transduction pathway activation of FAK and MyD88. J. Immunol. 2015, 195, 4999–5010. [Google Scholar] [CrossRef]
- Androulidaki, A.; Wachsmuth, L.; Polykratis, A.; Pasparakis, M. Differential role of MyD88 and TRIF signaling in myeloid cells in the pathogenesis of autoimmune diabetes. PLoS ONE 2018, 13, e0194048. [Google Scholar] [CrossRef]
- Blandino, G.; Inturri, R.; Lazzara, F.; Di Rosa, M.; Malaguarnera, L. Impact of gut microbiota on diabetes mellitus. Diabetes Metab. 2016, 42, 303–315. [Google Scholar] [CrossRef]
- Li, B.; Selmi, C.; Tang, R.; Gershwin, M.E.; Ma, X. The microbiome and autoimmunity: A paradigm from the gut-liver axis. Cell. Mol. Immunol. 2018, 15, 595–609. [Google Scholar] [CrossRef]
- Wen, L.; Ley, R.E.; Volchkov, P.Y.; Stranges, P.B.; Avanesyan, L.; Stonebraker, A.C.; Hu, C.; Wong, F.S.; Szot, G.L.; Bluestone, J.A.; et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 2008, 455, 1109–1113. [Google Scholar] [CrossRef]
- Mariño, E.; Richards, J.L.; McLeod, K.H.; Stanley, D.; Yap, Y.A.; Knight, J.; McKenzie, C.; Kranich, J.; Oliveira, A.C.; Rossello, F.J.; et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol. 2017, 18, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhou, H.Y.; Shen, H.H.; Lufumpa, E.; Li, X.M.; Guo, B.; Li, B.Z. Microbe-metabolite-host axis, two-way action in the pathogenesis and treatment of human autoimmunity. Autoimmun. Rev. 2019, 18, 455–475. [Google Scholar] [CrossRef] [PubMed]
- Kasubuchi, M.; Hasegawa, S.; Hiramatsu, T.; Ichimura, A.; Kimura, I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 2015, 7, 2839–2849. [Google Scholar] [CrossRef] [PubMed]
- Aljutaily, T.; Consuegra-Fernandez, M.; Aranda, F.; Lozano, F.; Huarte, E. Gut microbiota metabolites for sweetening type I diabetes. Cell. Mol. Immunol. 2018, 15, 92–95. [Google Scholar] [CrossRef]
- Al Nabhani, Z.; Dulauroy, S.; Marques, R.; Cousu, C.; Al Bounny, S.; Dejardin, F.; Sparwasser, T.; Berard, M.; Cerf-Bensussan, N.; Eberl, G. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity 2019, 50, 1276–1288. [Google Scholar] [CrossRef]
- Kim, C.H. Microbiota or short-chain fatty acids: Which regulates diabetes? Cell. Mol. Immunol. 2018, 15, 88–91. [Google Scholar] [CrossRef]
- Wen, L.; Wong, F.S. Dietary short-chain fatty acids protect against type 1 diabetes. Nat. Immunol. 2017, 18, 484–486. [Google Scholar] [CrossRef]
- Luu, M.; Visekruna, A. Short-chain fatty acids: Bacterial messengers modulating the immunometabolism of T cells. Eur. J. Immunol. 2019, 49, 842–848. [Google Scholar] [CrossRef]
- Sorini, C.; Cosorich, I.; Falcone, M. New therapeutic perspectives in Type 1 Diabetes: Dietary interventions prevent beta cell-autoimmunity by modifying the gut metabolic environment. Cell. Mol. Immunol. 2017, 14, 951–953. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Simon, M.C.; Reinbeck, A.L.; Wessel, C.; Heindirk, J.; Jelenik, T.; Kaul, K.; Arreguin-Cano, J.; Strom, A.; Blaut, M.; Backhed, F.; et al. Distinct alterations of gut morphology and microbiota characterize accelerated diabetes onset in nonobese diabetic mice. J. Biol. Chem. 2020, 295, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.P.; Wang, S.; Nagpal, R.; Miller, B.; Singh, R.; Taraphder, S.; Yadav, H. Probiotics and prebiotics for the amelioration of type 1 diabetes: Present and future perspectives. Microorganisms 2019, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Valladares, R.; Sankar, D.; Li, N.; Williams, E.; Lai, K.K.; Abdelgeliel, A.S.; Gonzalez, C.F.; Wasserfall, C.H.; Larkin, J.; Schatz, D.; et al. Lactobacillus johnsonii N6 2 mitigates the development of type 1 diabetes in BB-DP rats. PLoS ONE 2010, 5, e10507. [Google Scholar] [CrossRef]
- Jia, L.; Cao, M.; Chen, H.; Zhang, M.; Dong, X.; Ren, Z.; Sun, J.; Pan, L.L. Butyrate ameliorates antibiotic-driven type 1 diabetes in the female offspring of nonobese diabetic mice. J. Agric. Food Chem. 2020, 68, 3112–3120. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, L.; Hong, G.; Huang, C.; Qian, W.; Bai, T.; Song, J.; Song, Y.; Hou, X. Probiotic mixtures with aerobic constituent promoted the recovery of multi-barriers in DSS-induced chronic colitis. Life Sci. 2020, 240, 117089. [Google Scholar] [CrossRef]
- Uusitalo, U.; Liu, X.; Yang, J.; Aronsson, C.A.; Hummel, S.; Butterworth, M.; Lernmark, A.; Rewers, M.; Hagopian, W.; She, J.X.; et al. Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatrics 2016, 170, 20–28. [Google Scholar] [CrossRef]
- Mondanelli, G.; Orecchini, E.; Volpi, C.; Panfili, E.; Belladonna, M.L.; Pallotta, M.T.; Moretti, S.; Galarini, R.; Esposito, S.; Orabona, C. Effect of probiotic administration on serum tryptophan metabolites in pediatric type 1 diabetes patients. Int. J. Tryptophan Res. 2020, 13, 1178646920956646. [Google Scholar] [CrossRef]
- Torn, C.; Hadley, D.; Lee, H.S.; Hagopian, W.; Lernmark, A.; Simell, O.; Rewers, M.; Ziegler, A.; Schatz, D.; Akolkar, B.; et al. Role of type 1 diabetes-associated snps on risk of autoantibody positivity in the TEDDY study. Diabetes 2015, 64, 1818–1829. [Google Scholar] [CrossRef]
- Blanter, M.; Sork, H.; Tuomela, S.; Flodstrom-Tullberg, M. Genetic and environmental interaction in type 1 diabetes: A relationship between genetic risk alleles and molecular traits of enterovirus infection? Curr. Diabetes Rep. 2019, 19, 82. [Google Scholar] [CrossRef]
- Sharp, R.C.; Abdulrahim, M.; Naser, E.S.; Naser, S.A. Genetic variations of PTPN2 and PTPN22: Role in the pathogenesis of type 1 diabetes and Crohn’s disease. Front. Cell. Infect. Microbiol. 2015, 5, 95. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, B.; Spalinger, M.R.; Biedermann, L.; Franc, Y.; Fournier, N.; Rossel, J.B.; Juillerat, P.; Rogler, G.; Macpherson, A.J.; Scharl, M. The presence of genetic risk variants within PTPN2 and PTPN22 is associated with intestinal microbiota alterations in Swiss IBD cohort patients. PLoS ONE 2018, 13, e0199664. [Google Scholar] [CrossRef] [PubMed]
- Jerram, S.T.; Leslie, R.D. The genetic architecture of Type 1 diabetes. Genes 2017, 8, 209. [Google Scholar] [CrossRef] [PubMed]
- Cinek, O.; Tapia, G.; Witso, E.; Kramna, L.; Holkova, K.; Rasmussen, T.; Stene, L.C.; Ronningen, K.S. Enterovirus RNA in peripheral blood may be associated with the variants of rs1990760, a common type 1 diabetes associated polymorphism in IFIH1. PLoS ONE 2012, 7, e48409. [Google Scholar] [CrossRef]
- Jermendy, A.; Szatmari, I.; Korner, A.; Szabo, A.J.; Toth-Heyn, P.; Hermann, R. Association between interferon-induced helicase (IFIH1) rs1990760 polymorphism and seasonal variation in the onset of type 1 diabetes mellitus. Pediatric Diabetes 2018, 19, 300–304. [Google Scholar] [CrossRef]
- Domsgen, E.; Lind, K.; Kong, L.; Huhn, M.H.; Rasool, O.; van Kuppeveld, F.; Korsgren, O.; Lahesmaa, R.; Flodstrom-Tullberg, M. An IFIH1 gene polymorphism associated with risk for autoimmunity regulates canonical antiviral defence pathways in Coxsackievirus infected human pancreatic islets. Sci. Rep. 2016, 6, 39378. [Google Scholar] [CrossRef]
- Redondo, M.J.; Geyer, S.; Steck, A.K.; Sharp, S.; Wentworth, J.M.; Weedon, M.N.; Antinozzi, P.; Sosenko, J.; Atkinson, M.; Pugliese, A.; et al. A type 1 diabetes genetic risk score predicts progression of islet autoimmunity and development of type 1 diabetes in individuals at risk. Diabetes Care 2018, 41, 1887–1894. [Google Scholar] [CrossRef]
- Chen, B.; Sun, L.; Zhang, X. Integration of microbiome and epigenome to decipher the pathogenesis of autoimmune diseases. J. Autoimmun. 2017, 83, 31–42. [Google Scholar] [CrossRef]
- Miro-Blanch, J.; Yanes, O. Epigenetic regulation at the interplay between gut microbiota and host metabolism. Front. Genet. 2019, 10, 638. [Google Scholar] [CrossRef]
- Kala, R.; Peek, G.W.; Hardy, T.M.; Tollefsbol, T.O. MicroRNAs: An emerging science in cancer epigenetics. J. Clin. Bioinform. 2013, 3, 6. [Google Scholar] [CrossRef]
- Santin, I.; Eizirik, D.L. Candidate genes for type 1 diabetes modulate pancreatic islet inflammation and beta-cell apoptosis. Diabetes Obes. Metab. 2013, 15, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liu, Q.; Zhou, Z.; Ikeda, Y. PDX1, Neurogenin-3, and MAFA: Critical transcription regulators for beta cell development and regeneration. Stem Cell Res. Ther. 2017, 8, 240. [Google Scholar] [CrossRef] [PubMed]
- Salinno, C.; Cota, P.; Bastidas-Ponce, A.; Tarquis-Medina, M.; Lickert, H.; Bakhti, M. Beta-Cell Maturation and Identity in Health and Disease. Int. J. Mol. Sci. 2019, 20, 5417. [Google Scholar] [CrossRef]
- Alidjinou, E.K.; Engelmann, I.; Bossu, J.; Villenet, C.; Figeac, M.; Romond, M.B.; Sané, F.; Hober, D. Persistence of Coxsackievirus B4 in pancreatic ductal-like cells results in cellular and viral changes. Virulence 2017, 8, 1229–1244. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.; Barnes, S.; Demark-Wahnefried, W.; Morrow, C.; Salvador, C.; Skibola, C.; Tollefsbol, T.O. Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin. Epigenetics 2015, 7, 112. [Google Scholar] [CrossRef] [PubMed]
- Heintz-Buschart, A.; May, P.; Laczny, C.C.; Lebrun, L.A.; Bellora, C.; Krishna, A.; Wampach, L.; Schneider, J.G.; Hogan, A.; de Beaufort, C.; et al. Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes. Nat. Microbiol. 2016, 2, 16180. [Google Scholar] [CrossRef] [PubMed]
- Jialal, I.; Huet, B.A.; Kaur, H.; Chien, A.; Devaraj, S. Increased toll-like receptor activity in patients with metabolic syndrome. Diabetes Care 2012, 35, 900–904. [Google Scholar] [CrossRef]
- Jin, C.; Henao-Mejia, J.; Flavell, R.A. Innate immune receptors: Key regulators of metabolic disease progression. Cell Metab. 2013, 17, 873–882. [Google Scholar] [CrossRef]
- Dávila, L.A.; Pirela, V.B.; Díaz, W.; Villasmil, N.R.; León, S.C.; Contreras, M.C.E.; Bonacich, K.B.; Agüero, S.D.; Vergara, P.C.; Bonacich, R.B.; et al. The microbiome and the epigenetics of diabetes mellitus. In Diabetes Food Plan; Waisundara, V., Ed.; IntechOpen: Norderstedt, Germany, 2018; Available online: https://www.intechopen.com/books/diabetes-food-plan/the-microbiome-and-the-epigenetics-of-diabetes-mellitus (accessed on 22 November 2020). [CrossRef]
- Descamps, H.C.; Herrmann, B.; Wiredu, D.; Thaiss, C.A. The path toward using microbial metabolites as therapies. EBioMedicine 2019, 44, 747–754. [Google Scholar] [CrossRef]
- Khan, S.; Maremanda, K.P.; Jena, G. Butyrate, a Short-Chain Fatty Acid and Histone Deacetylases Inhibitor: Nutritional, Physiological, and Pharmacological Aspects in Diabetes. In Handbook of Nutrition, Diet, and Epigenetics; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 1–15. [Google Scholar]
- Khan, S.; Jena, G. The role of butyrate, a histone deacetylase inhibitor in diabetes mellitus: Experimental evidence for therapeutic intervention. Epigenomics 2015, 7, 669–680. [Google Scholar] [CrossRef]
- Khan, S.; Jena, G.B. Protective role of sodium butyrate, a HDAC inhibitor on beta-cell proliferation, function and glucose homeostasis through modulation of p38/ERK MAPK and apoptotic pathways: Study in juvenile diabetic rat. Chem. Biol. Interact. 2014, 213, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Luo, A.; Leach, S.T.; Barres, R.; Hesson, L.B.; Grimm, M.C.; Simar, D. The microbiota and epigenetic regulation of T helper 17/regulatory T cells: In search of a balanced immune system. Front. Immunol. 2017, 8, 417. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Shu, D.; Zheng, M.; Wang, J.; Luo, C.; Wang, Y.; Guo, F.; Zou, X.; Lv, X.; Li, Y.; et al. Microbial metabolite butyrate facilitates M2 macrophage polarization and function. Sci. Rep. 2016, 6, 24838. [Google Scholar] [CrossRef]
- Hur, K.Y.; Lee, M.S. Gut Microbiota and Metabolic Disorders. Diabetes Metab. J. 2015, 39, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Hesson, L.B. Gut microbiota and obesity-related gastrointestinal cancer: A focus on epigenetics. Transl. Gastrointest. Cancer 2013, 2, 204–210. [Google Scholar]
- Luu, M.; Pautz, S.; Kohl, V.; Singh, R.; Romero, R.; Lucas, S.; Hofmann, J.; Raifer, H.; Vachharajani, N.; Carrascosa, L.C.; et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat. Commun. 2019, 10, 760. [Google Scholar] [CrossRef]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef]
- Isobe, J.; Maeda, S.; Obata, Y.; Iizuka, K.; Nakamura, Y.; Fujimura, Y.; Kimizuka, T.; Hattori, K.; Kim, Y.G.; Morita, T.; et al. Commensal-bacteria-derived butyrate promotes the T cell-independent IgA response in the colon. Int. Immunol. 2019, 32, 243–258. [Google Scholar] [CrossRef]
- Elbarbary, N.S.; Ismail, E.A.R.; Zaki, M.A.; Darwish, Y.W.; Ibrahim, M.Z.; El-Hamamsy, M. Vitamin B complex supplementation as a homocysteine-lowering therapy for early stage diabetic nephropathy in pediatric patients with type 1 diabetes: A randomized controlled trial. Clin. Nutr. 2020, 39, 49–56. [Google Scholar] [CrossRef]
- Ghadimi, D.; Helwig, U.; Schrezenmeir, J.; Heller, K.J.; de Vrese, M. Epigenetic imprinting by commensal probiotics inhibits the IL-23/IL-17 axis in an in vitro model of the intestinal mucosal immune system. J. Leukoc. Biol. 2012, 92, 895–911. [Google Scholar] [CrossRef]
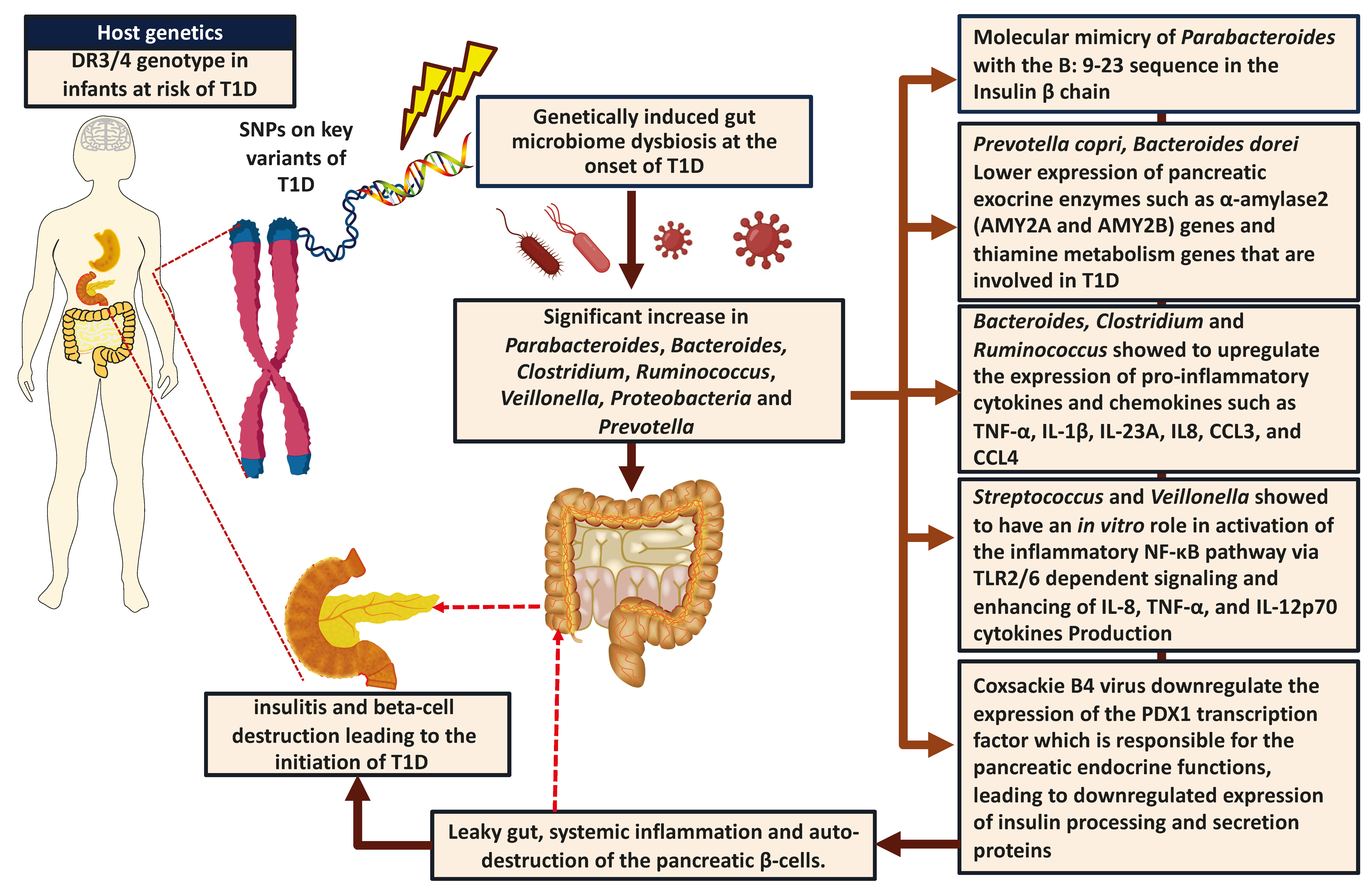

| Study Details | Age and HLA Genotype | Changes in Gut Microbiome | Findings Related to T1D and the Possible Role of Gut Microbiome | Reference |
|---|---|---|---|---|
| 16 cases 16 controls Caucasians | 7.48 ± 0.87 years | -Clostridium ↑ -Bacteroides ↑ -Veillonella ↑ -Actinobacteria ↓ -Firmicutes ↓ -Firmicutes to Bacteroidetes ratio ↓ -Lactobacillus ↓ -Bifidobacterium ↓ -Blautiacoccoides ↓ -Eubacteriumrectale group ↓ -Prevotella ↓ which was linked with the higher glycemic level in children with T1D | -Bacteroides and Clostridium showed to upregulate the expression of pro-inflammatory cytokines and chemokines such as TNF-α, IL-1β, IL-23A, IL8, CCL3, and CCL4 -Veillonella showed to have an in-vitro role in the activation of the inflammatory NF-κB pathway via TLR2/6 dependent signaling pathway -Lower number of the beneficial bacteria (Lactic acid bacteria) that plays a role in limiting the inflammatory response in the gut through enhancing the expression of IL-10 and TNF-α and maintaining the gut integrity | [54,76,78,79,80] |
| 15 cases 13 controls Caucasians | under 18 years old | -Bacteroides ↑ -Ruminococcus ↑ -Veillonella ↑ -Blautia ↑ -Streptococcus ↑ -Bifidobacterium ↓ -Roseburia ↓ -Faecalibacterium ↓ -Lachnospira ↓ | -Lower bacterial diversity and higher gut permeability -Bacteroides and Ruminococcus showed to upregulate the expression of different inflammatory cytokines and chemokines such as IL-1β, IL-23A, CCL3, and CCL4 that are involved in the recruitment of immune cells in human islets leading to oxidative stress and insulitis -Streptococcus and Veillonella showed to have an in vitro role in the activation of the inflammatory NF-κB pathway via TLR2/6 dependent signaling and enhancing of IL-8, TNF-α, and IL-12p70 cytokines Production | [55,76,78] |
| 20 cases 28 controls Caucasians and Afro-descendants | 23.1 ± 8.6 years | -Bacteroides vulgatus ↑ -Bacteroides rodentium ↑ -Prevotellacopri ↑ -Bacteroides xylanisolvens ↑ -Bifidobacterium ↓ -Lactobacillales ↓ | -Increase in the bacterial translocation through the epithelial barrier, leading to leaky gut, systemic inflammation, and destruction of the Pancreatic β cells -Bacteroides vulgatus, showed to activate the NF-κB inflammatory pathway in intestinal epithelial cells | [57,81] |
| 903 non-Hispanic Children with T1D risk | 3 to 46 months with DQB1, DQA1 or DQB1, DRB1 genotype positive | -Parabacteriodes ↑ -Streptococcus sp. ↑ -Lactococcus sp. ↑ Unclassified species ↓ -Ruminococcaceae ↓ -Lactococcus ↓ -Streptococcus ↓ -Akkermansia ↓ | -Molecular mimicry of Parabacteroides with the B:9–23 sequence in insulin B chain could be linked to the initiating of T1D | [3,71,72,82] |
| 11 cases T1D risk and 11 controls from Finland and Estonia | 0 to 77 months with HLA DR-DQ genotype positive | -Bacteroides ovatus ↑ -Bacteroides fragilis ↑ -Bacteroides vulgates ↑ -Lachnospiraceae ↓ -Veillonellaceae ↓ -Bifidobacterium ↓ Viral changes: -Circoviridae viruses ↓ -Microviridae ↓ -Myoviridae ↓ -Podoviridae ↓ | -Bacteroides vulgatus, showed to activate the NF-κB inflammatory pathway in intestinal epithelial cells -Bacteroides fragilis toxins (BFT) and colibactin cause DNA damage in the gut epithelial cells -Both are linked to leaky gut, systemic inflammation, and destruction of Pancreatic B cells -Bacteriophages changes that precede the seroconversion modulate the abundance of the dysbiotic T1D associated bacteria (mainly Bacteroides) suggesting the role of virome in triggering seroconversion | [59,81,83] |
| 28 cases 27 controls from Finland, Estonia, France, Greece or Lithuania | 1.3–4.6 years with HLA DR-DQ genotype positive | -Streptococci ↑ -Bacteriodes ↑ -non-butyrate-producing Clostridium species ↑ -butyrate-producing Clostridium clusters IV and XIVa ↓ | -Streptococci and Bacteriodes produce glutamate decarboxylase, which may stimulate GAD autoimmunity via molecular mimicry -Lower butyrate production and higher gut permeability | [75] |
| 10 cases 8 controls from Finland and Estonia | 0–3 years with HLA DR-DQ genotype positive | -Escherichia coli ↑ | -Releasing of LPS, DNA, and amyloid by bacteriophage infected E. coli may have a role in initiating autoimmunity -E. coli showed to have a role in DNA methylation, downregulating the expression of CDKN2A gene, which linked to the enhanced proinflammatory functions of CD14+ and CD16+ monocyte and the dysregulated functions of Treg cells in addition to the higher levels of HbA1C | [84,85] |
| 73 cases 103 controls From Azerbaijan Jordan, Nigeria, and Sudan | 3–19 years with HLADQ8, DQ2, or both genotype positive | -E. coli ↑ -Eubacterium ↓ -Roseburia ↓ -Clostridia clusters IV or XIVa ↓ | -LPS from Proteobacteria enhances the inflammation and endotoxemia in the gut, acting as an activation signal for M1 macrophage through enhancing the NF-κB signaling pathways and upregulating the expression of pro-inflammatory cytokines such as TNFα, IL-1β, IL-6, affecting the integrity of intestinal epithelia, leading to autoimmunity and T1D | [86,87,88] |
| 12 cases 10 controls (Han Chinese) | 12–33 years | -Bacteriodes/ Firmicutes ratio ↑ -Bilophila ↑ | -Higher HbA1c was associated with increased Bacteriodes -Higher number of anti-islet cell autoantibodies associated with decreased abundance of Faecalibacterium (butyrate-producing bacteria) and Ruminococcaceae | [86,89] |
| 47 children with islet autoimmunity or T1D | 5.3–16.3 years | -Prevotella ↓ -Butyricimonas ↓ -SCFA producing bacteria ↓ -bacterial diversity ↓ | -Gut microbiome dysbiosis was accompanied by higher intestinal permeability. Butyricimonas and Prevotella species are butyrate-producing bacteria that showed to have immunomodulatory properties in the gut -Butyrate enhances histone H3 acetylation in the promoter of the Foxp3 locus, promoting the differentiation of Treg cells and regulating the balance between Treg and Th17 -Butyrate signaling via Gpr109a receptor showed to enhance the anti-inflammatory functions in colonic macrophages and dendritic cells, which in turn enhances the differentiation of Treg cells and the production of IL-10 and IL-18 in colonic epithelium, in addition to suppression of TNF α production in monocytes | [77,90,91,92,93,94] |
| 41 controls | HLA-DR genotype positive |
| Animal Model Details | Changes in Gut Microbiome | Findings Related to T1D and the Possible Role of Gut Microbiome | Reference |
|---|---|---|---|
| SPF NOD mice with T1D | -Increased Ratio of G+/G− bacteria -α diversity ↓ -Firmicutes ↑ -Bacteroidetes ↑ -Erysipelotrichaceae ↑ | -Deficiency in the development of mucosal-associated lymphoid tissue -imbalance between Th1, Th17, and Treg in the intestine and increased intestinal permeability | [150,151,152] |
| Streptozotocin-induced T1D rats | -Firmicutes/Bacteroidetes ratio ↑ -Ruminococcaceae ↑ -Shigella ↑ -Enterococcus ↑ | -LPS enhances inflammatory response in the pancreatic β cells by upregulating the mRNA expression of various inflammatory cytokines including IL-1β, IL-18, and IL-12 in addition to enhanced expression of CD80 co-stimulatory molecule | [154] |
| Streptozotocin-induced T1D mice | -Bacteroidaceae ↑ -Alcaligenaceae ↑ -Ruminococcaceae ↑ -Bifidobacteriaceae ↑ | -Increased expression of NOD1 and NOD2 genes in the pancreatic lymph node -Increased IL-6, IL-12, IL-17, and IFN- γ -invasive insulitis in the pancreatic islets | [155] |
| ACE2 knockout Akita mice with T1D | -Firmicutes/Bacteroidetes ratio ↑ | Enhanced gut permeability, enhanced microbial translocation with higher amount of circulatory peptidoglycan which increases the risk of T1D associated retinopathy when transferred into HRECs via TLR 2 activation mechanism | [162] |
| SPF BDC2.5X NOD mice treated with DSS | -Firmicutes ↑ -Deferribacteres ↑ -Porphyromonadaceae ↑ -Bacteroidetes ↓ -Prevotellaceae ↓ -Rikenellaceae ↓ | -Lower gut integrity -Decrease in expression of immune-regulatory mucins Muc1 and Muc3 -Increase in inflammatory cytokines in the intestinal mucosa (TNF-α, IL1-β, IL-23p19, IL-17) | [162,163] |
| Antibiotic induced dysbiosis in SPF NOD mice | -Proteobacteria ↑ -Akkermansia mucinophila ↑ -Enterococcus ↑ -Blautia ↑ -Enterobacteriaceae sp ↑ -Clostridiales ↓ -Oscillospira ↓ -Ruminococcus ↓ | -Accelerated development of T1D -Lower numbers of RORγt+ Th 17 and FOXP3+ Treg Cells affecting the expression of genes that are associated with immunity and cholesterol synthesis -Increased intestinal permeability | [156,164] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elhag, D.A.; Kumar, M.; Al Khodor, S. Exploring the Triple Interaction between the Host Genome, the Epigenome, and the Gut Microbiome in Type 1 Diabetes. Int. J. Mol. Sci. 2021, 22, 125. https://doi.org/10.3390/ijms22010125
Elhag DA, Kumar M, Al Khodor S. Exploring the Triple Interaction between the Host Genome, the Epigenome, and the Gut Microbiome in Type 1 Diabetes. International Journal of Molecular Sciences. 2021; 22(1):125. https://doi.org/10.3390/ijms22010125
Chicago/Turabian StyleElhag, Duaa Ahmed, Manoj Kumar, and Souhaila Al Khodor. 2021. "Exploring the Triple Interaction between the Host Genome, the Epigenome, and the Gut Microbiome in Type 1 Diabetes" International Journal of Molecular Sciences 22, no. 1: 125. https://doi.org/10.3390/ijms22010125
APA StyleElhag, D. A., Kumar, M., & Al Khodor, S. (2021). Exploring the Triple Interaction between the Host Genome, the Epigenome, and the Gut Microbiome in Type 1 Diabetes. International Journal of Molecular Sciences, 22(1), 125. https://doi.org/10.3390/ijms22010125





