Gene Signatures of Early Response to Anti-TNF Drugs in Pediatric Inflammatory Bowel Disease
Abstract
1. Introduction
2. Results
2.1. Patients Characteristics
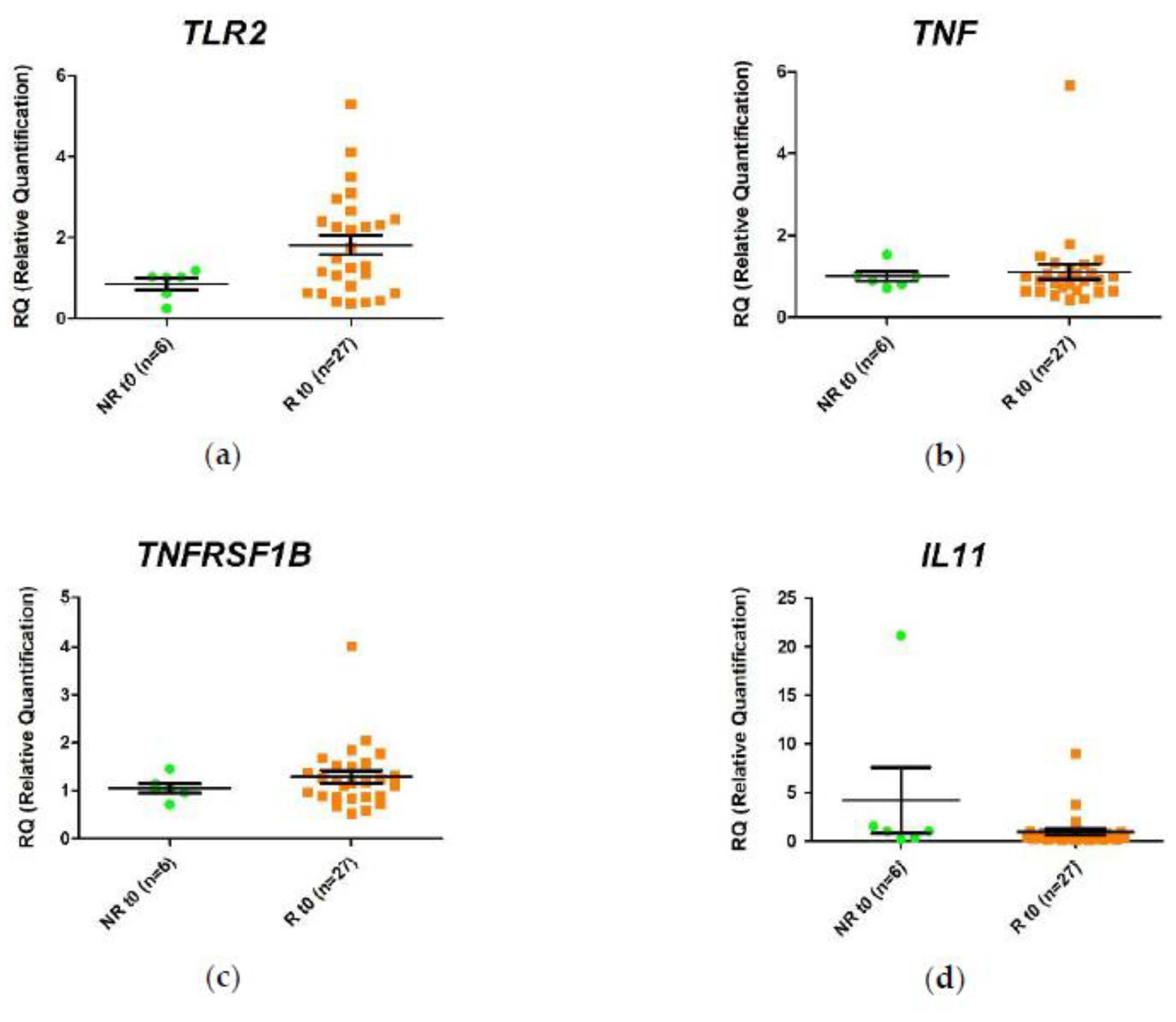
2.2. Differential Gene Expression in the Response of Anti-TNF Agents Prior to Starting Treatment
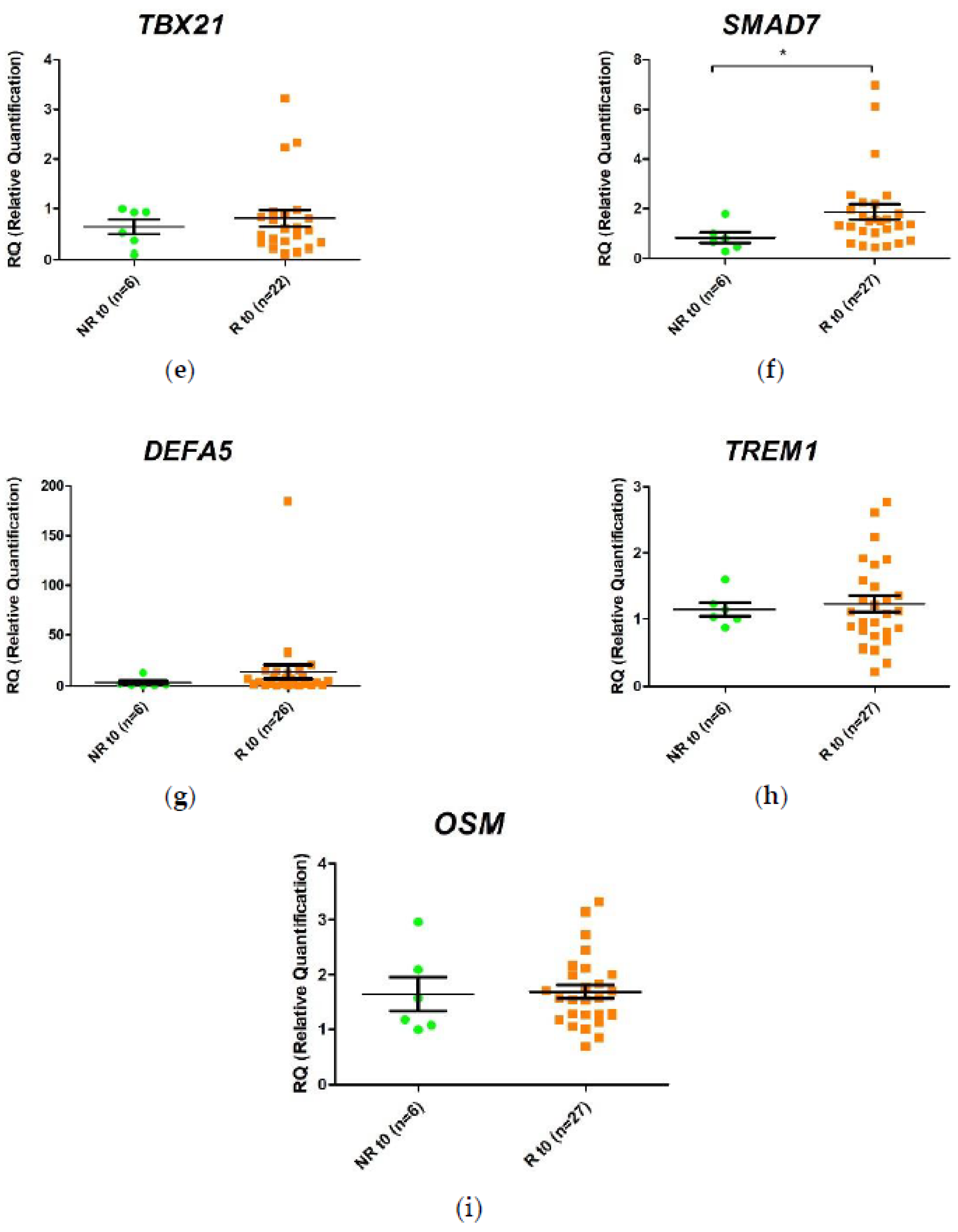
2.3. Differential Gene Expression in Response to Anti-TNF Agents at Week 2 Post-Treatment
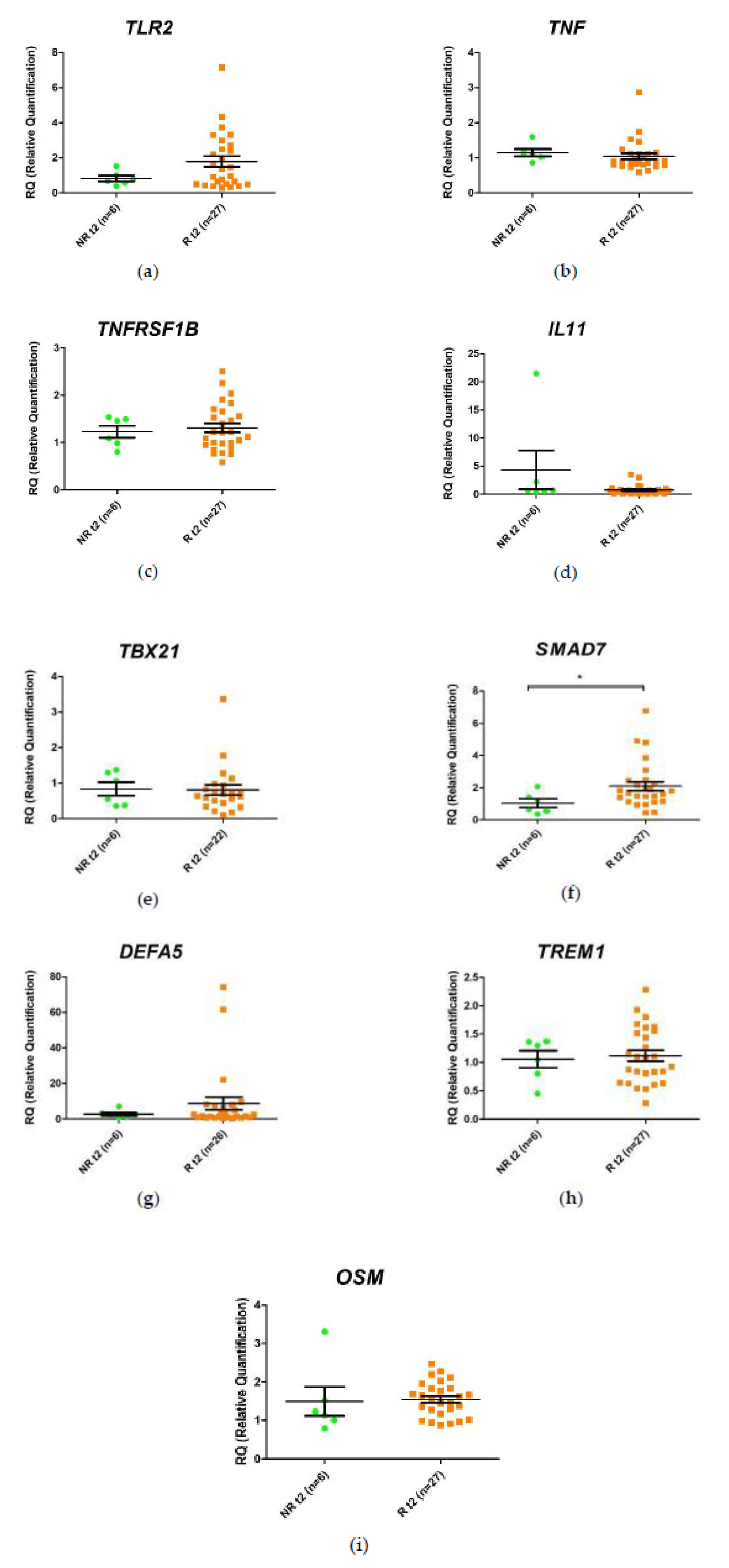
2.4. Differential Change in Gene Expression during the First 2 Weeks of Anti-TNF Treatment
2.5. Prediction of Response to Anti-TNF Therapy Based on SMAD7 Expression Prior to Starting Treatment
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. Ethics Statement
4.3. Extraction of Total RNA from Whole Blood
4.4. Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| IBD | Inflammatory bowel disease |
| pIBD | Pediatric inflammatory bowel disease |
| UC | Ulcerative colitis |
| CD | Crohn’s disease |
| SD | Standard deviation |
| IQR | Interquartile range |
| PCDAI | PCDAI. Pediatric Crohn’s Disease Activity Index |
| PUCAI | Pediatric Ulcerative Colitis Activity Index |
| SEM | Standard error of the mean |
| PPV | Positive predictive value |
| SEM | Standard error of the mean |
| PPV | Positive predictive value |
| NPV | Negative predictive value |
References
- Gu, P.; Feagins, L.A. Dining with Inflammatory Bowel Disease: A Review of the Literature on Diet in the Pathogenesis and Management of IBD. Inflamm. Bowel Dis. 2020, 26, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Kuhnen, A. Genetic and Environmental Considerations for Inflammatory Bowel Disease. Surg. Clin. N. Am. 2019, 99, 1197–1207. [Google Scholar] [CrossRef] [PubMed]
- Sawczenko, A.; Sandhu, B.K. Presenting features of inflammatory bowel disease in Great Britain and Ireland. Arch. Dis. Child. 2003, 88, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Henderson, P.; van Limbergen, J.E.; Wilson, D.C.; Satsangi, J.; Russell, R.K. Genetics of childhood-onset inflammatory bowel disease. Inflamm. Bowel Dis. 2011, 17, 346–361. [Google Scholar] [CrossRef]
- Hyams, J.; Crandall, W.; Kugathasan, S.; Griffiths, A.; Olson, A.; Johanns, J.; Liu, G.; Travers, S.; Heuschkel, R.; Markowitz, J.; et al. Induction and Maintenance Infliximab Therapy for the Treatment of Moderate-to-Severe Crohn’s Disease in Children. Gastroenterology 2007, 132, 863–873. [Google Scholar] [CrossRef]
- Shim, H.H.; Chan, P.W.; Chuah, S.W.; Schwender, B.J.; Kong, S.C.; Ling, K.L. A review of vedolizumab and ustekinumab for the treatment of inflammatory bowel diseases. JGH Open 2018, 2, 223–234. [Google Scholar] [CrossRef]
- Cholapranee, A.; Hazlewood, G.S.; Kaplan, G.G.; Peyrin-Biroulet, L.; Ananthakrishnan, A.N. Systematic review with meta-analysis: Comparative efficacy of biologics for induction and maintenance of mucosal healing in Crohn’s disease and ulcerative colitis controlled trials. Aliment. Pharmacol. Ther. 2017, 45, 1291–1302. [Google Scholar] [CrossRef]
- Hendy, P.; Hart, A.; Irving, P. Anti-TNF drug and antidrug antibody level monitoring in IBD: A practical guide. Frontline Gastroenterol. 2016, 7, 122–128. [Google Scholar] [CrossRef]
- Kelly, O.B.; Donnell, S.O.; Stempak, J.M.; Steinhart, A.H.; Silverberg, M.S. Therapeutic Drug Monitoring to Guide Infliximab Dose Adjustment is Associated with Better Endoscopic Outcomes than Clinical Decision Making Alone in Active Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2017, 23, 1202–1209. [Google Scholar] [CrossRef]
- Laserna-Mendieta, E.J.; Lucendo, A.J. Faecal calprotectin in inflammatory bowel diseases: A review focused on meta-analyses and routine usage limitations. Clin. Chem. Lab. Med. 2019, 57, 1295–1307. [Google Scholar] [CrossRef]
- Ma, C.; Battat, R.; Khanna, R.; Parker, C.E.; Feagan, B.G.; Jairath, V. What is the role of C-reactive protein and fecal calprotectin in evaluating Crohn’s disease activity? Best Pract. Res. Clin. Gastroenterol. 2019, 38–39, 101602. [Google Scholar] [CrossRef] [PubMed]
- Courbette, O.; Aupiais, C.; Viala, J.; Hugot, J.-P.; Roblin, X.; Candon, S.; Louveau, B.; Chatenoud, L.; Martinez-Vinson, C. Trough Levels of Infliximab at W6 Are Predictive of Remission at W14 in Pediatric Crohn Disease. J. Pediatr. Gastroenterol. Nutr. 2020, 70, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Balog, A.; Klausz, G.; Gal, J.; Molnar, T.; Nagy, F.; Ocsovszky, I.; Gyulai, Z.; Mandi, Y. Investigation of the prognostic value of TNF-alpha gene polymorphism among patients treated with infliximab, and the effects of infliximab therapy on TNF-α production and apoptosis. Pathobiology 2004, 71, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Matsukura, H.; Ikeda, S.; Yoshimura, N.; Takazoe, M.; Muramatsu, M. Genetic polymorphisms of tumour necrosis factor receptor superfamily 1A and 1B affect responses to infliximab in Japanese patients with Crohn’s disease. Aliment. Pharmacol. Ther. 2008, 27, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Bank, S.; Andersen, P.S.; Burisch, J.; Pedersen, N.; Roug, S.; Galsgaard, J.; Turino, S.Y.; Brodersen, J.B.; Rashid, S.; Rasmussen, B.K. Associations between functional polymorphisms in the NFκB signaling pathway and response to anti-TNF treatment in Danish patients with inflammatory bowel disease. Pharm. J. 2014, 14, 526–534. [Google Scholar] [CrossRef]
- Cravo, M.; Ferreira, P.; Sousa, P.; Moura-Santos, P.; Velho, S.; Tavares, L.; Deus, J.R.; Ministro, P.; da Silva, J.P.; Correia, L.; et al. Clinical and genetic factors predicting response to therapy in patients with Crohn’s disease. United Eur. Gastroenterol. J. 2014, 2, 47–56. [Google Scholar] [CrossRef]
- Bek, S.; Nielsen, J.V.; Bojesen, A.B.; Franke, A.; Bank, S.; Vogel, U.; Andersen, V. Systematic review: Genetic biomarkers associated with anti-TNF treatment response in inflammatory bowel diseases. Aliment. Pharmacol. Ther. 2016, 44, 554–567. [Google Scholar] [CrossRef]
- López-Hernández, R.; Valdés, M.; Campillo, J.A.; Martínez-García, P.; Salama, H.; Bolarin, J.M.; Martínez, H.; Moya-Quiles, M.R.; Minguela, A.; Sánchez-Torres, A.; et al. Pro- and anti-inflammatory cytokine gene single-nucleotide polymorphisms in inflammatory bowel disease. Int. J. Immunogenet. 2015, 42, 38–45. [Google Scholar] [CrossRef]
- Arijs, I.; Li, K.; Toedter, G.; Quintens, R.; Van Lommel, L.; Van Steen, K.; Leemans, P.; De Hertogh, G.; Lemaire, K.; Ferrante, M.; et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut 2009, 58, 1612–1619. [Google Scholar] [CrossRef]
- Arijs, I.; Quintens, R.; Van Lommel, L.; Van Steen, K.; De Hertogh, G.; Lemaire, K.; Schraenen, A.; Perrier, C.; Van Assche, G.; Vermeire, S.; et al. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm. Bowel Dis. 2010, 16, 2090–2098. [Google Scholar] [CrossRef]
- Julià, A.; Erra, A.; Palacio, C.; Tomas, C.; Sans, X.; Barceló, P.; Marsal, S. An eight-gene blood expression profile predicts the response to infliximab in rheumatoid arthritis. PLoS ONE 2009, 4, e7556. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Suzuki, K.; Iijima, H.; Hata, Y.; Lim, C.R.; Ishizawa, Y.; Kameda, H.; Amano, K.; Matsubara, K.; Matoba, R.; et al. Identification of baseline gene expression signatures predicting therapeutic responses to three biologic agents in rheumatoid arthritis: A retrospective observational study. Arthritis Res. Ther. 2016, 18, 159. [Google Scholar] [CrossRef] [PubMed]
- Toedter, G.; Li, K.; Marano, C.; Ma, K.; Sague, S.; Huang, C.C.; Song, X.-Y.; Rutgeerts, P.; Baribaud, F. Gene Expression Profiling and Response Signatures Associated With Differential Responses to Infliximab Treatment in Ulcerative Colitis. Am. J. Gastroenterol. 2011, 106, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, J.; Dabrowska, M.; Lazowska, I.; Paziewska, A.; Balabas, A.; Kluska, A.; Kulecka, M.; Karczmarski, J.; Ambrozkiewicz, F.; Piatkowska, M.; et al. Redefining the Practical Utility of Blood Transcriptome Biomarkers in Inflammatory Bowel Diseases. J. Crohn’s Colitis 2019, 13, 626–633. [Google Scholar] [CrossRef] [PubMed]
- Verstockt, B.; Verstockt, S.; Dehairs, J.; Ballet, V.; Blevi, H.; Wollants, W.-J.; Breynaert, C.; Van Assche, G.; Vermeire, S.; Ferrante, M. Low TREM1 expression in whole blood predicts anti-TNF response in inflammatory bowel disease. EBioMedicine 2019, 40, 733–742. [Google Scholar] [CrossRef]
- Toonen, E.J.M.; Gilissen, C.; Franke, B.; Kievit, W.; Eijsbouts, A.M.; den Broeder, A.A.; van Reijmersdal, S.V.; Veltman, J.A.; Scheffer, H.; Radstake, T.R.D.J.; et al. Validation Study of Existing Gene Expression Signatures for Anti-TNF Treatment in Patients with Rheumatoid Arthritis. PLoS ONE 2012, 7, e33199. [Google Scholar] [CrossRef]
- Abarca-Zabalia, J.; Garcia, M.I.; Lozano Ros, A.; Marin-Jimenez, I.; Martinez-Gines, M.L.; Lopez-Cauce, B.; Martin-Barbero, M.L.; Salvador-Martin, S.; Sanjurjo-Saez, M.; Garcia-Dominguez, J.M.; et al. Differential Expression of SMAD Genes and S1PR1 on Circulating CD4+ T Cells in Multiple Sclerosis and Crohn’s Disease. Int. J. Mol. Sci. 2020, 21, 676. [Google Scholar] [CrossRef]
- Dobre, M.; Manuc, T.E.; Milanesi, E.; Plesea, I.E.; Tieranu, E.N.; Popa, C.; Manuc, M.; Preda, C.M.; Tieranu, I.; Diculescu, M.M.; et al. Mucosal CCR1 gene expression as a marker of molecular activity in Crohn’s disease: Preliminary data. Rom. J. Morphol. Embryol. Rev. Roum. Morphol. Embryol. 2017, 58, 1263–1268. [Google Scholar]
- Nguyen, P.M.; Putoczki, T.L.; Ernst, M. STAT3-Activating Cytokines: A Therapeutic Opportunity for Inflammatory Bowel Disease? J. Interferon Cytokine Res. 2015, 35, 340–350. [Google Scholar] [CrossRef]
- Salvador-Martín, S.; López-Cauce, B.; Nuñez, O.; Laserna-Mendieta, E.J.; García, M.I.; Lobato, E.; Abarca-Zabalía, J.; Sanjurjo-Saez, M.; Lucendo, A.J.; Marín-Jiménez, I.; et al. Genetic predictors of long-term response and trough levels of infliximab in crohn’s disease. Pharmacol. Res. 2019, 149, 104478. [Google Scholar] [CrossRef]
- Arkteg, C.B.; Goll, R.; Gundersen, M.D.; Anderssen, E.; Fenton, C.; Florholmen, J. Mucosal gene transcription of ulcerative colitis in endoscopic remission. Scand. J. Gastroenterol. 2020, 55, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Gazouli, M.; Pachoula, I.; Panayotou, I.; Mantzaris, G.; Chrousos, G.; Anagnou, N.P.; Roma-Giannikou, E. NOD2/CARD15, ATG16L1 and IL23R gene polymorphisms and childhood-onset of Crohn’s disease. World J. Gastroenterol. 2010, 16, 1753–1758. [Google Scholar] [CrossRef] [PubMed]
- Moran, C.J.; Walters, T.D.; Guo, C.-H.; Kugathasan, S.; Klein, C.; Turner, D.; Wolters, V.M.; Bandsma, R.H.; Mouzaki, M.; Zachos, M.; et al. IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm. Bowel Dis. 2013, 19, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Vaiopoulou, A.; Gazouli, M.; Papadopoulou, A.; Anagnostopoulos, A.K.; Karamanolis, G.; Theodoropoulos, G.E.; M’Koma, A.; Tsangaris, G.T. Serum protein profiling of adults and children with Crohn disease. J. Pediatr. Gastroenterol. Nutr. 2015, 60, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Dubinsky, M.C.; Mei, L.; Friedman, M.; Dhere, T.; Haritunians, T.; Hakonarson, H.; Kim, C.; Glessner, J.; Targan, S.R.; McGovern, D.P.; et al. Genome wide association (GWA) predictors of anti-TNFalpha therapeutic responsiveness in pediatric inflammatory bowel disease. Inflamm. Bowel Dis. 2010, 16, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, J.; Wijbrandts, C.A.; van Baarsen, L.G.; Nader, G.; Klareskog, L.; Catrina, A.; Thurlings, R.; Vervoordeldonk, M.; Lundeberg, J.; Tak, P.P. The gene expression profile in the synovium as a predictor of the clinical response to infliximab treatment in rheumatoid arthritis. PLoS ONE 2010, 5, e11310. [Google Scholar] [CrossRef]
- Leal, R.F.; Planell, N.; Kajekar, R.; Lozano, J.J.; Ordás, I.; Dotti, I.; Esteller, M.; Masamunt, M.C.; Parmar, H.; Ricart, E.; et al. Identification of inflammatory mediators in patients with Crohn’s disease unresponsive to anti-TNFα therapy. Gut 2015, 64, 233–242. [Google Scholar] [CrossRef]
- Budi, E.H.; Duan, D.; Derynck, R. Transforming Growth Factor-β Receptors and Smads: Regulatory Complexity and Functional Versatility. Trends Cell Biol. 2017, 27, 658–672. [Google Scholar] [CrossRef]
- Akhurst, R.J.; Hata, A. Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 2012, 11, 790–811. [Google Scholar] [CrossRef]
- Liu, X.; Feng, A.; He, Y.; Li, Y.; Wu, Y.; Lian, X.; Hu, F.; Li, J.; Tu, Y.; Chen, S. Association of down-regulation of CD109 expression with up-expression of Smad7 in pathogenesis of psoriasis. J. Huazhong Univ. Sci. Technol. Med. Sci. 2016, 36, 132–136. [Google Scholar] [CrossRef]
- Monteleone, G.; Del Vecchio Blanco, G.; Monteleone, I.; Fina, D.; Caruso, R.; Gioia, V.; Ballerini, S.; Federici, G.; Bernardini, S.; Pallone, F.; et al. Post-transcriptional Regulation of Smad7 in the Gut of Patients With Inflammatory Bowel Disease. Gastroenterology 2005, 129, 1420–1429. [Google Scholar] [CrossRef] [PubMed]
- Cejas, P.J.; Walsh, M.C.; Pearce, E.L.; Han, D.; Harms, G.M.; Artis, D.; Turka, L.A.; Choi, Y. TRAF6 inhibits Th17 differentiation and TGF-beta-mediated suppression of IL-2. Blood 2010, 115, 4750–4757. [Google Scholar] [CrossRef] [PubMed]
- Achiron, A.; Gurevich, M.; Friedman, N.; Kaminski, N.; Mandel, M. Blood transcriptional signatures of multiple sclerosis: Unique gene expression of disease activity. Ann. Neurol. 2004, 55, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Bomprezzi, R.; Ringnér, M.; Kim, S.; Bittner, M.L.; Khan, J.; Chen, Y.; Elkahloun, A.; Yu, A.; Bielekova, B.; Meltzer, P.S.; et al. Gene expression profile in multiple sclerosis patients and healthy controls: Identifying pathways relevant to disease. Hum. Mol. Genet. 2003, 12, 2191–2199. [Google Scholar] [CrossRef]
- Lopez Fernandez, L.; Lozano Ros, A.; Marin Jimenez, I.; Garcia Garcia, M.I.; Martinez Gines, M.L.; Goicochea Briceno, H.; Lopez Cauce, B.; Romero Delgado, F.; Garcia Dominguez, J.M. Differential gene expression of SMAD family members and S1PR1 in circulating CD4+T cells in multiple sclerosis. Mult. Scler. J. 2017, 23, 777–778. [Google Scholar]
- Ardizzone, S.; Bevivino, G.; Monteleone, G. Mongersen, an oral Smad7 antisense oligonucleotide, in patients with active Crohn’s disease. Therap. Adv. Gastroenterol. 2016, 9, 527–532. [Google Scholar] [CrossRef]
- Wehkamp, J.; Stange, E.F. Paneth’s disease. J. Crohns. Colitis 2010, 4, 523–531. [Google Scholar] [CrossRef]
- Hu, X.; Deng, J.; Yu, T.; Chen, S.; Ge, Y.; Zhou, Z.; Guo, Y.; Ying, H.; Zhai, Q.; Chen, Y.; et al. ATF4 Deficiency Promotes Intestinal Inflammation in Mice by Reducing Uptake of Glutamine and Expression of Antimicrobial Peptides. Gastroenterology 2019, 156, 1098–1111. [Google Scholar] [CrossRef]
- Takeuchi, T.; Miyasaka, N.; Tatsuki, Y.; Yano, T.; Yoshinari, T.; Abe, T.; Koike, T. Baseline tumour necrosis factor alpha levels predict the necessity for dose escalation of infliximab therapy in patients with rheumatoid arthritis. Ann. Rheum. Dis. 2011, 70, 1208–1215. [Google Scholar] [CrossRef]
- Jessen, B.; Rodriguez-Sillke, Y.; Sonnenberg, E.; Schumann, M.; Kruglov, A.; Freise, I.; Schmidt, F.; Maul, J.; Kuhl, A.A.; Glauben, R.; et al. Level of Tumor Necrosis Factor Production by Stimulated Blood Mononuclear Cells Can be Used to Predict Response of Patients With Inflammatory Bowel Diseases to Infliximab. Clin. Gastroenterol. Hepatol. 2020. [Google Scholar] [CrossRef]
- Glas, A.S.; Lijmer, J.G.; Prins, M.H.; Bonsel, G.J.; Bossuyt, P.M.M. The diagnostic odds ratio: A single indicator of test performance. J. Clin. Epidemiol. 2003, 56, 1129–1135. [Google Scholar] [CrossRef]

| Characteristic | Overall (n = 33) | Responders (n = 27) | Non-Responders (n = 6) | p Value |
|---|---|---|---|---|
| Gender | ||||
| Male, n (%) | 16 (48.5%) | 12 (44.4%) | 4 (66.7%) | 0.398 |
| Female, n (%) | 17 (51.5%) | 15 (55.6%) | 2 (33.3%) | |
| Age (years) | ||||
| At diagnosis, median (IQR, range) | 10.4 (4.6; 0.7–17) | 10.5 (4.7; 6.5–17) | 10.1 (7.2; 0.7–13.0) | 0.276 |
| At start of treatment, median (IQR, range) | 12 (4; 1.1–17) | 12(4.8; 7.9–17) | 11.4 (6.2; 1.1–14.1) | 0.342 |
| Type of IBD | ||||
| CD, n (%) | 26 (78.8%) | 21 (77.8%) | 5 (83.3%) | 1 |
| UC, n (%) | 7 (21.1%) | 6 (22.2%) | 1 (16.2%) | |
| Type of Anti-TNF | ||||
| Infliximab, n (%) | 18 (54.5%) | 14 (51.9%) | 4 (66.7%) | 0.665 |
| Adalimumab, n (%) | 15 (45.5%) | 13 (48.1%) | 2 (33.3%) | |
| PCDAI at start of treatment, median (IQR, range) | 28.8 (25.6; 5–60) | 30 (28.8; 5–60) | 15 (12.5; 7.5–30) | 0.013 ** |
| PUCAI at start of treatment, median (IQR, range) | 45 (40; 5–60) | 50 (43.8; 5–60) | 45 * | - |
| C-reactive protein at start of treatment, median (IQR, range) | 22 (31.6; 0–150.3) | 22.6 (41.3; 0–105.3) | 10.6 (19.1; 6.1–27.5) | 0.054 |
| Other treatments | ||||
| Enteral nutrition | 14 (42.4%) | 12 (44.4%) | 2 (33.3%) | 0.682 |
| Corticosteroids | 12 (36.4%) | 10 (37%) | 2 (33.3%) | 1 |
| Azathioprine | 20 (60.6%) | 16 (59.3%) | 4 (66.7%) | 1 |
| Aminosalicylates | 12 (36.4%) | 11 (40.7%) | 2 (33.3%) | 0.379 |
| Methotrexate | 3 (9.1%) | 2 (7.4%) | 1 (16.7%) | 1 |
| Tacrolimus | 2 (6.1%) | 1 (3.7%) | 1 (16.7%) | 0.335 |
| Adalimumab | 1 (3%) | 0 | 1 (16.7%) | 0.182 |
| Infliximab | 1 (3%) | 0 | 1 (16.7%) | 0.182 |
| Gene | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| SMAD7 | ACCCGATGGATTTTCTCAA | AGGGGCCAGATAATTCGTTC |
| TNF | AGCCCATGTTGTAGCAAACC | TCTCAGTCTCACGCCATT |
| TLR2 | TGTCATTCTTTCTTCCTGCTAAGA | CTAGGTAGGACAGAGAATGCCTTT |
| TNFRSF1B | AGGCCACCATATTCAGTGCT | GCAGATTTCTAGTTAGAAGTGCGTTA |
| TBX21 | CCCAACTGTCAATTCCTTGG | GGGAACAGGATACTGGTTGG |
| DEFA5 | TCAGCTCTTTCCTGGAGTGAC | AGGACCATCGCCATCCTT |
| IL11 | GGACAGGGAAGGGTTAAAGG | GCTCAGCACGACCAGGAC |
| TREM1 | GATGCTCTTTGTCTCAGAAC | CTCTCCGTCCCTTATTATCTG |
| OSM | GTACTGCTCACACAGAGG | TATATAGGGGTCCAGGAGTC |
| GAPDH | AGCCACATCGCTCAGACAC | GCCCAATACGACCAAATCC |
| HPRT1 | GACCAGTCAACAGGGGACAT | GTGTCAATTATATCTTCCACAATCAAG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvador-Martín, S.; Raposo-Gutiérrez, I.; Navas-López, V.M.; Gallego-Fernández, C.; Moreno-Álvarez, A.; Solar-Boga, A.; Muñoz-Codoceo, R.; Magallares, L.; Martínez-Ojinaga, E.; Fobelo, M.J.; et al. Gene Signatures of Early Response to Anti-TNF Drugs in Pediatric Inflammatory Bowel Disease. Int. J. Mol. Sci. 2020, 21, 3364. https://doi.org/10.3390/ijms21093364
Salvador-Martín S, Raposo-Gutiérrez I, Navas-López VM, Gallego-Fernández C, Moreno-Álvarez A, Solar-Boga A, Muñoz-Codoceo R, Magallares L, Martínez-Ojinaga E, Fobelo MJ, et al. Gene Signatures of Early Response to Anti-TNF Drugs in Pediatric Inflammatory Bowel Disease. International Journal of Molecular Sciences. 2020; 21(9):3364. https://doi.org/10.3390/ijms21093364
Chicago/Turabian StyleSalvador-Martín, Sara, Irene Raposo-Gutiérrez, Víctor Manuel Navas-López, Carmen Gallego-Fernández, Ana Moreno-Álvarez, Alfonso Solar-Boga, Rosana Muñoz-Codoceo, Lorena Magallares, Eva Martínez-Ojinaga, María J. Fobelo, and et al. 2020. "Gene Signatures of Early Response to Anti-TNF Drugs in Pediatric Inflammatory Bowel Disease" International Journal of Molecular Sciences 21, no. 9: 3364. https://doi.org/10.3390/ijms21093364
APA StyleSalvador-Martín, S., Raposo-Gutiérrez, I., Navas-López, V. M., Gallego-Fernández, C., Moreno-Álvarez, A., Solar-Boga, A., Muñoz-Codoceo, R., Magallares, L., Martínez-Ojinaga, E., Fobelo, M. J., Millán-Jiménez, A., Rodriguez-Martinez, A., Vayo, C. A., Sánchez, C., Tolin, M., Bossacoma, F., Pujol-Muncunill, G., González de Caldas, R., Loverdos, I., ... López-Fernández, L. A. (2020). Gene Signatures of Early Response to Anti-TNF Drugs in Pediatric Inflammatory Bowel Disease. International Journal of Molecular Sciences, 21(9), 3364. https://doi.org/10.3390/ijms21093364







