DDX6 Helicase Behavior and Protein Partners in Human Adipose Tissue-Derived Stem Cells during Early Adipogenesis and Osteogenesis
Abstract
1. Introduction
2. Results
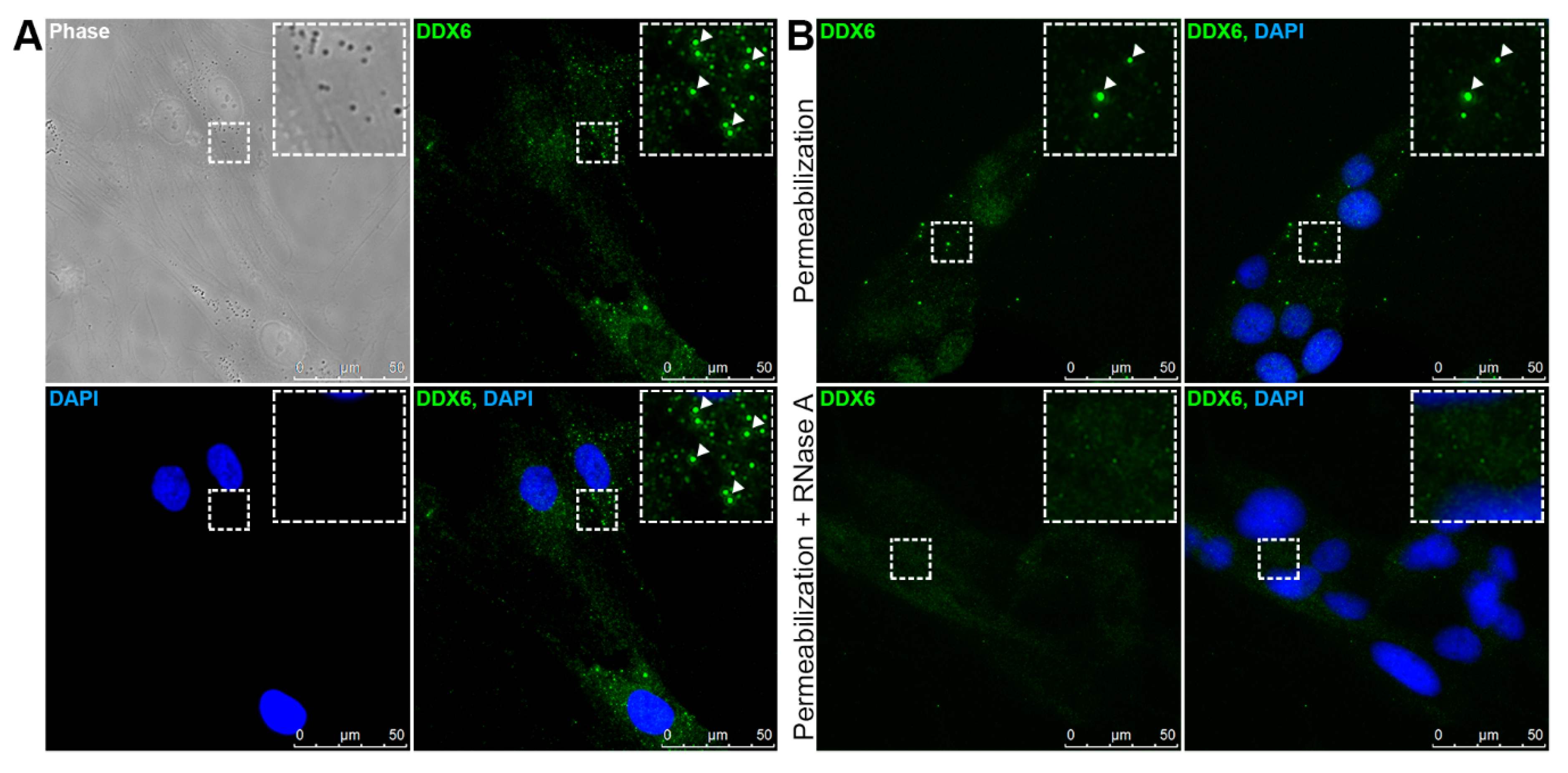
2.1. DDX6 Is Localized with RNA-Dependent Granules in the hASCs
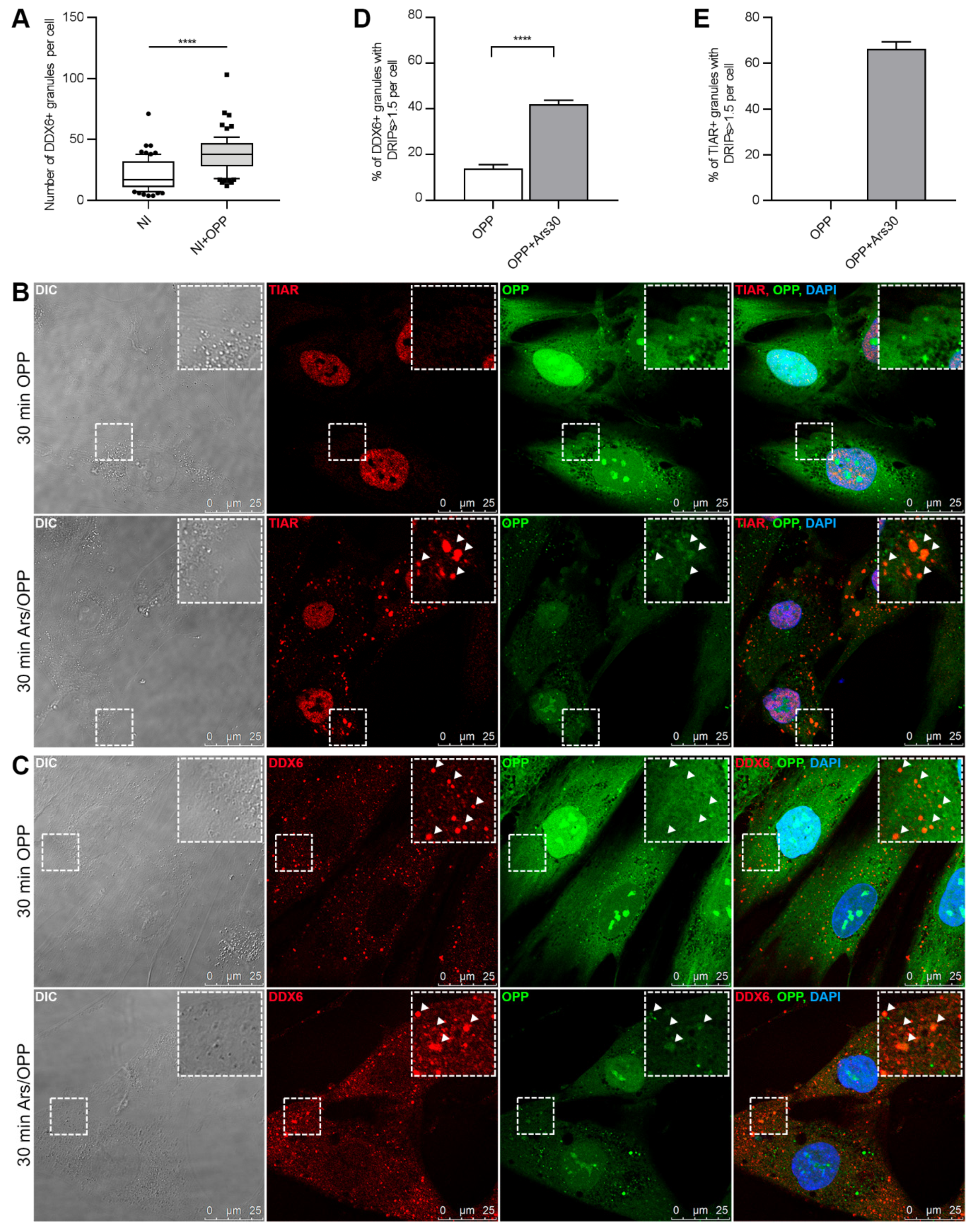
2.2. DDX6 Granules Observed in the hASCs Have P-Body-Like Behavior
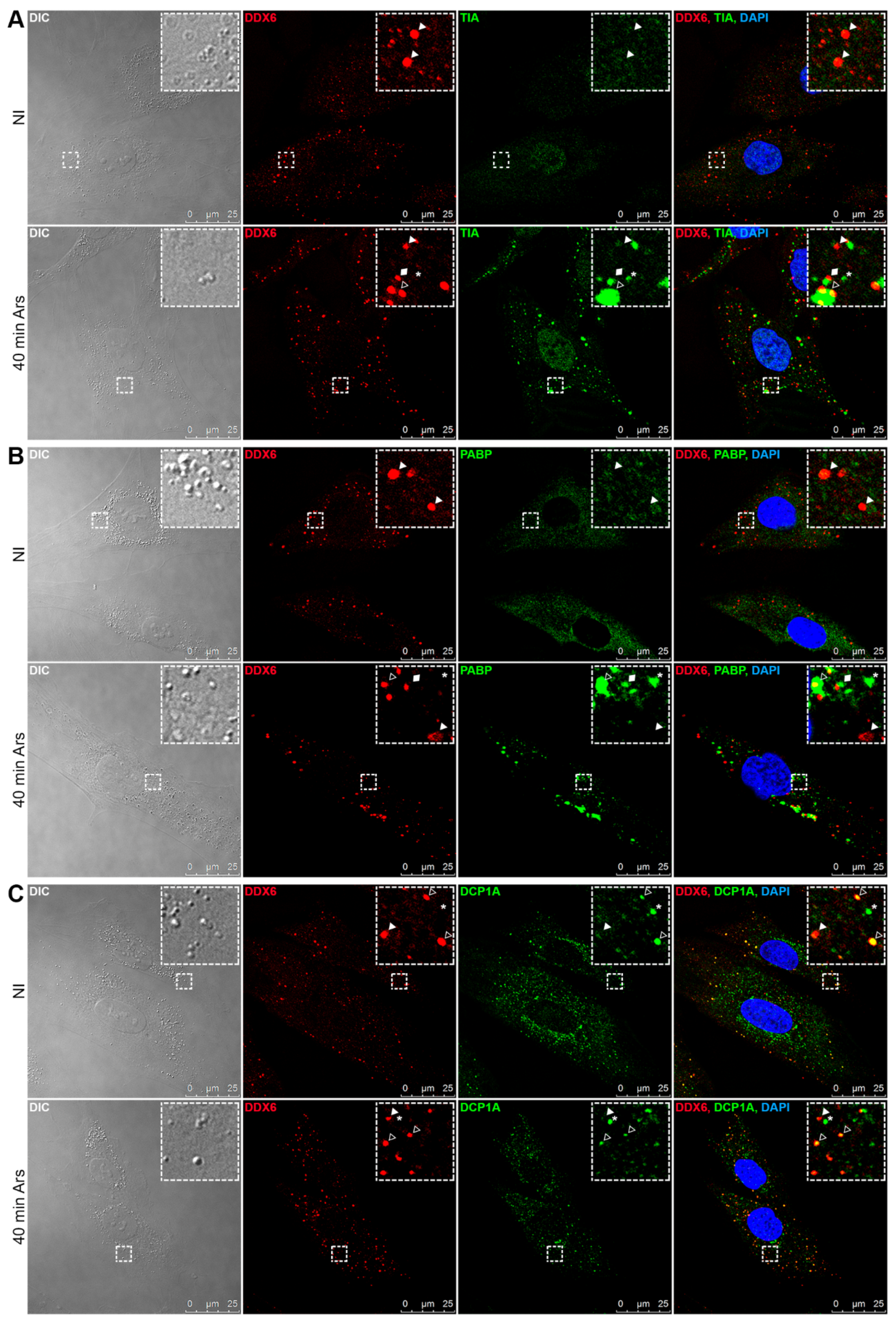
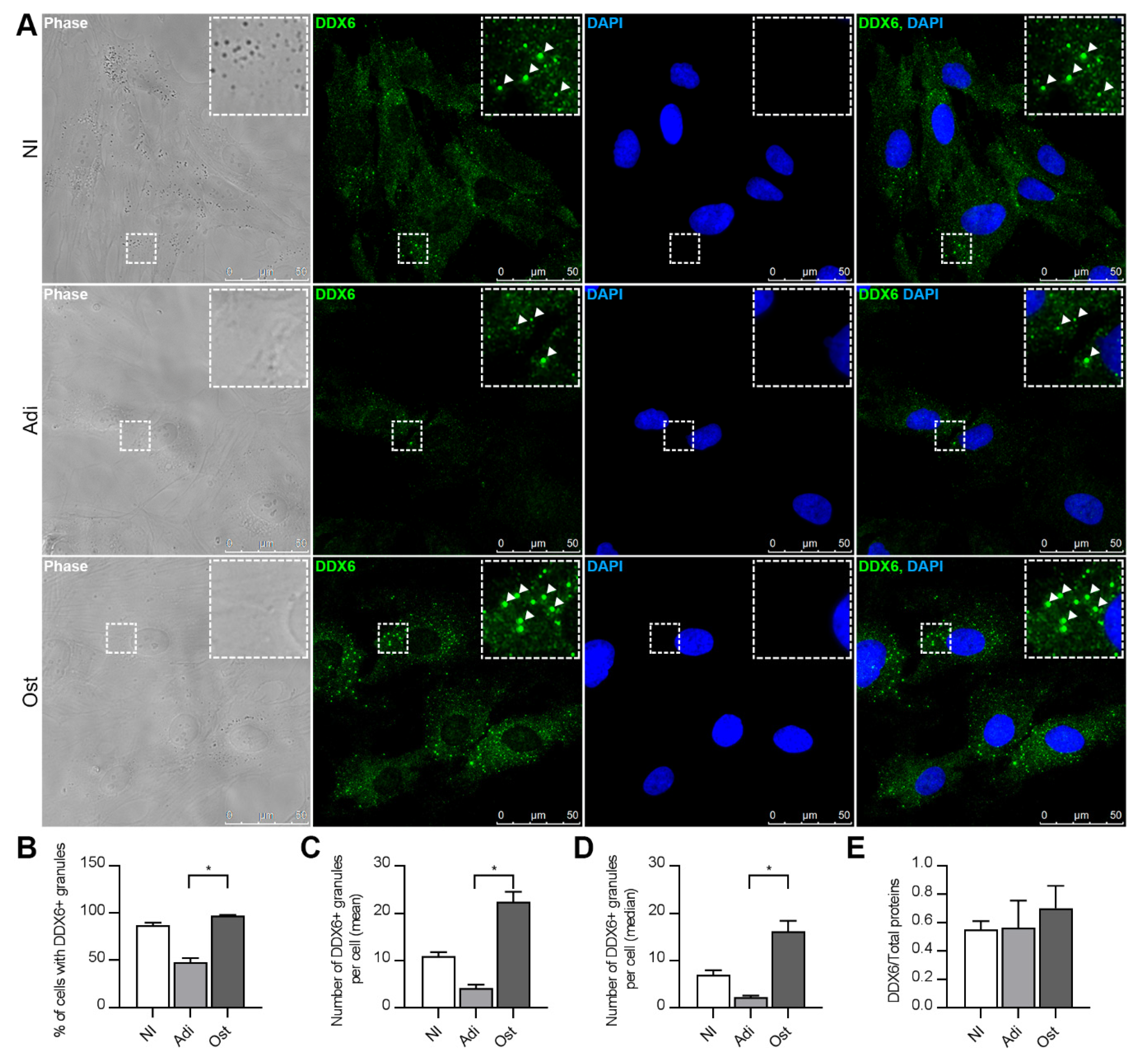
2.3. DDX6 Distribution Changes upon Adipogenic or Osteogenic Induction
2.4. DDX6 Granules Observed in the hASCs after Adipogenic and Osteogenic Induction Exhibit P-Body-Like Behavior
2.5. Complexes Associated with DDX6 after 24 h of Adipogenic and Osteogenic Induction Have Different Sedimentation Patterns
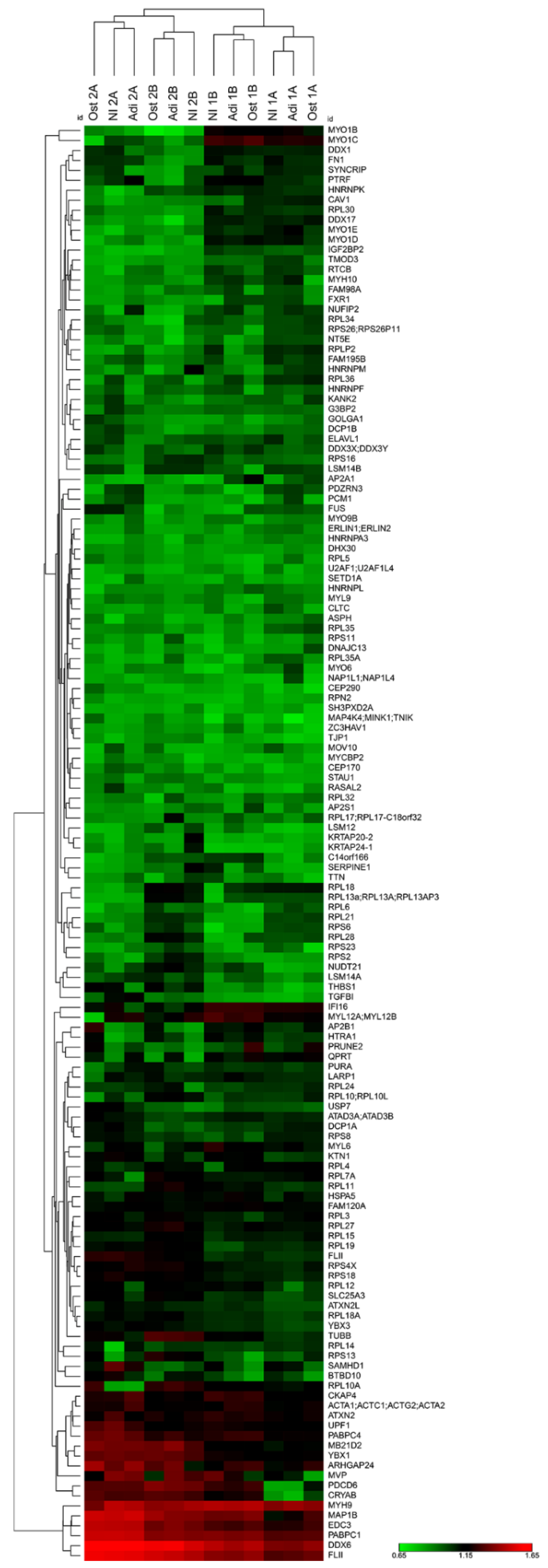
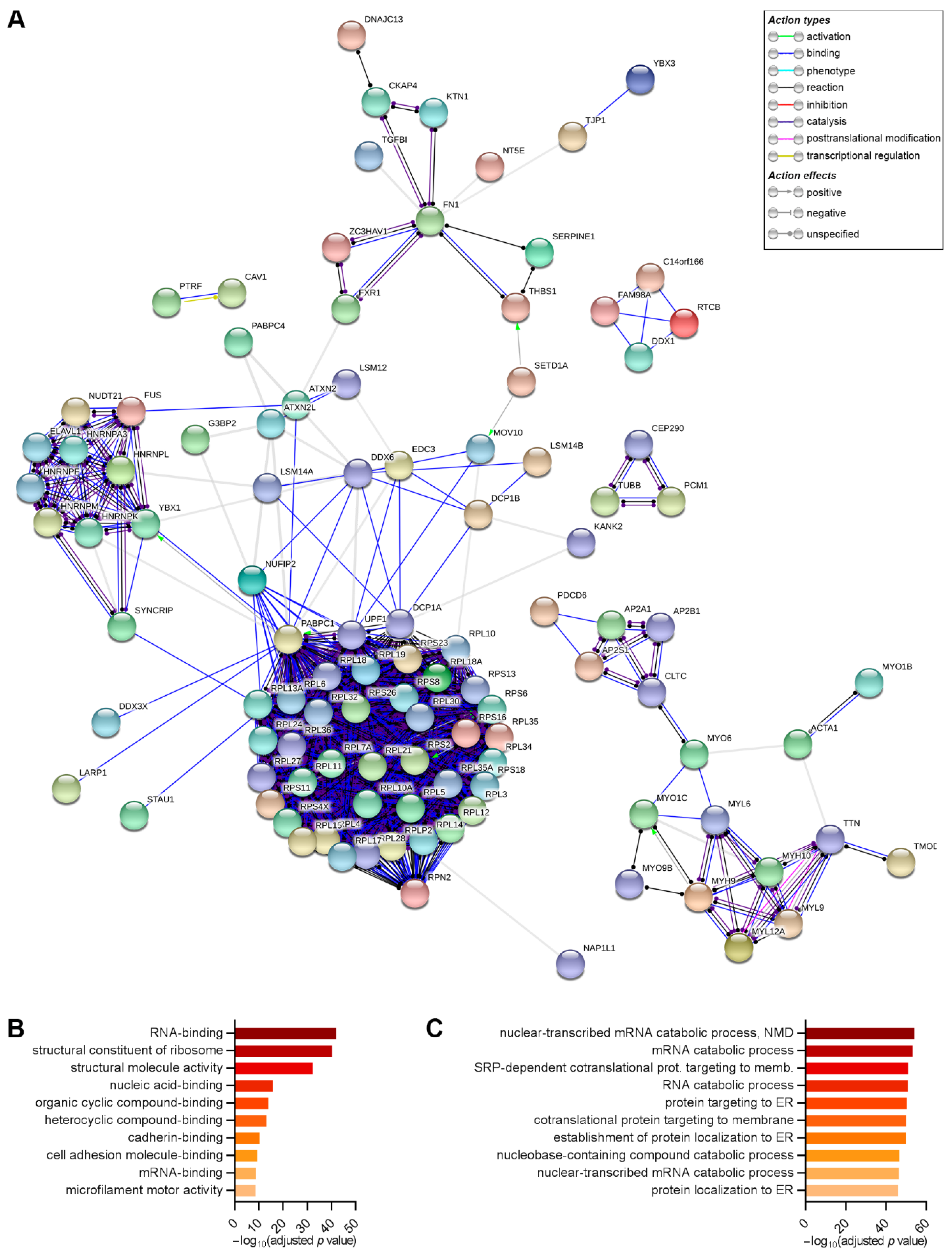
2.6. DDX6 Is Associated with Regulators of the mRNA Life Cycle in the hASCs That Were Not Induced and Those Induced to Adipogenesis or Osteogenesis for 24 h
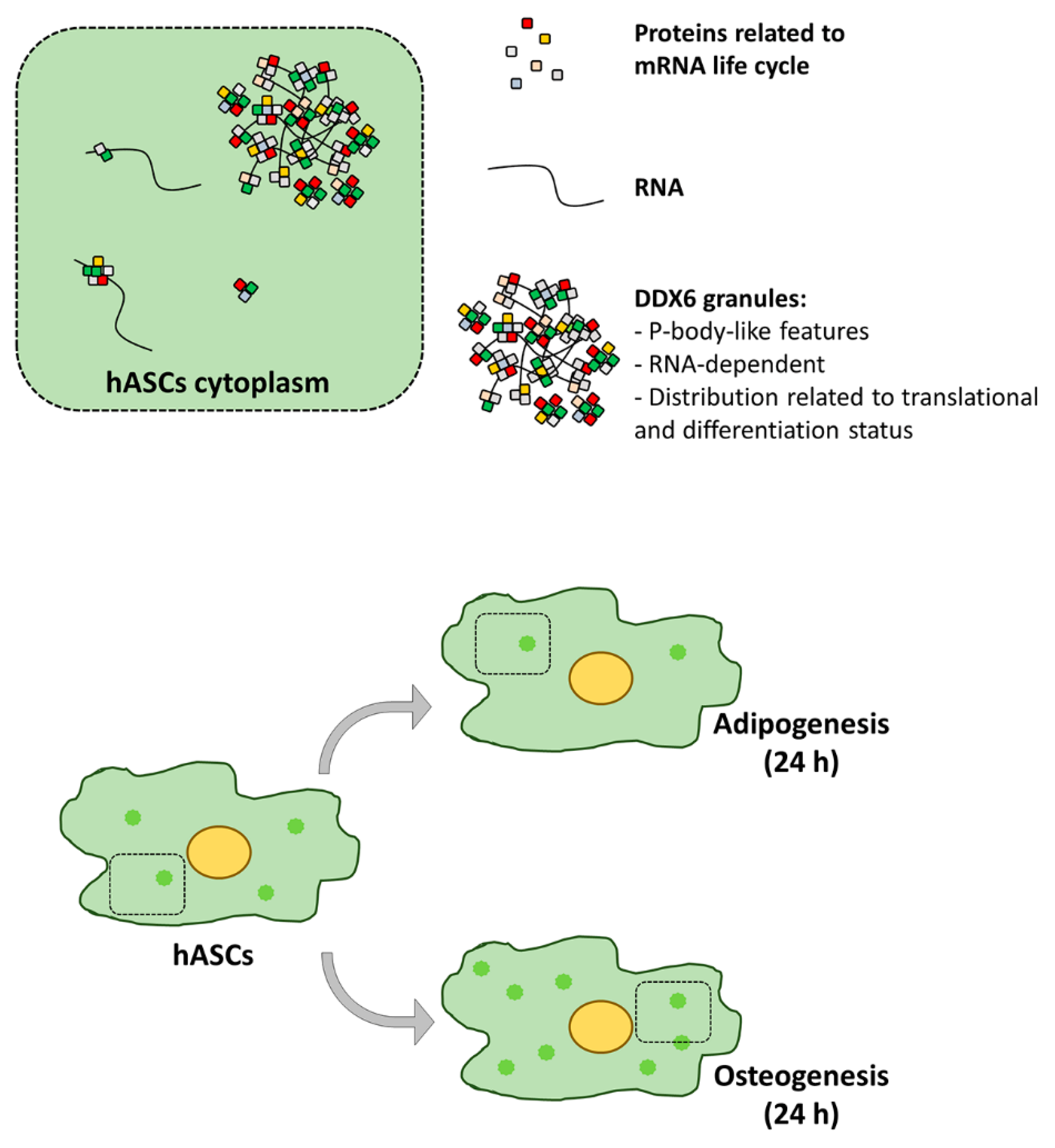
3. Discussion
4. Materials and Methods
4.1. Subjects and Cell Culture
4.2. Western Blot Analysis
4.3. Immunofluorescence and DDX6 Granule Quantification
4.4. Assessment of the Dependence on RNA for DDX6 Granule Maintenance
4.5. Treatment with o-propargyl Puromycin and Localization of Nascent Peptides
4.6. Quantification of DRIPs
4.7. Sucrose Density Gradient Separation
4.8. Immunoprecipitation of DDX6
4.9. Proteomic Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ESC | Embryonic stem cell |
| FBS | Fetal bovine serum |
| hASC | Human adipose tissue-derived stem cell |
| hIPSC | Human-induced pluripotent stem cell |
| IFF | Immunofluorescence |
| IP | Immunoprecipitation |
| PBS | Phosphate-buffered saline |
| SG | Stress granule |
References
- Wang, Y.; Arribas-Layton, M.; Chen, Y.; Lykke-Andersen, J.; Sen, G.L. DDX6 Orchestrates Mammalian Progenitor Function through the mRNA Degradation and Translation Pathways. Mol. Cell 2015, 60, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Coller, J.M.; Tucker, M.; Sheth, U.; Valencia-Sanchez, M.A.; Parker, R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 2001, 7, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Ernoult-Lange, M.; Baconnais, S.; Harper, M.; Minshall, N.; Souquere, S.; Boudier, T.; Bénard, M.; Andrey, P.; Pierron, G.; Kress, M.; et al. Multiple binding of repressed mRNAs by the P-body protein Rck/p54. RNA 2012, 18, 1702–1715. [Google Scholar] [CrossRef] [PubMed]
- Freimer, J.W.; Hu, T.; Blelloch, R. Decoupling the impact of microRNAs on translational repression versus RNA degradation in embryonic stem cells. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, B.; Luo, E.-C.; Haggerty, C.; Aigner, S.; Charlton, J.; Brumbaugh, J.; Ji, F.; Rabano Jiménez, I.; Clowers, K.J.; Huebner, A.J.; et al. The RNA Helicase DDX6 Controls Cellular Plasticity by Modulating P-Body Homeostasis. Cell Stem Cell 2019, 25, 622–638. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, S.; Okawa, S.; Hillje, A.-L.; González-Cano, L.; del Sol, A.; Schwamborn, J.C. The RNA helicase DDX6 regulates cell-fate specification in neural stem cells via miRNAs. Nucleic Acids Res. 2015, 43, 2638–2654. [Google Scholar] [CrossRef]
- Segre, J.A.; Bauer, C.; Fuchs, E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 1999, 22, 356–400. [Google Scholar] [CrossRef]
- Ayache, J.; Bénard, M.; Ernoult-Lange, M.; Minshall, N.; Standart, N.; Kress, M.; Weil, D. P-body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Mol. Biol. Cell 2015, 26, 2579–2595. [Google Scholar] [CrossRef]
- Nonhoff, U.; Ralser, M.; Welzel, F.; Piccini, I.; Balzereit, D.; Yaspo, M.-L.; Lehrach, H.; Krobitsch, S. Ataxin-2 Interacts with the DEAD/H-Box RNA Helicase DDX6 and Interferes with P-Bodies and Stress Granules. Mol. Biol. Cell 2007, 18, 1385–1396. [Google Scholar] [CrossRef]
- Wilczynska, A.; Aigueperse, C.; Kress, M.; Dautry, F.; Weil, D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 2005, 118, 981–992. [Google Scholar] [CrossRef]
- Chu, C.; Rana, T.M. Translation Repression in Human Cells by MicroRNA-Induced Gene Silencing Requires RCK/p54. PLoS Biol. 2006, 4, e210. [Google Scholar] [CrossRef] [PubMed]
- Coller, J.; Parker, R. General Translational Repression by Activators of mRNA Decapping. Cell 2005, 122, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Hubstenberger, A.; Courel, M.; Bénard, M.; Souquere, S.; Ernoult-Lange, M.; Chouaib, R.; Yi, Z.; Morlot, J.-B.; Munier, A.; Fradet, M.; et al. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol. Cell 2017, 68, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Sheth, U.; Parker, R. Decapping and Decay of Messenger RNA Occur in Cytoplasmic Processing Bodies. Science 2003, 300, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Elbaum-Garfinkle, S.; Kim, Y.; Szczepaniak, K.; Chen, C.C.-H.; Eckmann, C.R.; Myong, S.; Brangwynne, C.P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 7189–7194. [Google Scholar] [CrossRef]
- Jain, S.; Wheeler, J.R.; Walters, R.W.; Agrawal, A.; Barsic, A.; Parker, R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016, 164, 487–498. [Google Scholar] [CrossRef]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Julicher, F.; Hyman, A.A. Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef]
- Standart, N.; Weil, D. P-Bodies: Cytosolic Droplets for Coordinated mRNA Storage. Trends Genet. 2018, 34, 612–626. [Google Scholar] [CrossRef]
- Lin, Y.; Protter, D.S.W.; Rosen, M.K.; Parker, R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell 2015, 60, 208–219. [Google Scholar] [CrossRef]
- Kedersha, N.; Cho, M.R.; Li, W.; Yacono, P.W.; Chen, S.; Gilks, N.; Golan, D.E.; Anderson, P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000, 151, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Stoecklin, G.; Ayodele, M.; Yacono, P.; Lykke-Andersen, J.; Fritzler, M.J.; Scheuner, D.; Kaufman, R.J.; Golan, D.E.; Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005, 169, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.L.; Gupta, M.; Li, W.; Miller, I.; Anderson, P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999, 147, 1431–1442. [Google Scholar] [CrossRef]
- Arribere, J.A.; Doudna, J.A.; Gilbert, W.V. Reconsidering Movement of Eukaryotic mRNAs between Polysomes and P Bodies. Mol. Cell 2011, 44, 745–758. [Google Scholar] [CrossRef]
- Brengues, M.; Teixeira, D.; Parker, R. Movement of Eukaryotic mRNAs Between Polysomes and Cytoplasmic Processing Bodies. Science 2005, 310, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Serman, A.; Le Roy, F.; Aigueperse, C.; Kress, M.; Dautry, F.; Weil, D. GW body disassembly triggered by siRNAs independently of their silencing activity. Nucleic Acids Res. 2007, 35, 4715–4727. [Google Scholar] [CrossRef]
- Minshall, N.; Kress, M.; Weil, D.; Standart, N. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol. Biol. Cell 2009, 20, 2464–2472. [Google Scholar] [CrossRef]
- Kocan, B.; Maziarz, A.; Tabarkiewicz, J.; Ochiya, T.; Banaś-Ząbczyk, A. Trophic Activity and Phenotype of Adipose Tissue-Derived Mesenchymal Stem Cells as a Background of Their Regenerative Potential. Stem Cells Int. 2017, 2017, 1653254. [Google Scholar] [CrossRef]
- Marcon, B.H.; Shigunov, P.; Spangenberg, L.; Pereira, I.T.; de Aguiar, A.M.; Amorín, R.; Rebelatto, C.K.; Correa, A.; Dallagiovanna, B. Cell cycle genes are downregulated after adipogenic triggering in human adipose tissue-derived stem cells by regulation of mRNA abundance. Sci. Rep. 2019, 9, 5611. [Google Scholar] [CrossRef]
- Robert, A.W.; Angulski, A.B.B.; Spangenberg, L.; Shigunov, P.; Pereira, I.T.; Bettes, P.S.L.; Naya, H.; Correa, A.; Dallagiovanna, B.; Stimamiglio, M.A. Gene expression analysis of human adipose tissue-derived stem cells during the initial steps of in vitro osteogenesis. Sci. Rep. 2018, 8, 4739. [Google Scholar] [CrossRef]
- Teixeira, D.; Sheth, U.; Valencia-Sanchez, M.A.; Brengues, M.; Parker, R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 2005, 11, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Ganassi, M.; Mateju, D.; Bigi, I.; Mediani, L.; Poser, I.; Lee, H.O.; Seguin, S.J.; Morelli, F.F.; Vinet, J.; Leo, G.; et al. A Surveillance Function of the HSPB8-BAG3-HSP70 Chaperone Complex Ensures Stress Granule Integrity and Dynamism. Mol. Cell 2016, 63, 796–810. [Google Scholar] [CrossRef]
- Liu, J.; Xu, Y.; Stoleru, D.; Salic, A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc. Natl. Acad. Sci. USA 2012, 109, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Marcon, B.H.; Holetz, F.B.; Eastman, G.; Origa-Alves, A.C.; Amorós, M.A.; de Aguiar, A.M.; Rebelatto, C.K.; Brofman, P.R.S.; Sotelo-Silveira, J.; Dallagiovanna, B. Downregulation of the protein synthesis machinery is a major regulatory event during early adipogenic differentiation of human adipose-derived stromal cells. Stem Cell Res. 2017, 25, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, A.; Chen, Y.-H.; Martin, S.; Alhusaini, N.; Green, R.; Coller, J. The DEAD-Box Protein Dhh1p Couples mRNA Decay and Translation by Monitoring Codon Optimality. Cell 2016, 167, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Sweet, T.; Kovalak, C.; Coller, J. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 2012, 10, e1001342. [Google Scholar] [CrossRef] [PubMed]
- Kami, D.; Kitani, T.; Nakamura, A.; Wakui, N.; Mizutani, R.; Ohue, M.; Kametani, F.; Akimitsu, N.; Gojo, S. The DEAD-box RNA-binding protein DDX6 regulates parental RNA decay for cellular reprogramming to pluripotency. PLoS ONE 2018, 13, e0203708. [Google Scholar] [CrossRef]
- Yang, Z.; Jakymiw, A.; Wood, M.R.; Eystathioy, T.; Rubin, R.L.; Fritzler, M.J.; Chan, E.K.L. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 2004, 117, 5567–5578. [Google Scholar] [CrossRef]
- Aizer, A.; Kafri, P.; Kalo, A.; Shav-Tal, Y. The P Body Protein Dcp1a Is Hyper-phosphorylated during Mitosis. PLoS ONE 2013, 8, e49783. [Google Scholar] [CrossRef]
- Baer, P.C.; Geiger, H. Adipose-Derived Mesenchymal Stromal/Stem Cells: Tissue Localization, Characterization, and Heterogeneity. Stem Cells Int. 2012, 2012, 1–11. [Google Scholar] [CrossRef]
- Şovrea, A.S.; Boşca, A.B.; Constantin, A.M.; Dronca, E.; Ilea, A. State of the art in human adipose stem cells and their role in therapy. Rom. J. Morphol. Embryol. 2019, 60, 7–31. [Google Scholar]
- Spangenberg, L.; Shigunov, P.; Abud, A.P.R.; Cofré, A.R.; Stimamiglio, M.A.; Kuligovski, C.; Zych, J.; Schittini, A.V.; Costa, A.D.T.; Rebelatto, C.K.; et al. Polysome profiling shows extensive posttranscriptional regulation during human adipocyte stem cell differentiation into adipocytes. Stem Cell Res. 2013, 11, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Scheideler, M.; Elabd, C.; Zaragosi, L.E.; Chiellini, C.; Hackl, H.; Sanchez-Cabo, F.; Yadav, S.; Duszka, K.; Friedl, G.; Papak, C.; et al. Comparative transcriptomics of human multipotent stem cells during adipogenesis and osteoblastogenesis. BMC Genomics 2008, 9, 340. [Google Scholar] [CrossRef]
- Eulalio, A.; Behm-Ansmant, I.; Schweizer, D.; Izaurralde, E. P-Body Formation Is a Consequence, Not the Cause, of RNA-Mediated Gene Silencing. Mol. Cell. Biol. 2007, 27, 3970–3981. [Google Scholar] [CrossRef]
- Decker, C.J.; Parker, R. P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012, 4, a012286. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Romfo, C.M.; Nilsen, T.W.; Green, M.R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 1999, 402, 832–835. [Google Scholar] [CrossRef] [PubMed]
- Ozeki, K.; Sugiyama, M.; Akter, K.A.; Nishiwaki, K.; Asano-Inami, E.; Senga, T. FAM98A is localized to stress granules and associates with multiple stress granule-localized proteins. Mol. Cell. Biochem. 2019, 451, 107–115. [Google Scholar] [CrossRef]
- Tatsumi, Y.; Takano, R.; Islam, M.S.; Yokochi, T.; Itami, M.; Nakamura, Y.; Nakagawara, A. BMCC1, which is an interacting partner of BCL2, attenuates AKT activity, accompanied by apoptosis. Cell Death Dis. 2015, 6, e1607. [Google Scholar] [CrossRef]
- Baker, N.; Sohn, J.; Tuan, R.S. Promotion of human mesenchymal stem cell osteogenesis by PI3-kinase/Akt signaling, and the influence of caveolin-1/cholesterol homeostasis. Stem Cell Res. Ther. 2015, 6, 238. [Google Scholar] [CrossRef]
- Mukherjee, A.; Rotwein, P. Akt promotes BMP2-mediated osteoblast differentiation and bone development. J. Cell Sci. 2009, 122, 716–726. [Google Scholar] [CrossRef]
- Fujiwara, T.; Takahashi, R.; Kosaka, N.; Nezu, Y.; Kawai, A.; Ozaki, T.; Ochiya, T. RPN2 Gene Confers Osteosarcoma Cell Malignant Phenotypes and Determines Clinical Prognosis. Mol. Ther.-Nucleic Acids 2014, 3, e189. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Wu, L.; Wang, Q.; Jiang, Z.; Li, Y.; Ma, N.; Chen, W.; Hou, Z.; Gan, W.; Chen, S. MicroRNA-128 targeting RPN2 inhibits cell proliferation and migration through the Akt-p53-cyclin pathway in colorectal cancer cells. Oncol. Lett. 2018, 16, 6940–6949. [Google Scholar] [CrossRef] [PubMed]
- Das, D.; Wang, Y.-H.; Hsieh, C.-Y.; Suzuki, Y.J. Major vault protein regulates cell growth/survival signaling through oxidative modifications. Cell. Signal. 2016, 28, 12–18. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rebelatto, C.K.; Aguiar, A.M.; Moretão, M.P.; Senegaglia, A.C.; Hansen, P.; Barchiki, F.; Oliveira, J.; Martins, J.; Kuligovski, C.; Mansur, F.; et al. Dissimilar differentiation of mesenchymal stem cells from bone marrow, umbilical cord blood, and adipose tissue. Exp. Biol. Med. 2008, 233, 901–913. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F..; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Reimand, U.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. g:Profiler––A web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
| Donor | Sex | Age (years) | Weight (kg) | Height (m) | BMI (kg/m2) | Analysis |
|---|---|---|---|---|---|---|
| A | Female | 46 | 61 | 1.59 | 24.1 | IP, IFF |
| B | Female | 17 | 64 | 1.58 | 25.6 | IP, IFF |
| C | Female | 46 | 57 | 1.58 | 22.8 | IP, IFF |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcon, B.H.; Rebelatto, C.K.; Cofré, A.R.; Dallagiovanna, B.; Correa, A. DDX6 Helicase Behavior and Protein Partners in Human Adipose Tissue-Derived Stem Cells during Early Adipogenesis and Osteogenesis. Int. J. Mol. Sci. 2020, 21, 2607. https://doi.org/10.3390/ijms21072607
Marcon BH, Rebelatto CK, Cofré AR, Dallagiovanna B, Correa A. DDX6 Helicase Behavior and Protein Partners in Human Adipose Tissue-Derived Stem Cells during Early Adipogenesis and Osteogenesis. International Journal of Molecular Sciences. 2020; 21(7):2607. https://doi.org/10.3390/ijms21072607
Chicago/Turabian StyleMarcon, Bruna Hilzendeger, Carmen K. Rebelatto, Axel R. Cofré, Bruno Dallagiovanna, and Alejandro Correa. 2020. "DDX6 Helicase Behavior and Protein Partners in Human Adipose Tissue-Derived Stem Cells during Early Adipogenesis and Osteogenesis" International Journal of Molecular Sciences 21, no. 7: 2607. https://doi.org/10.3390/ijms21072607
APA StyleMarcon, B. H., Rebelatto, C. K., Cofré, A. R., Dallagiovanna, B., & Correa, A. (2020). DDX6 Helicase Behavior and Protein Partners in Human Adipose Tissue-Derived Stem Cells during Early Adipogenesis and Osteogenesis. International Journal of Molecular Sciences, 21(7), 2607. https://doi.org/10.3390/ijms21072607





