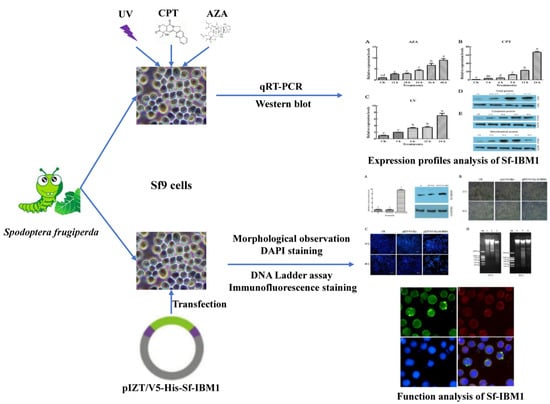
Pro-Apoptotic Function Analysis of the Reaper Homologue IBM1 in Spodoptera frugiperda
Abstract
1. Introduction
2. Results
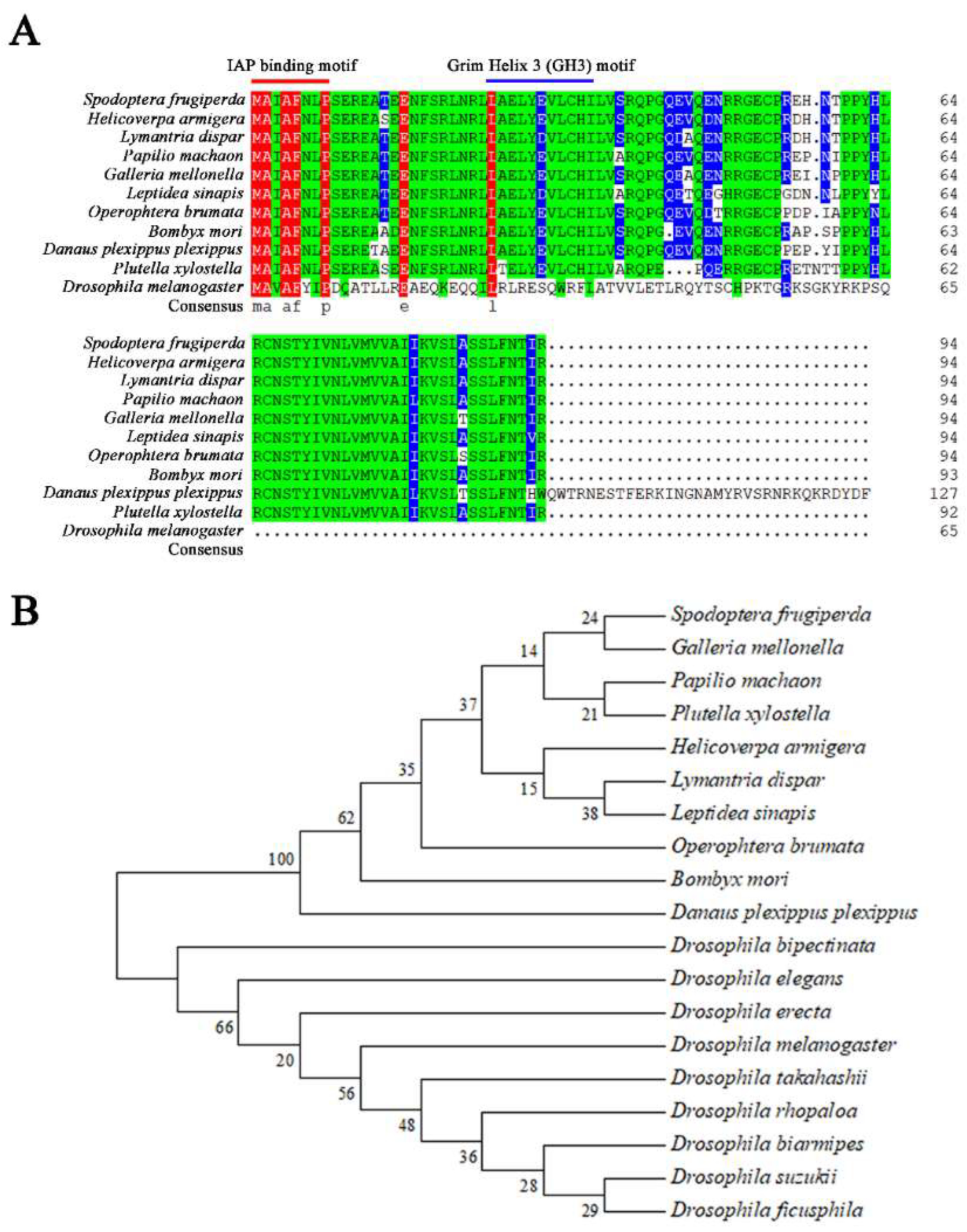
2.1. Cloning and Sequencing Sf-IBM1
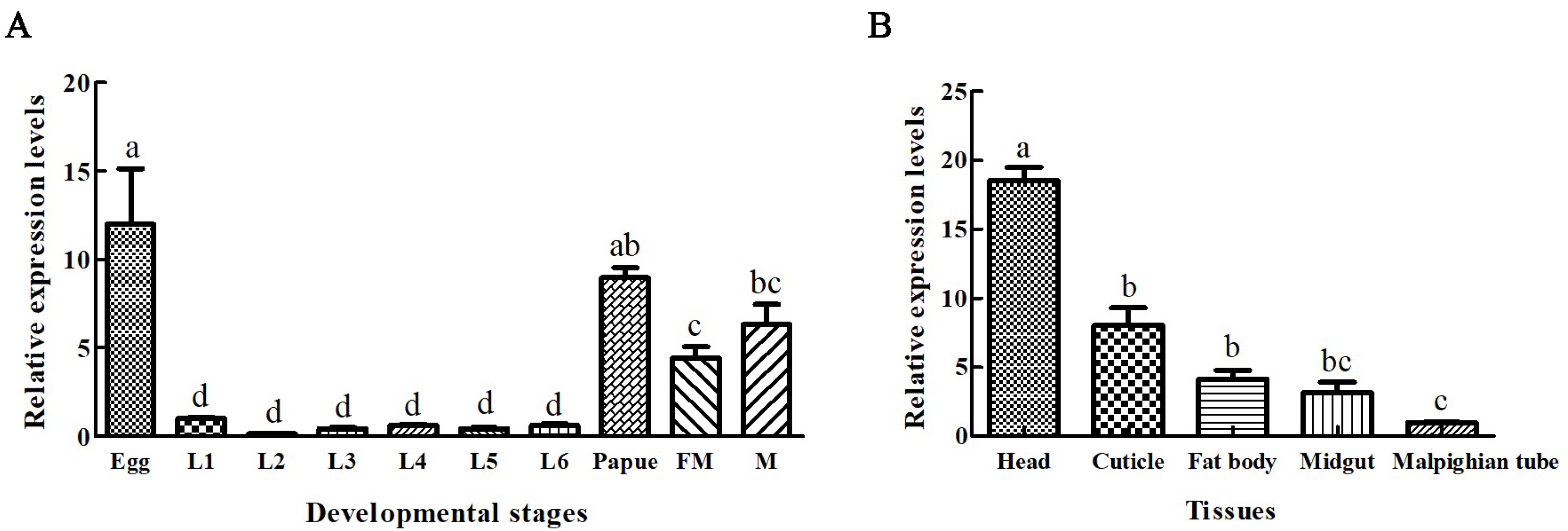
2.2. Expression Patterns of Sf-IBM1 among Different Developmental Stages and Tissues
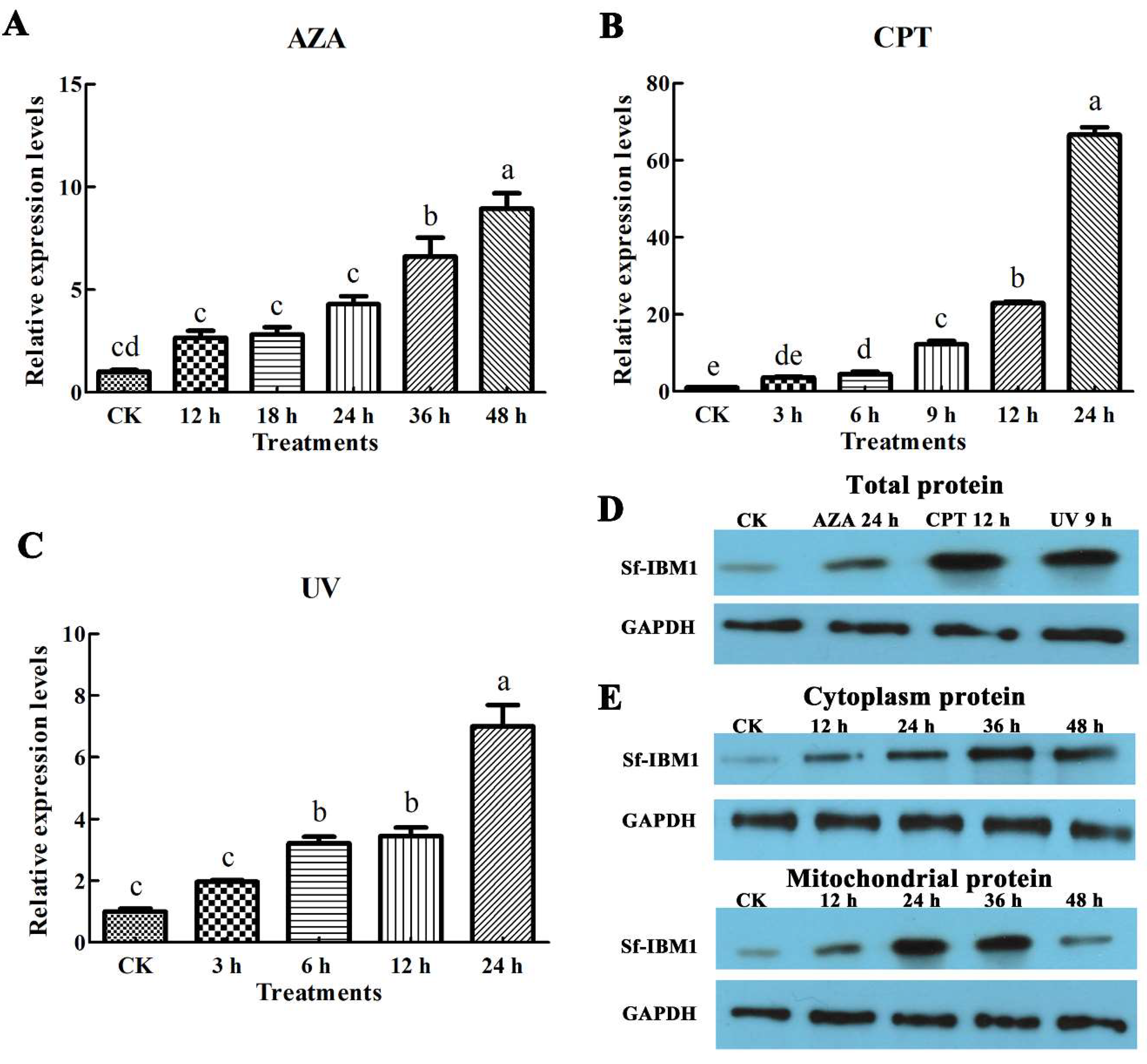
2.3. Apoptotic Stimuli Induced the Up-Regulation of Sf-IBM1
2.4. Transient Expression of Sf-IBM1 Induced Apoptosis in Sf9 Cells
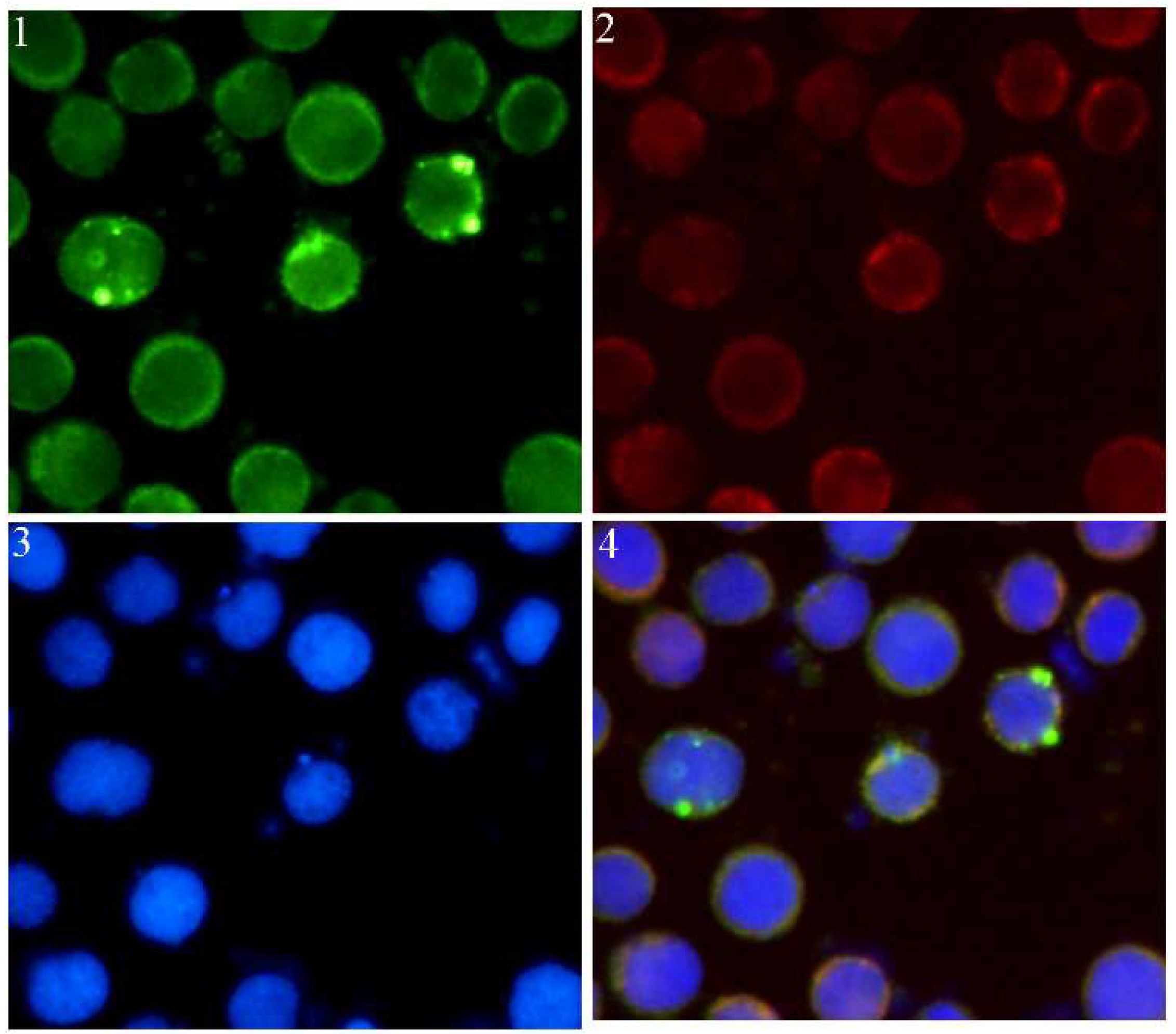
2.5. Sf-IBM1 Localized to Mitochondria
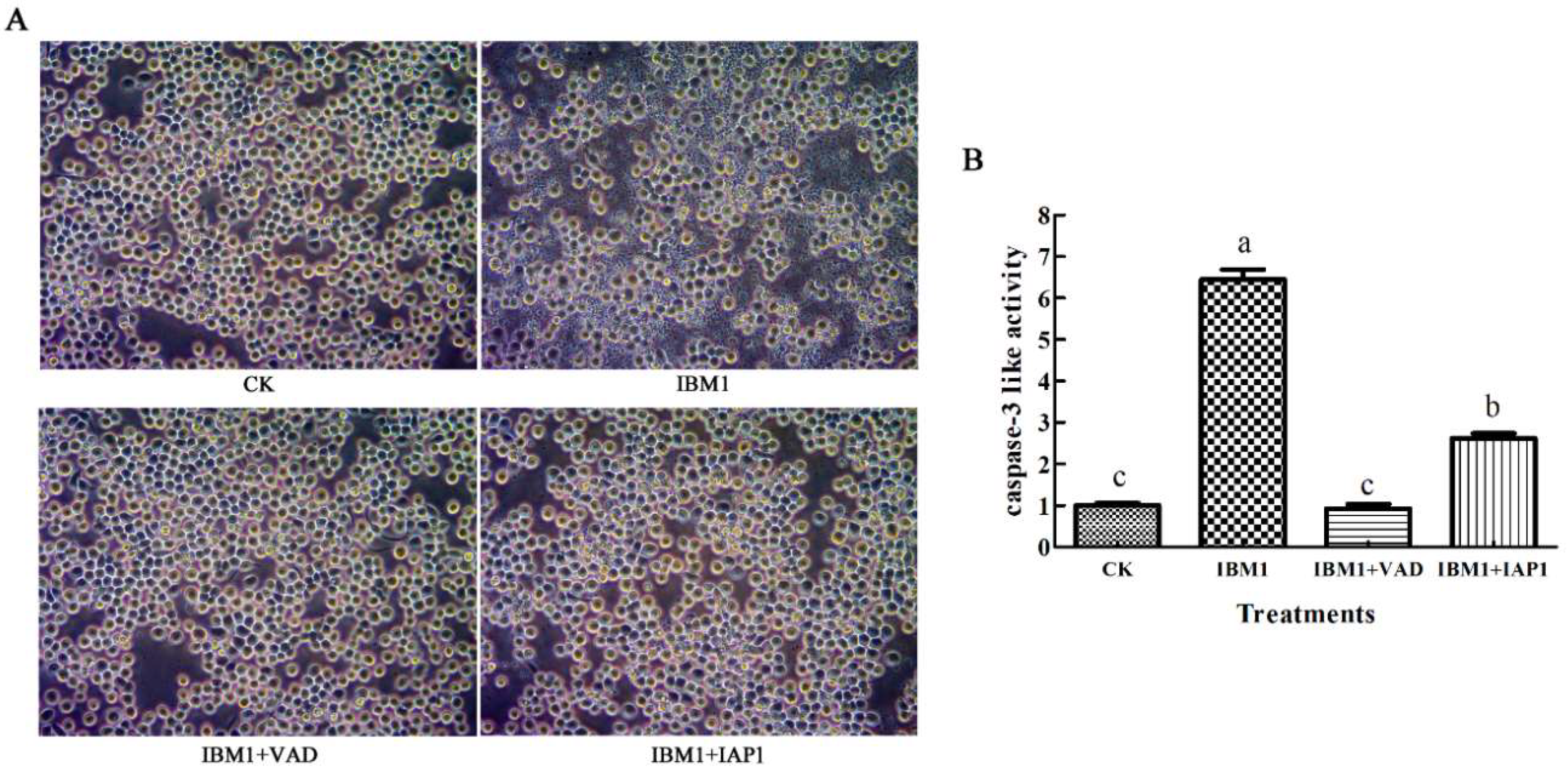
2.6. The Apoptosis Induced by Sf-IBM1 Could Be Blocked by Z-VAD-FMK and Sf-IAP1
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Insect Rearing
4.2. Cloning and Sequencing Sf-IBM1
4.3. qRT-PCR
4.4. Western Blot
4.5. Recombination Plasmid Construction
4.6. Cell Transfection and Morphological Observation
4.7. DAPI Staining
4.8. DNA Ladder Assay
4.9. Caspase-3 Activity Assay
4.10. Immunofluorescence Staining
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Waldron, J.A.; Jones, C.I.; Towler, B.P.; Pashler, A.L.; Grima, D.P.; Hebbes, S.; Crossman, S.H.; Zabolotskaya, M.V.; Newbury, S.F. Xrn1/Pacman affects apoptosis and regulates expression of hid and reaper. Biol. Open 2015, 4, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Nirmala, X.; Schetelig, M.F.; Zimowska, G.J.; Zhou, L.; Handler, A.M. Pro-apoptotic gene regulation and its activation by gamma-irradiation in the Caribbean fruit fly, Anastrepha suspensa. Apoptosis 2015, 20, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hwangbo, D.S.; Biteau, B.; Rath, S.; Kim, J.; Jasper, H. Control of apoptosis by Drosophila DCAF12. Dev. Biol. 2016, 413, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Civciristov, S.; Hawkins, C.J.; Clem, R.J. SfDronc, an initiator caspase involved in apoptosis in the fall armyworm Spodoptera frugiperda. Insect Biochem. Mol. Biol. 2013, 43, 444–454. [Google Scholar] [CrossRef]
- Tajbakhsh, A.; Kovanen, P.T.; Rezaee, M.; Banach, M.; Moallem, S.A.; Sahebkar, A. Regulation of efferocytosis by caspase-dependent apoptotic cell death in atherosclerosis. Int. J. Biochem. Cell Biol. 2020, 120, 105684. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Akey, C.W. Apoptosome structure, assembly, and procaspase activation. Structure 2013, 21, 501–515. [Google Scholar] [CrossRef]
- Kang, Y.; Neuman, S.D.; Bashirullah, A. Tango7 regulates cortical activity of caspases during reaper-triggered changes in tissue elasticity. Nat. Commun. 2017, 8, 603. [Google Scholar] [CrossRef]
- Ito, H.; Bando, H.; Shimada, T.; Katsuma, S. The BIR and BIR-like domains of Bombyx mori nucleopolyhedrovirus IAP2 protein are required for efficient viral propagation. Biochem. Biophys. Res. Commun. 2014, 454, 581–587. [Google Scholar] [CrossRef]
- Hamajima, R.; Iwamoto, A.; Tomizaki, M.; Suganuma, I.; Kitaguchi, K.; Kobayashi, M.; Yamada, H.; Ikeda, M. Functional analysis of inhibitor of apoptosis 1 of the silkworm Bombyx mori. Insect Biochem, Mol. Biol. 2016, 79, 97–107. [Google Scholar] [CrossRef]
- Lee, T.V.; Fan, Y.; Wang, S.; Srivastava, M.; Broemer, M.; Meier, P.; Bergmann, A. Drosophila IAP1-mediated ubiquitylation controls activation of the initiator caspase DRONC independent of protein degradation. PLoS Genet. 2011, 7, e1002261. [Google Scholar] [CrossRef]
- Kamezaki, M.; Yokoi, K.; Miurarna, K. Interference mediated knockdown of an inhibitor of apoptosis protein induces apoptosis in Mythimna separata (Lepidoptera: Noctuidae). Eur. J. Entomol. 2018, 115, 223–231. [Google Scholar] [CrossRef]
- Vasudevan, D.; Ryoo, H.D. Regulation of Cell Death by IAPs and their Antagonists. Curr. Top. Dev. Biol. 2015, 114, 185–208. [Google Scholar] [PubMed]
- Wu, Y.; Wu, Y.; Hui, T.; Wu, H.; Wu, Y.; Wang, W. Reaper homologue IBM1 in silkworm Bombyx mori induces apoptosis upon baculovirus infection. FEBS Lett. 2013, 587, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Wolf, B.B.; Green, D.R. Apoptosis: Letting slip the dogs of war. Curr. Biol. 2002, 12, R177–R179. [Google Scholar] [CrossRef]
- Wang, H.; Clem, R.J. The role of IAP antagonist proteins in the core apoptosis pathway of the mosquito disease vector Aedes aegypti. Apoptosis 2011, 16, 235–248. [Google Scholar] [CrossRef]
- Yoo, S.J.; Huh, J.R.; Muro, I.; Yu, H.; Wang, L.; Wang, S.L.; Feldman, R.M.R.; Clem, R.J.; Muüller, H.A.J.; Hay, B.A. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat. Cell Biol. 2002, 4, 416–424. [Google Scholar] [CrossRef]
- Zachariou, A.; Tenev, T.; Goyal, L.; Agapite, J.; Steller, H.; Meier, P. IAP-antagonists exhibit nonredundant modes of action through differential DIAP1 binding. EMBO J. 2003, 22, 6642–6652. [Google Scholar] [CrossRef]
- Freel, C.D.; Richardson, D.A.; Thomenius, M.J.; Gan, E.C.; Horn, S.R.; Olson, M.R.; Kornbluth, S. Mitochondrial localization of Reaper to promote inhibitors of apoptosis protein degradation conferred by GH3 domain-lipid interactions. J. Biol. Chem. 2008, 283, 367–379. [Google Scholar] [CrossRef]
- Sandu, C.; Ryoo, H.D.; Steller, H. Drosophila IAP antagonists form multimeric complexes to promote cell death. J. Cell Biol. 2010, 190, 1039–1052. [Google Scholar] [CrossRef]
- Liu, B.; Becnel, J.J.; Zhang, Y.; Zhou, L. Induction of reaper ortholog mx in mosquito midgut cells following baculovirus infection. Cell Death Differ. 2011, 18, 1337–1345. [Google Scholar] [CrossRef][Green Version]
- Schetelig, M.F.; Nirmala, X.; Handler, A.M. Pro-apoptotic cell death genes, hid and reaper, from the tephritid pest species, Anastrepha suspensa. Apoptosis 2011, 16, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Bryant, B.; Zhang, Y.; Zhang, C.; Santos, C.P.; Clem, R.J.; Zhou, L. A lepidopteran orthologue of reaper reveals functional conservation and evolution of IAP antagonists. Insect Mol. Biol. 2009, 18, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Joazeiro, C.A.; Bonfoco, E.; Kamada, S.; Leverson, J.D.; Hunter, T. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J. Biol. Chem. 2000, 275, 26661–26664. [Google Scholar] [PubMed]
- Clavier, A.; Rincheval-Arnold, A.; Colin, J.; Mignotte, B.; Guénal, I. Apoptosis in Drosophila: Which role for mitochondria? Apoptosis 2016, 21, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Pan, M.H.; Sun, Z.Y.; Huang, S.J.; Yu, Z.S.; Liu, D.; Zhao, D.H.; Lu, C. The genomic underpinnings of apoptosis in the silkworm, Bombyx mori. BMC Genom. 2010, 11, 611. [Google Scholar] [CrossRef]
- Tettamanti, G.; Casartelli, M. Cell death during complete metamorphosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20190065. [Google Scholar] [CrossRef]
- Bao, X.; Chen, P.; Liu, T.; Wang, L.; Liu, W.; Pan, M.; Lu, C. Advances in apoptosis-related genes in the silkworm, Bombyx mori. Acta Entomol. Sin. 2017, 60, 487–498. [Google Scholar]
- Ying, Q.Q.; Liu, T.; Gu, W. Effect of ultraviolet irradiation on the expressions of apoptosis related genes reaper, grim, hid in Drosophila. Chin. J. Gerontol. 2012, 32, 2307–2309. [Google Scholar]
- Shu, B.; Zhang, J.; Cui, G.; Sun, R.; Yi, X.; Zhong, G. Azadirachtin affects the growth of Spodoptera litura Fabricius by inducing apoptosis in larval midgut. Front. Physiol. 2018, 9, 137. [Google Scholar] [CrossRef]
- Thomenius, M.; Kornbluth, S. Multifunctional reaper: Sixty-five amino acids of fury. Cell Death Differ. 2006, 13, 1305–1309. [Google Scholar] [CrossRef]
- Claveria, C.; Caminero, E.; Martinez, A.C.; Campuzano, S.; Torres, M. GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. EMBO J. 2002, 21, 3327–3336. [Google Scholar] [CrossRef] [PubMed]
- Haining, W.N.; Carboy-Newcomb, C.; Wei, C.L.; Steller, H. The proapoptotic function of Drosophila Hid is conserved in mammalian cells. Proc. Natl. Acad. Sci. USA 1999, 96, 4936–4941. [Google Scholar] [CrossRef] [PubMed]
- Abdelwahid, E.; Yokokura, T.; Krieser, R.J.; Balasundaram, S.; Fowle, W.H.; White, K. Mitochondrial disruption in Drosophila apoptosis. Dev. Cell 2007, 12, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Morishita, J.; Kang, M.J.; Fidelin, K.; Ryoo, H.D. CDK7 regulates the mitochondrial localization of a tail-anchored proapoptotic protein, Hid. Cell Rep. 2013, 5, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Vucic, D.; Seshagiri, S.; Miller, L.K. Characterization of reaper- and FADD-induced apoptosis in a lepidopteran cell line. Mol. Cell Biol. 1997, 17, 667–676. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tait, S.W.; Werner, A.B.; de Vries, E.; Borst, J. Mechanism of action of Drosophila Reaper in mammalian cells: Reaper globally inhibits protein synthesis and induces apoptosis independent of mitochondrial permeability. Cell Death Differ. 2004, 11, 800–811. [Google Scholar] [CrossRef]
- Yu, H.; Li, Z.Q.; Ou-Yang, Y.Y.; Huang, G.H. Identification of four caspase genes from Spodoptera exigua (Lepidoptera: Noctuidae) and their regulations towards different apoptotic stimulations. Insect Sci. 2019. [Google Scholar] [CrossRef]
- Chai, J.J.; Yan, N.; Huh, J.R.; Wu, J.W.; Li, W.Y.; Hay, B.A.; Shi, Y.G. Molecular mechanism of Reaper-Grim-Hid mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat. Struct. Biol. 2003, 10, 892–898. [Google Scholar] [CrossRef]
- Shu, B.; Zhang, J.; Sethuraman, V.; Cui, G.; Yi, X.; Zhong, G. Transcriptome analysis of Spodoptera frugiperda Sf9 cells reveals putative apoptosis-related genes and a preliminary apoptosis mechanism induced by azadirachtin. Sci. Rep. 2017, 7, 13231. [Google Scholar] [CrossRef]
- Betz, A.; Ryoo, H.D.; Steller, H.; Darnell, J.E., Jr. STAT92E is a positive regulator of Drosophila inhibitor of apoptosis 1 (DIAP/1) and protects against radiation-induced apoptosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13805–13810. [Google Scholar] [CrossRef]
- Shu, B.; Jia, J.; Zhang, J.; Sethuraman, V.; Yi, X.; Zhong, G. DnaJ homolog subfamily A member1 (DnaJ1) is a newly discovered anti-apoptotic protein regulated by azadirachtin in Sf9 cells. BMC Genom. 2018, 19, 413. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shu, B.; Zhang, J.; Veeran, S.; Zhong, G. Pro-Apoptotic Function Analysis of the Reaper Homologue IBM1 in Spodoptera frugiperda. Int. J. Mol. Sci. 2020, 21, 2729. https://doi.org/10.3390/ijms21082729
Shu B, Zhang J, Veeran S, Zhong G. Pro-Apoptotic Function Analysis of the Reaper Homologue IBM1 in Spodoptera frugiperda. International Journal of Molecular Sciences. 2020; 21(8):2729. https://doi.org/10.3390/ijms21082729
Chicago/Turabian StyleShu, Benshui, Jingjing Zhang, Sethuraman Veeran, and Guohua Zhong. 2020. "Pro-Apoptotic Function Analysis of the Reaper Homologue IBM1 in Spodoptera frugiperda" International Journal of Molecular Sciences 21, no. 8: 2729. https://doi.org/10.3390/ijms21082729
APA StyleShu, B., Zhang, J., Veeran, S., & Zhong, G. (2020). Pro-Apoptotic Function Analysis of the Reaper Homologue IBM1 in Spodoptera frugiperda. International Journal of Molecular Sciences, 21(8), 2729. https://doi.org/10.3390/ijms21082729