S-Adenosyl-l-Methionine Overcomes uL3-Mediated Drug Resistance in p53 Deleted Colon Cancer Cells
Abstract
1. Introduction
2. Results
2.1. AdoMet Inhibits Proliferation in HCT 116p53−/− and uL3ΔHCT 116p53−/− Colon Cancer Cells
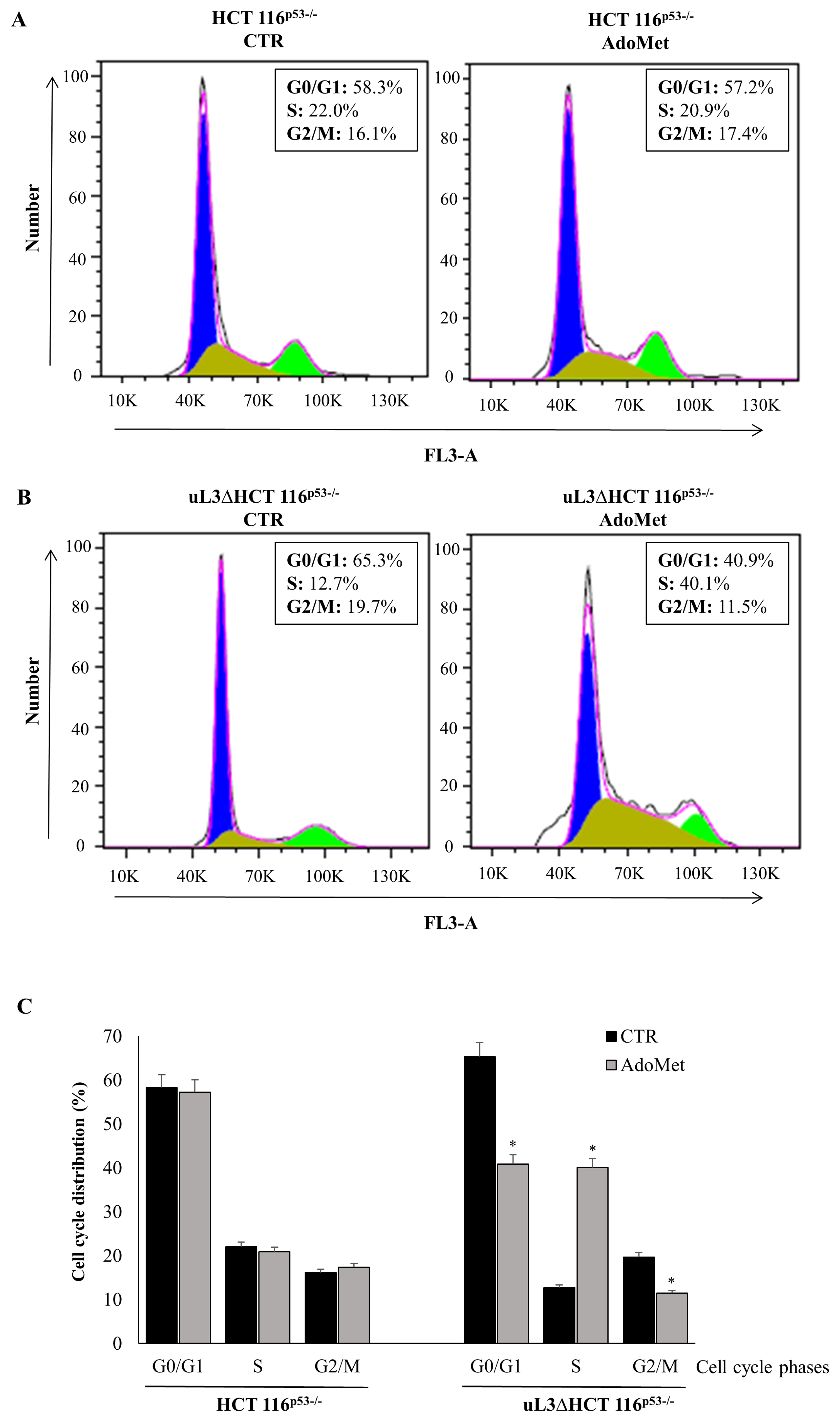
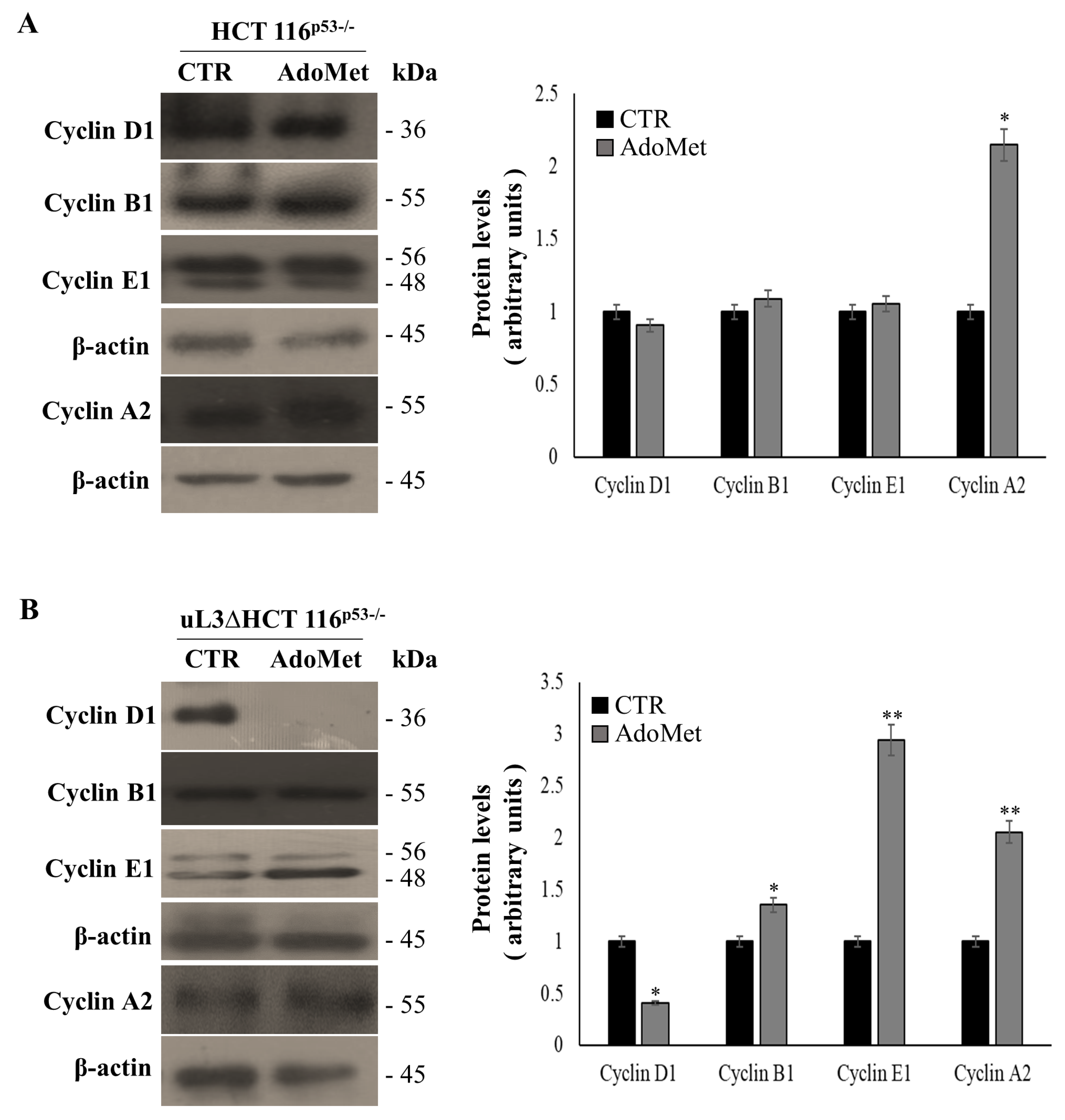
2.2. AdoMet Causes Cell Cycle Arrest in S Phase in uL3ΔHCT 116p53−/−
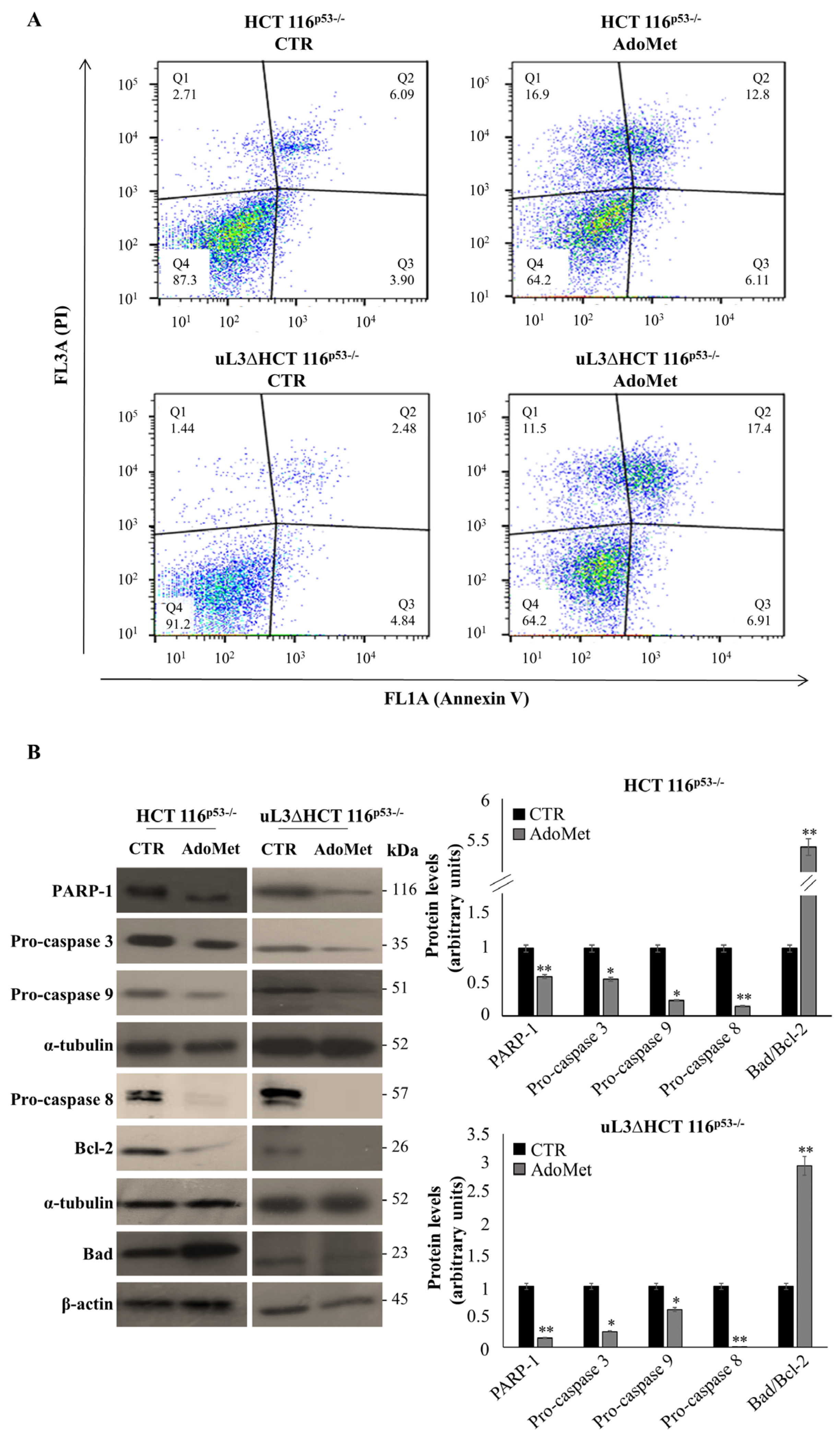
2.3. AdoMet Induces Apoptosis in HCT 116p53−/− and uL3ΔHCT 116p53−/− Colon Cancer Cells
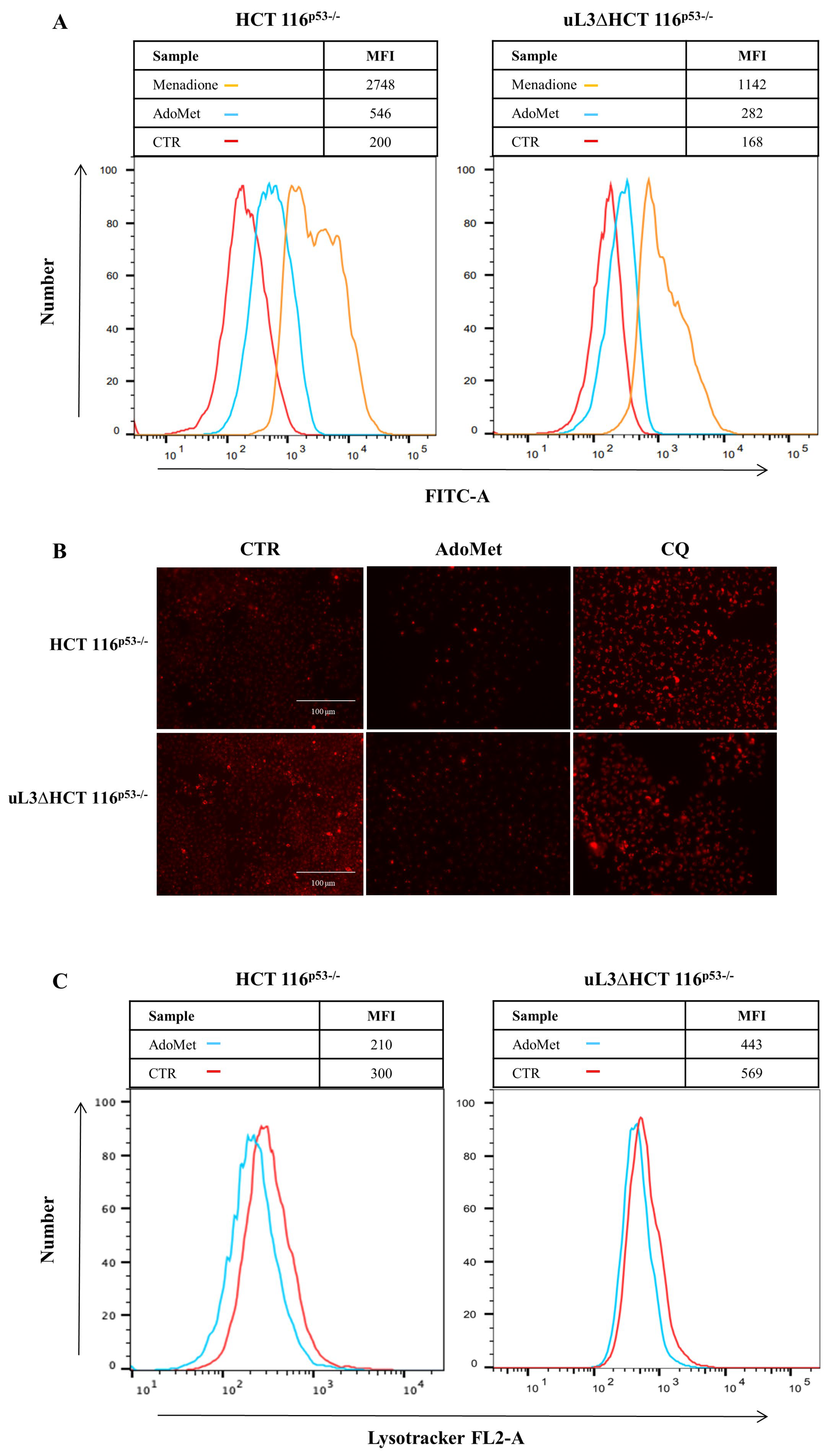
2.4. AdoMet Increases ROS Production in HCT 116p53−/− and uL3ΔHCT 116p53−/− Colon Cancer Cells
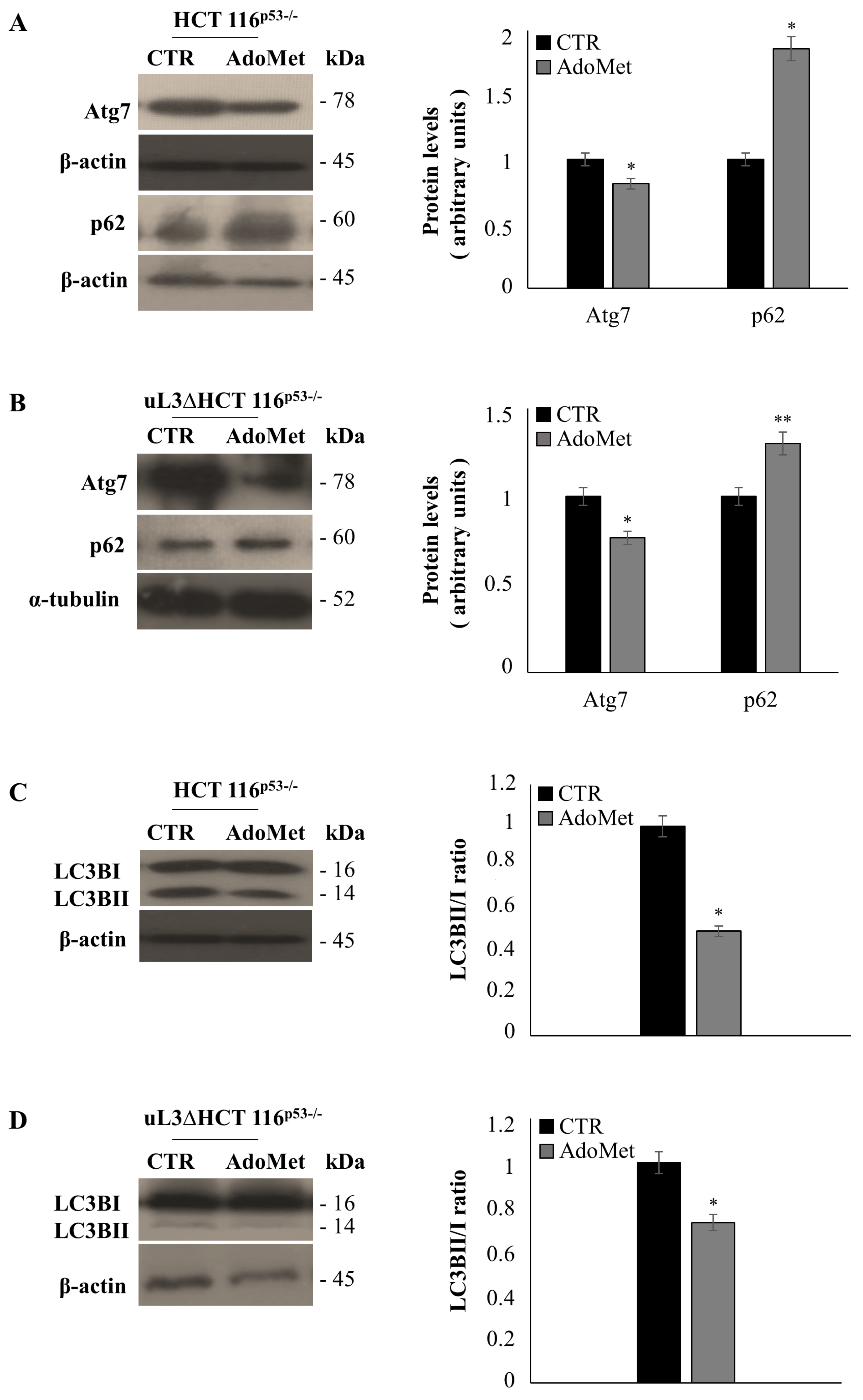
2.5. AdoMet Inhibits Autophagy in HCT 116p53−/− and uL3ΔHCT 116p53−/− Colon Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Cultures and Drug Treatments
4.3. Cell Viability Assays
4.4. Cell Cycle Analysis
4.5. Cell Death Assay
4.6. Determination of ROS by the 2′,7′-dichlorofluorescein Diacetate (DCFH-DA) Assay
4.7. LysoTracker-Red Staining
4.8. Western Blot Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AdoMet | S-adenosyl-l-methionine |
| MDR | multidrug resistance |
| 5-FU | 5-fluorouracil |
| ROS | reactive oxygen species |
| DCF-DA | 2′,7′-dichlorofluorescein diacetate |
| MFI | median intensity fluorescence |
| LTR | LysoTracker |
| CTR | untreated cells |
| CQ | chloroquine |
| FBS | fetal bovine serum |
| DMEM | Dulbecco’s modified Eagle’s medium |
| MTT | 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide |
| PI | Propidium iodide |
| PBS | phosphate-buffered saline |
| Annexin V-FITC | Annexin V-fluorescein isothiocyanate |
References
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef]
- Arruebo, M.; Vilaboa, N.; Sáez-Gutierrez, B.; Lambea, J.; Tres, A.; Valladares, M.; González-Fernández, A. Assessment of the evolution of cancer treatment therapies. Cancers 2011, 3, 3279–3330. [Google Scholar] [CrossRef]
- Van der Jeught, K.; Xu, H.C.; Li, Y.J.; Lu, X.B.; Ji, G. Drug resistance and new therapies in colorectal cancer. World J. Gastroenterol. 2018, 24, 3834–3848. [Google Scholar] [CrossRef]
- Bertram, J.; Palfner, K.; Hiddemann, W.; Kneba, M. Overexpression of ribosomal proteins L4 and L5 and the putative alternative elongation factor PTI-1 in the doxorubicin resistant human colon cancer cell line LoVoDxR. Eur. J. Cancer 1998, 34, 731–736. [Google Scholar] [CrossRef]
- Johnsson, A.; Zeelenberg, I.; Min, Y.; Hilinski, J.; Berry, C.; Howell, S.B.; Los, G. Identification of genes differentially expressed in association with acquired cisplatin resistance. Br. J. Cancer 2000, 83, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; You, H.; Liu, F.; An, H.; Shi, Y.; Yu, Q.; Fan, D. Differentially expressed gene profiles between multidrug resistant gastric adenocarcinoma cells and their parental cells. Cancer Lett. 2002, 185, 211–218. [Google Scholar] [CrossRef]
- Shi, Y.; Zhai, H.; Wang, X.; Han, Z.; Liu, C.; Lan, M.; Du, J.; Guo, C.; Zhang, Y.; Wu, K.; et al. Ribosomal proteins S13 and L23 promote multidrug resistance in gastric cancer cells by suppressing drug-induced apoptosis. Exp. Cell Res. 2004, 296, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Russo, G. Ribosomal Proteins Control or Bypass p53 during Nucleolar Stress. Int. J. Mol. Sci. 2017, 18, 140. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Saide, A.; Smaldone, S.; Faraonio, R.; Russo, G. Role of uL3 in Multidrug Resistance in p53-Mutated Lung Cancer Cells. Int. J. Mol. Sci. 2017, 18, 547. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, A.; Carotenuto, P.; Russo, G.; Russo, A. Ribosomal protein uL3 targets E2F1 and Cyclin D1 in cancer cell response to nucleolar stress. Sci. Rep. 2019, 9, 15431. [Google Scholar] [CrossRef]
- Pecoraro, A.; Carotenuto, P.; Franco, B.; De Cegli, R.; Russo, G.; Russo, A. Role of uL3 in the Crosstalk between Nucleolar Stress and Autophagy in Colon Cancer Cells. Int. J. Mol. Sci. 2020, 21, 2143. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Cirulli, C.; Amoresano, A.; Pucci, P.; Pietropaolo, C.; Russo, G. cis-acting sequences and trans-acting factors in the localization of mRNA for mitochondrial ribosomal proteins. Biochim. Biophys. Acta 2008, 1779, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Siciliano, G.; Catillo, M.; Giangrande, C.; Amoresano, A.; Pucci, P.; Pietropaolo, C.; Russo, G. hnRNP H1 and intronic G runs in the splicing control of the human rpL3 gene. Biochim. Biophys. Acta 2010, 1799, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Catillo, M.; Esposito, D.; Briata, P.; Pietropaolo, C.; Russo, G. Autoregulatory circuit of human rpL3 expression requires hnRNP H1, NPM and KHSRP. Nucleic. Acids Res. 2011, 39, 7576–7585. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Maiolino, S.; Pagliara, V.; Ungaro, F.; Tatangelo, F.; Leone, A.; Scalia, G.; Budillon, A.; Quaglia, F.; Russo, G. Enhancement of 5-FU sensitivity by the proapoptotic rpL3 gene in p53 null colon cancer cells through combined polymer nanoparticles. Oncotarget 2016, 7, 79670–79687. [Google Scholar] [CrossRef]
- Russo, A.; Esposito, D.; Catillo, M.; Pietropaolo, C.; Crescenzi, E.; Russo, G. Human rpL3 induces G₁/S arrest or apoptosis by modulating p21 (waf1/cip1) levels in a p53-independent manner. Cell Cycle 2013, 12, 76–87. [Google Scholar] [CrossRef]
- Pagliara, V.; Saide, A.; Mitidieri, E.; d’Emmanuele di Villa Bianca, R.; Sorrentino, R.; Russo, G.; Russo, A. 5-FU targets rpL3 to induce mitochondrial apoptosis via cystathionine-β-synthase in colon cancer cells lacking p53. Oncotarget 2016, 7, 50333–50348. [Google Scholar] [CrossRef]
- Esposito, D.; Crescenzi, E.; Sagar, V.; Loreni, F.; Russo, A.; Russo, G. Human rpL3 plays a crucial role in cell response to nucleolar stress induced by 5-FU and L-OHP. Oncotarget 2014, 5, 11737–11751. [Google Scholar] [CrossRef]
- Russo, A.; Pagliara, V.; Albano, F.; Esposito, D.; Sagar, V.; Loreni, F.; Irace, C.; Santamaria, R.; Russo, G. Regulatory role of rpL3 in cell response to nucleolar stress induced by Act D in tumor cells lacking functional p53. Cell Cycle 2016, 15, 41–51. [Google Scholar] [CrossRef]
- Russo, A.; Pellosi, D.S.; Pagliara, V.; Milone, M.R.; Pucci, B.; Caetano, W.; Hioka, N.; Budillon, A.; Ungaro, F.; Russo, G.; et al. Biotin-targeted Pluronic(®) P123/F127 mixed micelles delivering niclosamide: A repositioning strategy to treat drug-resistant lung cancer cells. Int. J. Pharm. 2016, 511, 127–139. [Google Scholar] [CrossRef]
- Lu, S.C. S-Adenosylmethionine. Int. J. Biochem. Cell Biol. 2000, 32, 391–395. [Google Scholar] [CrossRef]
- Porcelli, M.; Ilisso, C.P.; Mosca, L.; Cacciapuoti, G. A thermostable archaeal S-adenosylmethionine synthetase: A promising tool to improve the synthesis of adenosylmethionine analogs of biotechnological interest. Bioengineered 2015, 6, 184–186. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Minici, C.; Mosca, L.; Ilisso, C.P.; Cacciapuoti, G.; Porcelli, M.; Degano, M. Structures of catalytic cycle intermediates of the Pyrococcus furiosus methionine adenosyltransferase demonstrate negative cooperativity in the archaeal orthologues. J. Struct. Biol. 2020, 210, 107462. [Google Scholar] [CrossRef] [PubMed]
- Mato, J.M.; Corrales, F.J.; Lu, S.C.; Avila, M.A. S-Adenosylmethionine: A control switch that regulates liver function. FASEB J. 2002, 16, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Fontecave, M.; Atta, M.; Mulliez, E. S-adenosylmethionine: Nothing goes to waste. Trends Biochem. Sci. 2004, 29, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C.; Mato, J.M. S-Adenosylmethionine in cell growth, apoptosis and liver cancer. J. Gastroenterol. Hepatol. 2008, 23 (Suppl. 1), S73–S77. [Google Scholar] [CrossRef]
- Martínez-López, N.; Varela-Rey, M.; Ariz, U.; Embade, N.; Vazquez-Chantada, M.; Fernandez-Ramos, D.; Gomez-Santos, L.; Lu, S.C.; Mato, J.M.; Martinez-Chantar, M.L. S-adenosylmethionine and proliferation: New pathways, new targets. Biochem. Soc. Trans. 2008, 36, 848–852. [Google Scholar] [CrossRef]
- Cave, D.D.; Ilisso, C.P.; Mosca, L.; Pagano, M.; Martino, E.; Porcelli, M.; Cacciapuoti, G. The anticancer effects of S-adenosylmethionine on breast cancer cells. JSM Chem. 2017, 5, 1049. [Google Scholar]
- Cave, D.D.; Desiderio, V.; Mosca, L.; Ilisso, C.P.; Mele, L.; Caraglia, M.; Cacciapuoti, G.; Porcelli, M. S-Adenosylmethionine-mediated apoptosis is potentiated by autophagy inhibition induced by chloroquine in human breast cancer cells. J. Cell. Physiol. 2018, 233, 1370–1383. [Google Scholar] [CrossRef]
- Ilisso, C.P.; Delle Cave, D.; Mosca, L.; Pagano, M.; Coppola, A.; Mele, L.; Caraglia, M.; Cacciapuoti, G.; Porcelli, M. S-Adenosylmethionine regulates apoptosis and autophagy in MCF-7 breast cancer cells through the modulation of specific microRNAs. Cancer Cell Int. 2018, 18, 197. [Google Scholar] [CrossRef]
- Mosca, L.; Pagano, M.; Ilisso, C.P.; Cave, D.D.; Desiderio, V.; Mele, L.; Caraglia, M.; Cacciapuoti, G.; Porcelli, M. AdoMet triggers apoptosis in head and neck squamous cancer by inducing ER stress and potentiates cell sensitivity to cisplatin. J. Cell. Physiol. 2019, 234, 13277–13291. [Google Scholar] [CrossRef] [PubMed]
- Mosca, L.; Minopoli, M.; Pagano, M.; Vitiello, F.; Carriero, M.V.; Cacciapuoti, G.; Porcelli, M. Effects of S-adenosyl-L-methionine on the invasion and migration of head and neck squamous cancer cells and analysis of the underlying mechanisms. Int. J. Oncol. 2020, 56, 1212–1224. [Google Scholar] [CrossRef] [PubMed]
- Mosca, L.; Vitiello, F.; Coppola, A.; Borzacchiello, L.; Ilisso, C.P.; Pagano, M.; Caraglia, M.; Cacciapuoti, G.; Porcelli, M. Therapeutic Potential of the Natural Compound S-Adenosylmethionine as a Chemoprotective Synergistic Agent in Breast, and Head and Neck Cancer Treatment: Current Status of Research. Int. J. Mol. Sci. 2020, 21, 8547. [Google Scholar] [CrossRef]
- Ekberg, J.; Brunhoff, C.; Järås, M.; Fan, X.; Landberg, G.; Persson, J.L. Increased expression of cyclin A1 protein is associated with all-trans retinoic acid-induced apoptosis. Int. J. Biochem. Cell Biol. 2006, 38, 1330–1339. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Terrasi, M.; Agnese, V.; Santini, D.; Bazan, V. Apoptosis: A relevant tool for anticancer therapy. Ann. Oncol. 2006, 17 (Suppl. 7), vii115-23. [Google Scholar] [CrossRef]
- Pecoraro, A.; Pagano, M.; Russo, G.; Russo, A. Role of Autophagy in Cancer Cell Response to Nucleolar and Endoplasmic Reticulum Stress. Int. J. Mol. Sci. 2020, 21, 7334. [Google Scholar] [CrossRef]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int. J. Mol. Sci. 2020, 21, 3233. [Google Scholar] [CrossRef]
- Fiore, D.; Piscopo, C.; Proto, M.C.; Vasaturo, M.; Dal Piaz, F.; Fusco, B.M.; Pagano, C.; Laezza, C.; Bifulco, M.; Gazzerro, P. N6-Isopentenyladenosine Inhibits Colorectal Cancer and Improves Sensitivity to 5-Fluorouracil-Targeting FBXW7 Tumor Suppressor. Cancers 2019, 11, 1456. [Google Scholar] [CrossRef]
- Carotenuto, P.; Pecoraro, A.; Palma, G.; Russo, G.; Russo, A. Therapeutic Approaches Targeting Nucleolus in Cancer. Cells 2019, 8, 1090. [Google Scholar] [CrossRef]
- Moyer, M.P.; Mato, J.M.; Aw, T.Y.; Lu, S.C. S-Adenosylmethionine and methylthioadenosine inhibit cellular FLICE inhibitory protein expression and induce apoptosis in colon cancer cells. Mol. Pharm. 2009, 76, 192–200. [Google Scholar] [CrossRef]
- Luo, J.; Li, Y.N.; Wang, F.; Zhang, W.M.; Geng, X. S-adenosylmethionine inhibits the growth of cancer cells by reversing the hypomethylation status of c-myc and H-ras in human gastric cancer and colon cancer. Int. J. Biol. Sci. 2010, 6, 784–795. [Google Scholar] [CrossRef] [PubMed]
- Li, T.W.; Yang, H.; Peng, H.; Xia, M.; Mato, J.M.; Lu, S.C. Effects of S-adenosylmethionine and methylthioadenosine on inflammation-induced colon cancer in mice. Carcinogenesis 2012, 33, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Hussain, Z.; Khan, M.I.; Shahid, M.; Almajhdi, F.N. S-adenosylmethionine, a methyl donor, up regulates tissue inhibitor of metalloproteinase-2 in colorectal cancer. Genet. Mol. Res. 2013, 12, 1106–1118. [Google Scholar] [CrossRef]
- Li, T.W.; Peng, H.; Yang, H.; Kurniawidjaja, S.; Panthaki, P.; Zheng, Y.; Mato, J.M.; Lu, S.C. S-Adenosylmethionine and methylthioadenosine inhibit β-catenin signaling by multiple mechanisms in liver and colon cancer. Mol. Pharm. 2015, 87, 77–86. [Google Scholar] [CrossRef] [PubMed]
- John, R.R.; Malathi, N.; Ravindran, C.; Anandan, S. Mini review: Multifaceted role played by cyclin D1 in tumor behavior. Indian J. Dent. Res. 2017, 28, 187–192. [Google Scholar] [CrossRef]
- Indran, I.R.; Tufo, G.; Pervaiz, S.; Brenner, C. Recent advances in apoptosis, mitochondria and drug resistance in cancer cells. Biochim. Biophys. Acta 2011, 1807, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Julien, O.; Wells, J.A. Caspases and their substrates. Cell Death Differ. 2017, 24, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Chong, S.J.; Low, I.C.; Pervaiz, S. Mitochondrial ROS and involvement of Bcl-2 as a mitochondrial ROS regulator. Mitochondrion 2014, 19, 39–48. [Google Scholar] [CrossRef]
- Kongara, S.; Karantza, V. The interplay between autophagy and ROS in tumorigenesis. Front. Oncol. 2012, 2, 171. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Okamoto, K.; Yu, C.; Sinicrope, F.A. p62/sequestosome-1 up-regulation promotes ABT-263-induced caspase-8 aggregation/activation on the autophagosome. J. Biol. Chem. 2013, 288, 33654–33666. [Google Scholar] [CrossRef] [PubMed]
- Esposito, V.; Russo, A.; Amato, T.; Varra, M.; Vellecco, V.; Bucci, M.; Russo, G.; Virgilio, A.; Galeone, A. Backbone modified TBA analogues endowed with antiproliferative activity. Biochim. Biophys. Acta Gen. Subj. 2017, 1861 Pt B, 1213–1221. [Google Scholar] [CrossRef]
- Pecoraro, A.; Virgilio, A.; Esposito, V.; Galeone, A.; Russo, G.; Russo, A. uL3 mediated nucleolar stress pathway as a new mechanism of action of antiproliferative g-quadruplex TBA derivatives in colon cancer cells. Biomolecules 2020, 10, 583. [Google Scholar] [CrossRef] [PubMed]
- Mele, L.; Del Vecchio, V.; Marampon, F.; Regad, T.; Wagner, S.; Mosca, L.; Bimonte, S.; Giudice, A.; Liccardo, D.; Prisco, C.; et al. β2-AR blockade potentiates MEK1/2 inhibitor effect on HNSCC by regulating Nrf2-mediated defense mechanism. Cell Death Dis. 2020, 11, 850. [Google Scholar] [CrossRef] [PubMed]
- Pagano, C.; Siauciunaite, R.; Idda, M.; Ruggiero, G.; Ceinos, R.M.; Pagano, M.; Frigato, E.; Bertolucci, C.; Foulkes, N.S.; Vallone, D. Evolution shapes the responsiveness of the D-box enhancer element to light and reactive oxygen species in vertebrates. Sci. Rep. 2018, 4, 13180. [Google Scholar] [CrossRef] [PubMed]
- Russo, G.; Cuccurese, M.; Monti, G.; Russo, A.; Amoresano, A.; Pucci, P.; Pietropaolo, C. Ribosomal protein L7a binds RNA through two distinct RNA-binding domains. Biochem. J. 2005, 385 Pt 1, 289–299. [Google Scholar] [CrossRef]
- Fernández-Ramos, D.; Lopitz-Otsoa, F.; Delacruz-Villar, L.; Bilbao, J.; Pagano, M.; Mosca, L.; Bizkarguenaga, M.; Serrano-Macia, M.; Azkargorta, M.; Iruarrizaga-Lejarreta, M.; et al. Aramchol improves liver glucose and lipid homeostasis in NASH via AMPK and mTOR regulation. World J. Gastroenterol. 2020, 26, 5101–5117. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mosca, L.; Pagano, M.; Pecoraro, A.; Borzacchiello, L.; Mele, L.; Cacciapuoti, G.; Porcelli, M.; Russo, G.; Russo, A. S-Adenosyl-l-Methionine Overcomes uL3-Mediated Drug Resistance in p53 Deleted Colon Cancer Cells. Int. J. Mol. Sci. 2021, 22, 103. https://doi.org/10.3390/ijms22010103
Mosca L, Pagano M, Pecoraro A, Borzacchiello L, Mele L, Cacciapuoti G, Porcelli M, Russo G, Russo A. S-Adenosyl-l-Methionine Overcomes uL3-Mediated Drug Resistance in p53 Deleted Colon Cancer Cells. International Journal of Molecular Sciences. 2021; 22(1):103. https://doi.org/10.3390/ijms22010103
Chicago/Turabian StyleMosca, Laura, Martina Pagano, Annalisa Pecoraro, Luigi Borzacchiello, Luigi Mele, Giovanna Cacciapuoti, Marina Porcelli, Giulia Russo, and Annapina Russo. 2021. "S-Adenosyl-l-Methionine Overcomes uL3-Mediated Drug Resistance in p53 Deleted Colon Cancer Cells" International Journal of Molecular Sciences 22, no. 1: 103. https://doi.org/10.3390/ijms22010103
APA StyleMosca, L., Pagano, M., Pecoraro, A., Borzacchiello, L., Mele, L., Cacciapuoti, G., Porcelli, M., Russo, G., & Russo, A. (2021). S-Adenosyl-l-Methionine Overcomes uL3-Mediated Drug Resistance in p53 Deleted Colon Cancer Cells. International Journal of Molecular Sciences, 22(1), 103. https://doi.org/10.3390/ijms22010103




