Hypermethylation of the TSPOAP1-AS1 Promoter May Be Associated with Obesity in Overweight/Obese Korean Subjects
Abstract
1. Introduction
2. Results
2.1. Characteristics of Subjects
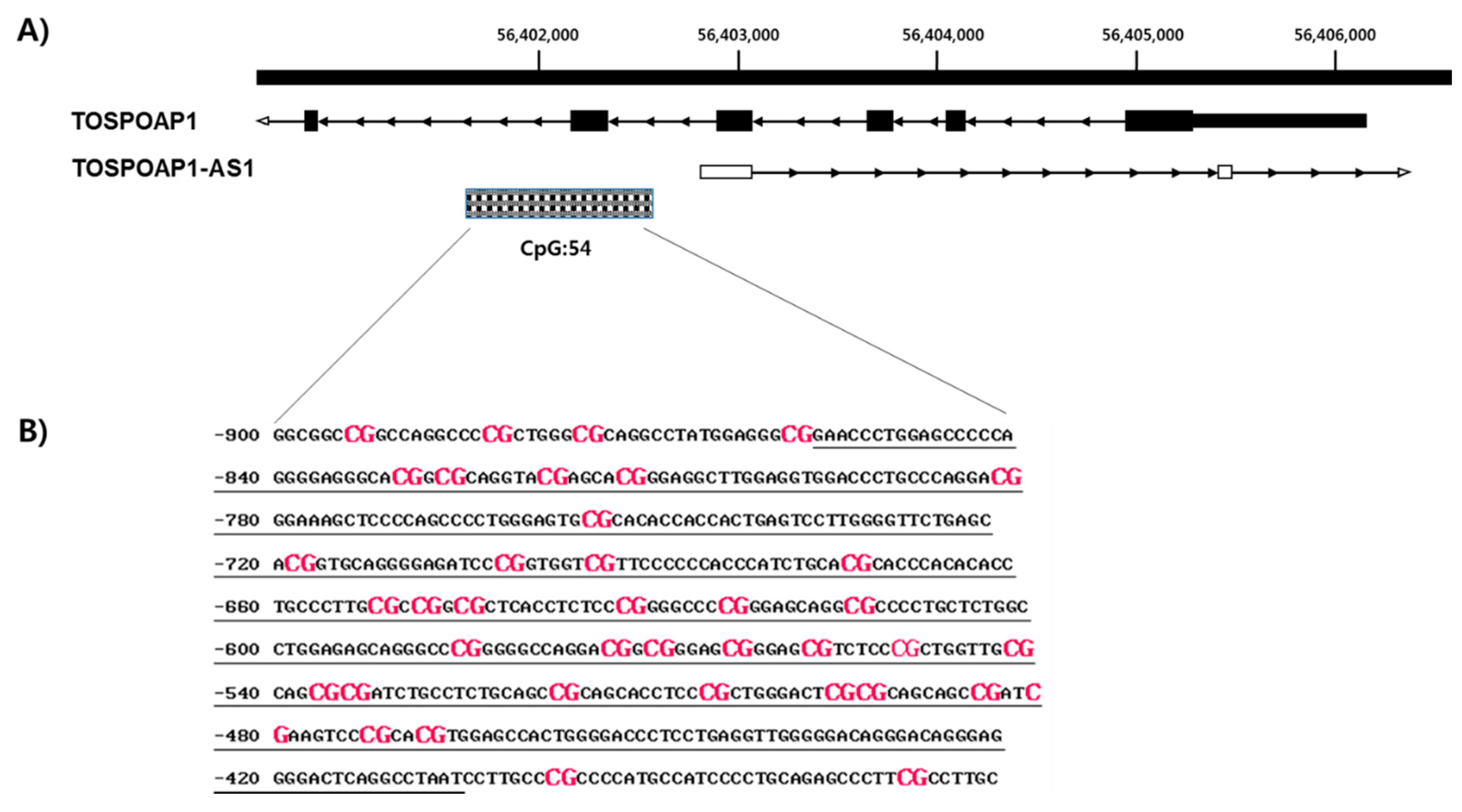
2.2. GpG Island in TSPOAP1-AS1 Promoter Region
2.3. Comparison of TSPOAP1-AS1 Methylation between Normal and Overweight/Obese Subjects
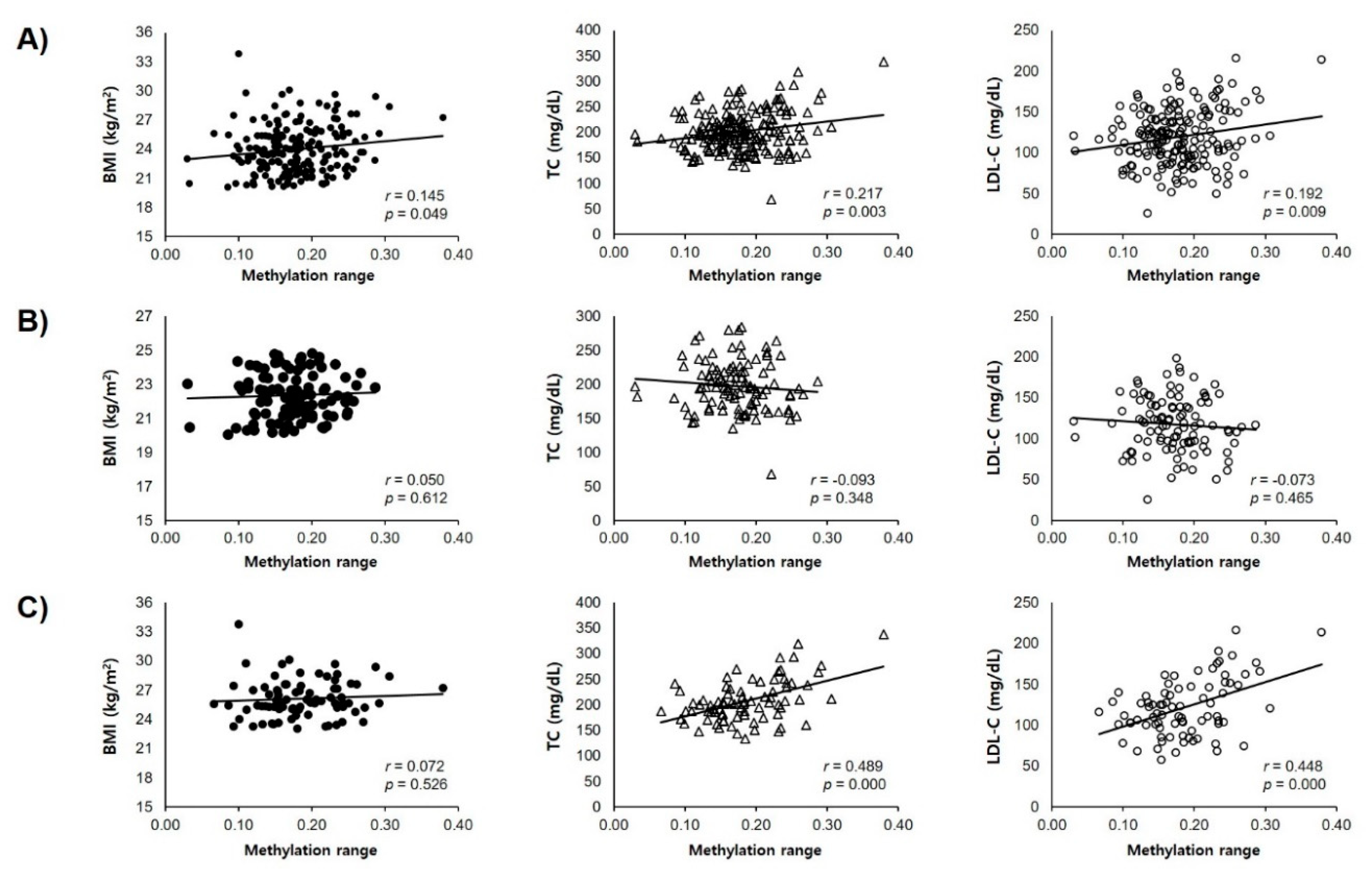
2.4. Comparison of TSPOAP1-AS1 Methylation Associated with Obesity-Related Phenotypes between Normal and Overweight/Obese Subjects
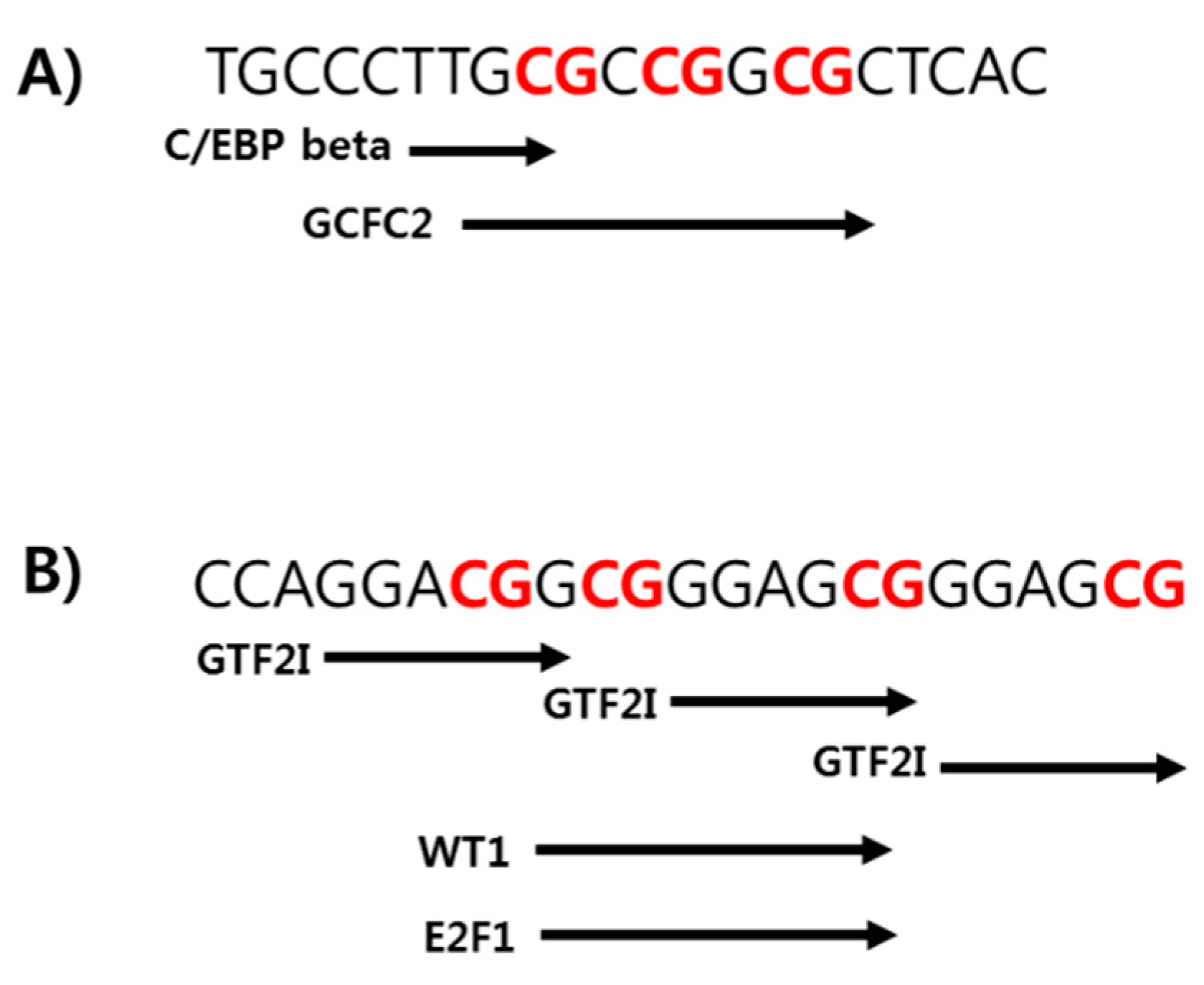
2.5. Prediction of the Binding Affinity with Transcription Factors at the Specific CpG Sites in TSPOAP1-AS1 Promoter Region
3. Discussion
4. Methods
4.1. Study Participants
4.2. DNA Methylation Analysis
4.3. Prediction of Transcription Factor Binding
4.4. Statistical Analysis
4.5. Availability of Data and Materials
4.6. Ethics Approval and Consent to Participate
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Dietz, W.H. The response of the US Centers for Disease Control and Prevention to the obesity epidemic. Annu. Rev. Public Health 2015, 36, 575–596. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, A.; Snehalatha, C. Rising burden of obesity in Asia. J. Obes. 2010, 2010, 868573. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, S.J.; Molloy, P.L.; Varinli, H.; Morrison, J.L.; Muhlhausler, B.S.; Members of Epi, S. Epigenetics and human obesity. Int. J. Obes. 2015, 39, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.R.; Lal, N.; Giridharan, N.V. Genetic & epigenetic approach to human obesity. Indian J. Med. Res. 2014, 140, 589–603. [Google Scholar]
- Soubry, A.; Guo, L.; Huang, Z.; Hoyo, C.; Romanus, S.; Price, T.; Murphy, S.K. Obesity-related DNA methylation at imprinted genes in human sperm: Results from the TIEGER study. Clin. Epigenet. 2016, 8, 51. [Google Scholar] [CrossRef]
- Guay, S.P.; Brisson, D.; Lamarche, B.; Biron, S.; Lescelleur, O.; Biertho, L.; Marceau, S.; Vohl, M.C.; Gaudet, D.; Bouchard, L. ADRB3 gene promoter DNA methylation in blood and visceral adipose tissue is associated with metabolic disturbances in men. Epigenomics 2014, 6, 33–43. [Google Scholar] [CrossRef]
- Garcia-Cardona, M.C.; Huang, F.; Garcia-Vivas, J.M.; Lopez-Camarillo, C.; Del Rio Navarro, B.E.; Navarro Olivos, E.; Hong-Chong, E.; Bolanos-Jimenez, F.; Marchat, L.A. DNA methylation of leptin and adiponectin promoters in children is reduced by the combined presence of obesity and insulin resistance. Int. J. Obes. 2014, 38, 1457–1465. [Google Scholar] [CrossRef]
- Brunet, A.; Berger, S.L. Epigenetics of aging and aging-related disease. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69 (Suppl. 1), S17–S20. [Google Scholar] [CrossRef]
- Mittelstaedt, T.; Schoch, S. Structure and evolution of RIM-BP genes: Identification of a novel family member. Gene 2007, 403, 70–79. [Google Scholar] [CrossRef]
- Witoelar, A.; Rongve, A.; Almdahl, I.S.; Ulstein, I.D.; Engvig, A.; White, L.R.; Selbaek, G.; Stordal, E.; Andersen, F.; Braekhus, A.; et al. Meta-analysis of Alzheimer’s disease on 9751 samples from Norway and IGAP study identifies four risk loci. Sci. Rep. 2018, 8, 18088. [Google Scholar] [CrossRef]
- Jun, G.R.; Chung, J.; Mez, J.; Barber, R.; Beecham, G.W.; Bennett, D.A.; Buxbaum, J.D.; Byrd, G.S.; Carrasquillo, M.M.; Crane, P.K.; et al. Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement. 2017, 13, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, W.; Xia, P.; Wan, L.; Zhang, L.; Yu, L.; Wang, L.; Chen, X.; Xiao, Y.; Xu, C. Identification of a five-lncRNA signature for predicting the risk of tumor recurrence in patients with breast cancer. Int. J. Cancer 2018, 143, 2150–2160. [Google Scholar] [CrossRef] [PubMed]
- Blasi, C. Influence of benzodiazepines on body weight and food intake in obese and lean Zucker rats. Prog. Neuropsychopharmacol. Biol. Psychiatry 2000, 24, 561–577. [Google Scholar] [CrossRef]
- Thompson, M.M.; Manning, H.C.; Ellacott, K.L.J. Translocator Protein 18 kDa (TSPO) Is Regulated in White and Brown Adipose Tissue by Obesity. PLoS ONE 2013, 8, e79980. [Google Scholar] [CrossRef] [PubMed]
- Galiegue, S.; Jbilo, O.; Combes, T.; Bribes, E.; Carayon, P.; Le Fur, G.; Casellas, P. Cloning and characterization of PRAX-1. A new protein that specifically interacts with the peripheral benzodiazepine receptor. J. Biol. Chem. 1999, 274, 2938–2952. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Zhu, H.D.; Snieder, H.; Su, S.Y.; Munn, D.; Harshfield, G.; Maria, B.L.; Dong, Y.B.; Treiber, F.; Gutin, B.; et al. Obesity related methylation changes in DNA of peripheral blood leukocytes. BMC Med. 2010, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.J.; Su, S.Y.; Barnes, V.A.; De Miguel, C.; Pollock, J.; Ownby, D.; Shi, H.D.; Zhu, H.D.; Snieder, H.; Wang, X.L. A genome-wide methylation study on obesity Differential variability and differential methylation. Epigenetics 2013, 8, 522–533. [Google Scholar] [CrossRef]
- Demerath, E.W.; Guan, W.H.; Grove, M.L.; Aslibekyan, S.; Mendelson, M.; Zhou, Y.H.; Hedman, A.K.; Sandling, J.K.; Li, L.A.; Irvin, M.R.; et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum. Mol. Genet. 2015, 24, 4464–4479. [Google Scholar] [CrossRef]
- Wahl, S.; Drong, A.; Lehne, B.; Loh, M.; Scott, W.R.; Kunze, S.; Tsai, P.C.; Ried, J.S.; Zhang, W.H.; Yang, Y.W.; et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 2017, 541, 81–86. [Google Scholar] [CrossRef]
- Dick, K.J.; Nelson, C.P.; Tsaprouni, L.; Sandling, J.K.; Aissi, D.; Wahl, S.; Meduri, E.; Morange, P.E.; Gagnon, F.; Grallert, H.; et al. DNA methylation and body-mass index: A genome-wide analysis. Lancet 2014, 383, 1990–1998. [Google Scholar] [CrossRef]
- Shah, S.; Bonder, M.J.; Marioni, R.E.; Zhu, Z.H.; McRae, A.F.; Zhernakova, A.; Harris, S.E.; Liewald, D.; Henders, A.K.; Mendelson, M.M.; et al. Improving Phenotypic Prediction by Combining Genetic and Epigenetic Associations. Am. J. Hum. Genet. 2015, 97, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Sayols-Baixeras, S.; Subirana, I.; Fernandez-Sanles, A.; Senti, M.; Lluis-Ganella, C.; Marrugat, J.; Elosua, R. DNA methylation and obesity traits: An epigenome-wide association study. The REGICOR study. Epigenetics 2017, 12, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Mendelson, M.M.; Marioni, R.E.; Joehanes, R.; Liu, C.Y.; Hedman, A.K.; Aslibekyan, S.; Demerath, E.W.; Guan, W.H.; Zhi, D.H.; Yao, C.; et al. Association of Body Mass Index with DNA Methylation and Gene Expression in Blood Cells and Relations to Cardiometabolic Disease: A Mendelian Randomization Approach. PLoS Med. 2017, 14, e1002215. [Google Scholar] [CrossRef] [PubMed]
- Richmond, R.C.; Sharp, G.C.; Ward, M.E.; Fraser, A.; Lyttleton, O.; McArdle, W.L.; Ring, S.M.; Gaunt, T.R.; Lawlor, D.A.; Smith, G.D.; et al. DNA Methylation and BMI: Investigating Identified Methylation Sites at HIF3A in a Causal Framework. Diabetes 2016, 65, 1231–1244. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, V.; Stocco, D.M. The changing landscape in translocator protein (TSPO) function. Trends Endocrin Met. 2015, 26, 341–348. [Google Scholar] [CrossRef]
- Biswas, L.; Farhan, F.; Reilly, J.; Bartholomew, C.; Shu, X. TSPO Ligands Promote Cholesterol Efflux and Suppress Oxidative Stress and Inflammation in Choroidal Endothelial Cells. Int. J. Mol. Sci. 2018, 19, 3740. [Google Scholar] [CrossRef]
- Giulietti, M.; Righetti, A.; Principato, G.; Piva, F. LncRNA co-expression network analysis reveals novel biomarkers for pancreatic cancer. Carcinogenesis 2018, 39, 1016–1025. [Google Scholar] [CrossRef]
- Tan, J.F.; Jin, X.F.; Wang, K.C. Integrated Bioinformatics Analysis of Potential Biomarkers for Prostate Cancer. Pathol. Oncol. Res. 2019, 25, 455–460. [Google Scholar] [CrossRef]
- Tang, Q.Q.; Lane, M.D. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 1999, 13, 2231–2241. [Google Scholar] [CrossRef]
- Tang, Q.Q.; Lane, M.D. Adipogenesis: From stem cell to adipocyte. Annu. Rev. Biochem. 2012, 81, 715–736. [Google Scholar] [CrossRef]
- Guo, L.; Li, X.; Tang, Q.Q. Transcriptional regulation of adipocyte differentiation: A central role for CCAAT/enhancer-binding protein (C/EBP) beta. J. Biol. Chem. 2015, 290, 755–761. [Google Scholar] [CrossRef]
- Hastie, N.D. Wilms’ tumour 1 (WT1) in development, homeostasis and disease. Development 2017, 144, 2862–2872. [Google Scholar] [CrossRef] [PubMed]
- Ndisang, J.F.; Tiwari, S. Mechanisms by which heme oxygenase rescue renal dysfunction in obesity. Redox Biol. 2014, 2, 1029–1037. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, K.Y.; Luong, Q.; Sharma, R.; Dreyfuss, J.M.; Ussar, S.; Kahn, C.R. Developmental and functional heterogeneity of white adipocytes within a single fat depot. EMBO J. 2019, 38, e99291. [Google Scholar] [CrossRef]
- Milagro, F.I.; Campion, J.; Cordero, P.; Goyenechea, E.; Gomez-Uriz, A.M.; Abete, I.; Zulet, M.A.; Martinez, J.A. A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J. 2011, 25, 1378–1389. [Google Scholar] [CrossRef]
- Abete, I.; Gomez-Uriz, A.M.; Mansego, M.L.; De Arce, A.; Goyenechea, E.; Blazquez, V.; Martinez-Zabaleta, M.T.; Gonzalez-Muniesa, P.; De Munain, A.L.; Martinez, J.A.; et al. Epigenetic Changes in the Methylation Patterns of KCNQ1 and WT1 after a Weight Loss Intervention Program in Obese Stroke Patients. Curr. Neurovasc. Res. 2015, 12, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.K.; Moon, T.W.; Lee, J.A.; Park, T.Y.; Ko, M.M.; Lee, M.S. The fundamental study for the standardisation and objectification of pattern identification in traditional Korean medicine for stroke (SOPI-Stroke): Development and interobserver agreement of the Korean standard pattern identification for stroke (K-SPI-Stroke) tool. Eur. J. Integr. Med. 2012, 4, E133–E139. [Google Scholar]
- WHO Expert Consultation. Appropriate body-mass index in Asian populations and its implications for policy and intervention strategies. Lancet 2004, 363, 157–163. [Google Scholar]
- Jiang, H.S.; Lei, R.; Ding, S.W.; Zhu, S.F. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinform. 2014, 15, 182. [Google Scholar] [CrossRef]
- Messeguer, X.; Escudero, R.; Farre, D.; Nunez, O.; Martinez, J.; Alba, M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef]

| Characteristics | Normal | Obesity | p-Value |
|---|---|---|---|
| n | 104 | 80 | |
| Anthropometric characteristics | |||
| Sex (M/F) | 64/40 * | 31/49 | 0.003 a |
| Age (years) | 59.84 ±10.68 # | 64.04 ±9.87 | 0.007 b |
| Smoking (none/stop/active) | 55/31/18 | 56/16/8 | 0.061 |
| Alcohol consumption (none/former/active) | 50/9/45 | 47/5/27 | 0.307 |
| Weight (kg) | 59.15 ± 7.84 | 65.16 ± 9.22 | <0.001 |
| BMI (kg/m2) | 22.35 ± 1.33 | 26.12 ± 1.96 | <0.001 |
| Waist circumference (cm) | 80.24 ± 6.93 | 89.02 ± 6.06 | <0.001 |
| WHR | 0.869 ± 0.056 | 0.895 ± 0.046 | 0.002 |
| Medical history | |||
| Depression (Yes, %) | 0 (0.0) * | 5 (6.3) | 0.013 |
| Migraine (Yes, %) | 9 (8.7) | 12 (15.2) | 0.241 |
| Blood parameters | |||
| WBC (×103) | 5.72 ± 1.49 | 5.82 ± 1.22 | 0.348 c |
| RBC (×106) | 4.51 ± 0.45 | 4.46 ± 0.40 | 0.306 |
| Hg (g/dL) | 13.90 ± 1.45 | 13.75 ± 1.31 | 0.103 |
| Hct (%) | 41.54 ± 4.15 | 40.86 ± 3.73 | 0.223 |
| Platelet (×103/µL) | 184.77 ± 75.92 | 208.01 ± 78.61 | 0.199 |
| GOT (U/dL) | 23.40 ± 6.96 | 25.84 ± 18.23 | 0.107 |
| GPT (U/dL) | 19.76 ± 8.45 | 25.19 ± 20.53 | 0.003 |
| Total cholesterol (mg/dL) | 197.59 ± 36.84 | 206.92 ± 40.90 | 0.285 |
| Triglyceride (mg/dL) | 125.63 ± 70.26 | 153.57 ± 75.21 | 0.011 |
| HDL-cholesterol (mg/dL) | 56.24 ± 12.80 | 55.29 ± 12.18 | 0.110 |
| LDL-cholesterol (mg/dL) | 117.53 ± 33.41 | 120.92 ± 34.65 | 0.772 |
| FBS (mg/dL) | 97.88 ± 9.76 | 97.95 ± 11.62 | 0.399 |
| CpG No. | Loci at Chromosome | BMI | Total Cholesterol | LDL-Cholesterol | |||
|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | ||
| 11 | 56402159 | 0.138 | 0.062 | 0.205 | 0.005 | 0.188 | 0.011 |
| 12 | 56402162 | 0.183 | 0.013 | 0.244 | 0.001 | 0.197 | 0.008 |
| 13 | 56402165 | 0.186 | 0.012 | 0.226 | 0.002 | 0.193 | 0.009 |
| 18 | 56402238 | 0.180 | 0.015 | 0.266 | <0.001 | 0.232 | 0.002 |
| 19 | 56402241 | 0.185 | 0.012 | 0.217 | 0.003 | 0.212 | 0.004 |
| 20 | 56402247 | 0.214 | 0.004 | 0.227 | 0.002 | 0.195 | 0.008 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yim, N.-H.; Cha, M.H.; Kim, M.S. Hypermethylation of the TSPOAP1-AS1 Promoter May Be Associated with Obesity in Overweight/Obese Korean Subjects. Int. J. Mol. Sci. 2020, 21, 3307. https://doi.org/10.3390/ijms21093307
Yim N-H, Cha MH, Kim MS. Hypermethylation of the TSPOAP1-AS1 Promoter May Be Associated with Obesity in Overweight/Obese Korean Subjects. International Journal of Molecular Sciences. 2020; 21(9):3307. https://doi.org/10.3390/ijms21093307
Chicago/Turabian StyleYim, Nam-Hui, Min Ho Cha, and Myung Sunny Kim. 2020. "Hypermethylation of the TSPOAP1-AS1 Promoter May Be Associated with Obesity in Overweight/Obese Korean Subjects" International Journal of Molecular Sciences 21, no. 9: 3307. https://doi.org/10.3390/ijms21093307
APA StyleYim, N.-H., Cha, M. H., & Kim, M. S. (2020). Hypermethylation of the TSPOAP1-AS1 Promoter May Be Associated with Obesity in Overweight/Obese Korean Subjects. International Journal of Molecular Sciences, 21(9), 3307. https://doi.org/10.3390/ijms21093307




