Role of Deubiquitinases in Human Cancers: Potential Targeted Therapy
Abstract
1. Introduction
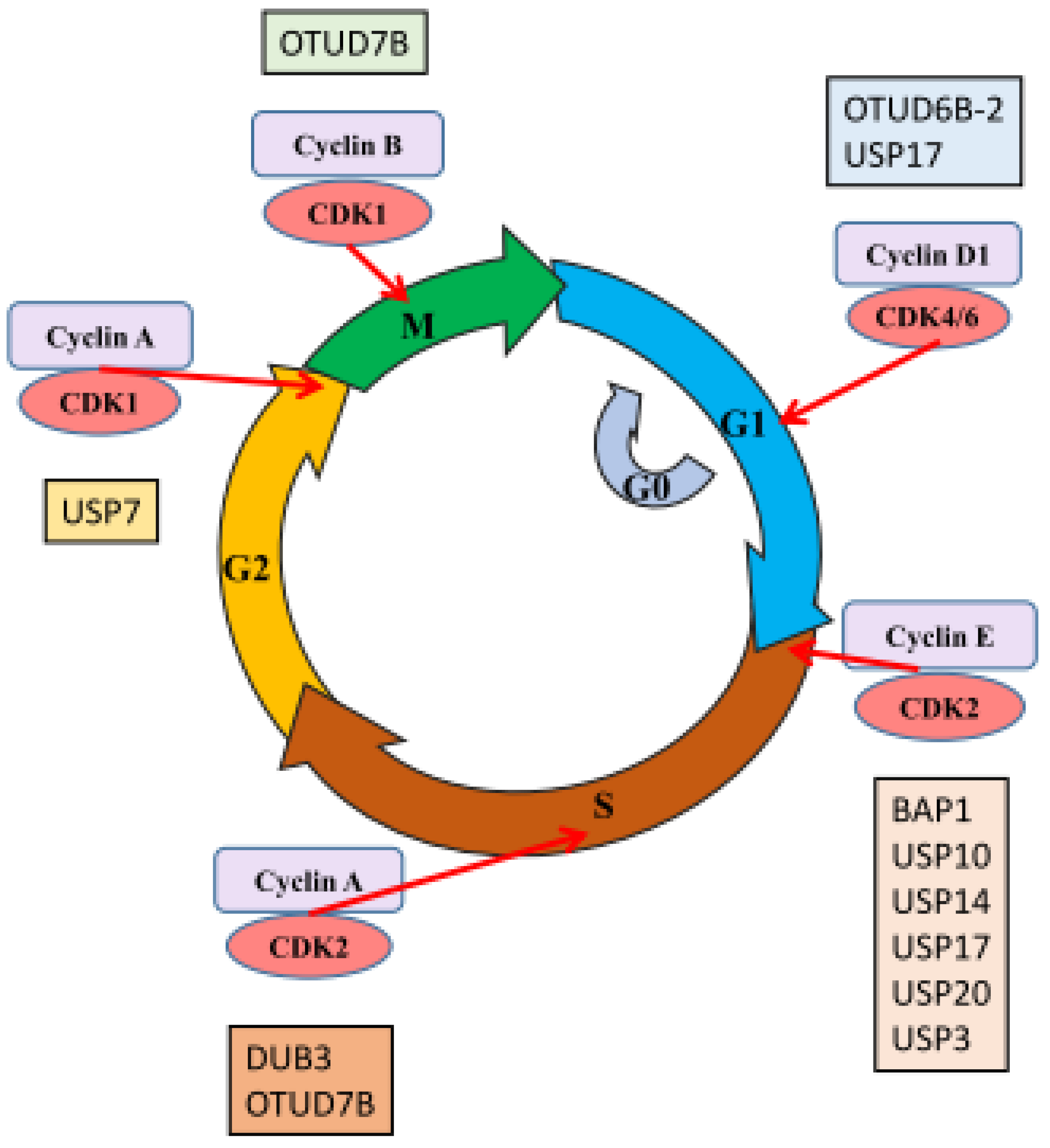
2. DUBs and Cell Cycle Control
3. DUBs and Cell Proliferation
4. DUBs and Apoptosis
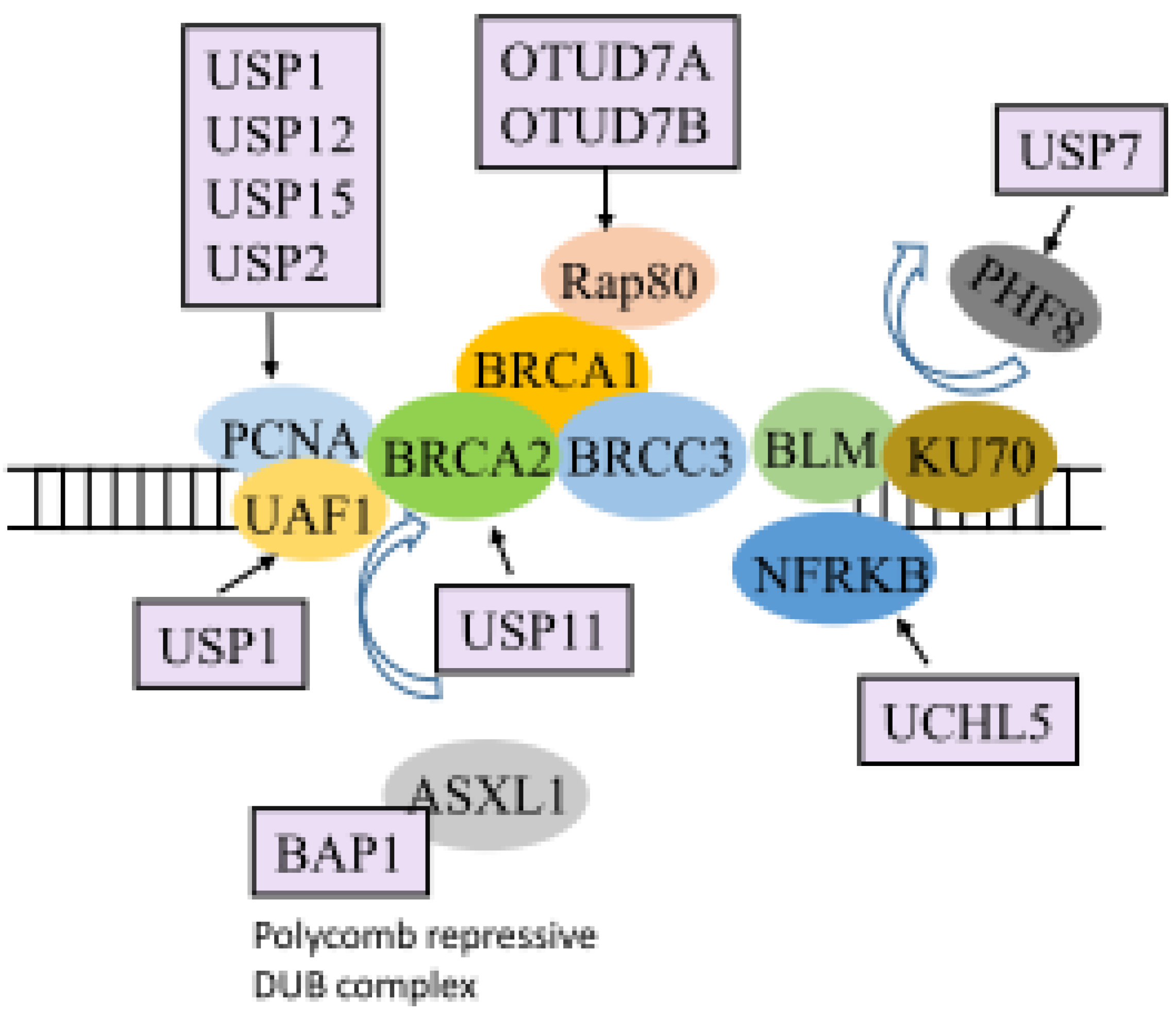
5. DUBs and the DDR
6. DUBs and Tumor Suppressors/Oncogenes
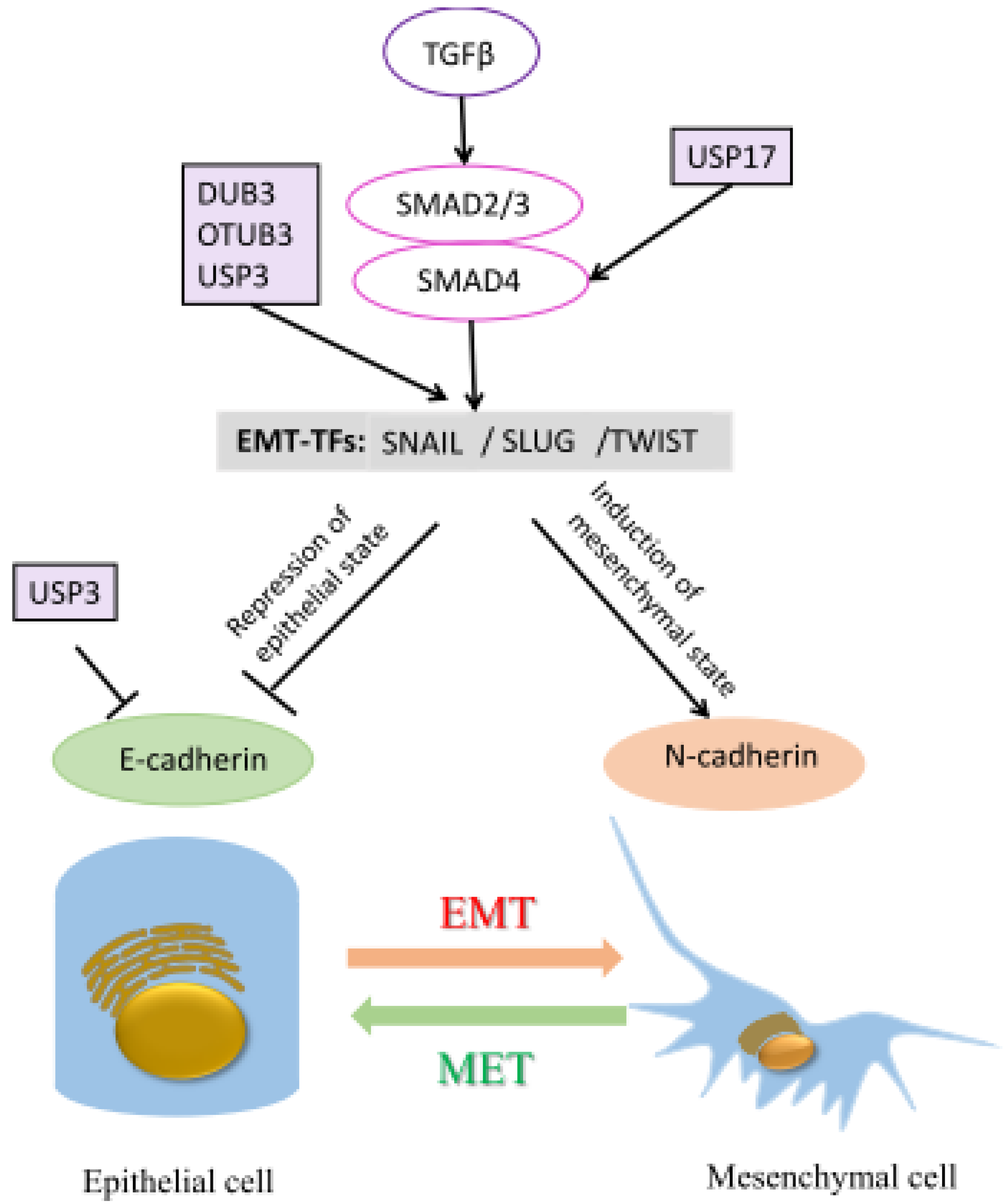
7. DUBs and Metastasis
8. DUBs as Therapeutic Targets for Cancer Treatment
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 53BP1 | p53-binding protein 1 |
| Akt | protein kinase B |
| APC/C | anaphase-promoting complex/cyclosome |
| AR | androgen receptor |
| BAP1 | BRCA1 associated protein 1 |
| BRCA | breast-cancer susceptibility gene |
| BRCC3 | BRCA1/BRCA2-containing complex 3 |
| CDK | cyclin-dependent kinase |
| CRC | colorectal cancer |
| DDR | DNA damage response |
| DSB | double-strand break |
| DUB | Deubiquitinase |
| ELK-1 | ETS like-1 protein |
| EMT | epithelial-mesenchymal transition |
| FBW7 | F-box and WD repeat domain-containing 7 |
| FKBP51 | FK506-binding protein 51 |
| GC | gastric cancer |
| GRB2 | growth factor receptor bound protein 2 |
| HCC | hepatocellular carcinoma |
| JOSD1 | Josephin domain containing 1 |
| KLF5 | Krüppel-like factor 5 |
| MDM2 | mouse double minute 2 |
| mTORC1 | mammalian target of rapamycin complex 1 |
| NK-κB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| NSCLC | non-small cell lung cancer |
| OTUB1 | otubain 1 |
| OTU | otubain protease |
| PBX1 | pre-B cell leukemia homeobox-1 |
| PCa | prostate cancer |
| PCNA | proliferating-cell nuclear antigen |
| PHF8 | PHD finger protein 8 |
| PHLPP | PH domain leucine-rich-repeats protein phosphatase |
| PML | promyelocytic leukemia |
| PSMD14 | 26S proteasome non-ATPase regulatory subunit 14 |
| PTEN | phosphatase and tensin homolog deleted on chromosome 10 |
| RNF | ring finger proteins |
| SKP2 | S-phase kinase associated protein 2. |
| TGF-β | transforming growth factor beta |
| TRAIL | tumor necrosis factor alpha apoptosis-inducing ligand |
| UBQ | ubiquitin |
| UBR5 | ubiquitin protein ligase E3 component N-recognin 5 |
| UCH | ubiquitin C-terminal hydrolases |
| UCHL | ubiquitin C-terminal hydrolases like |
| USP | ubiquitin-specific protease |
| WM | Waldenström macroglobulinemia |
References
- Nijman, S.M.B.; Luna-Vargas, M.P.A.; Velds, A.; Brummelkamp, T.R.; Dirac, A.M.G.; Sixma, T.K.; Bernards, R. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005, 123, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Abdul Rehman, S.A.; Kristariyanto, Y.A.; Choi, S.Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. Mindy-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell 2016, 63, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Saad, Y.; Lei, T.; Wang, J.; Qi, D.; Yang, Q.; Kolattukudy, P.E.; Fu, M. Mcp-induced protein 1 deubiquitinates traf proteins and negatively regulates jnk and nf-kappab signaling. J. Exp. Med. 2010, 207, 2959–2973. [Google Scholar] [CrossRef] [PubMed]
- Hermanns, T.; Pichlo, C.; Woiwode, I.; Klopffleisch, K.; Witting, K.F.; Ovaa, H.; Baumann, U.; Hofmann, K. A family of unconventional deubiquitinases with modular chain specificity determinants. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Clague, M.J.; Barsukov, I.; Coulson, J.M.; Liu, H.; Rigden, D.J.; Urbe, S. Deubiquitylases from genes to organism. Physiol. Rev. 2013, 93, 1289–1315. [Google Scholar] [CrossRef]
- Sowa, M.E.; Bennett, E.J.; Gygi, S.P.; Harper, J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell 2009, 138, 389–403. [Google Scholar] [CrossRef]
- Komander, D.; Clague, M.J.; Urbe, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef]
- Urbe, S.; Liu, H.; Hayes, S.D.; Heride, C.; Rigden, D.J.; Clague, M.J. Systematic survey of deubiquitinase localization identifies usp21 as a regulator of centrosome- and microtubule-associated functions. Mol. Biol Cell 2012, 23, 1095–1103. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Pickart, C.M. Ubiquitin enters the new millennium. Mol. Cell 2001, 8, 499–504. [Google Scholar] [CrossRef]
- Ventii, K.H.; Wilkinson, K.D. Protein partners of deubiquitinating enzymes. Biochem. J. 2008, 414, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Mennerich, D.; Kubaichuk, K.; Kietzmann, T. Dubs, hypoxia, and cancer. Trends Cancer 2019, 5, 632–653. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhou, Z.; Wu, G.; Chen, Q.; Wan, Y. Emerging role of dubs in tumor metastasis and apoptosis: Therapeutic implication. Pharmacol. Ther. 2017, 177, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Zhang, Y.; Galardy, P.J. Dubs and cancer: The role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle 2009, 8, 1688–1697. [Google Scholar] [CrossRef]
- Bednash, J.S.; Mallampalli, R.K. Targeting deubiquitinases in cancer. Methods Mol. Biol. 2018, 1731, 295–305. [Google Scholar]
- Fraile, J.M.; Quesada, V.; Rodriguez, D.; Freije, J.M.; Lopez-Otin, C. Deubiquitinases in cancer: New functions and therapeutic options. Oncogene 2012, 31, 2373–2388. [Google Scholar] [CrossRef]
- Cheng, J.; Guo, J.; North, B.J.; Wang, B.; Cui, C.P.; Li, H.; Tao, K.; Zhang, L.; Wei, W. Functional analysis of deubiquitylating enzymes in tumorigenesis and development. Biochim. Et Biophys. Acta. Rev. Cancer 2019, 1872, 188312. [Google Scholar] [CrossRef]
- Clague, M.J.; Urbe, S.; Komander, D. Breaking the chains: Deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 2019, 20, 338–352. [Google Scholar] [CrossRef]
- Venuto, S.; Merla, G. E3 ubiquitin ligase trim proteins, cell cycle and mitosis. Cells 2019, 8, 510. [Google Scholar] [CrossRef]
- Vodermaier, H.C. Apc/c and scf: Controlling each other and the cell cycle. Curr. Biol. 2004, 14, R787–R796. [Google Scholar] [CrossRef]
- Sivakumar, S.; Gorbsky, G.J. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 2015, 16, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Zhou, Z.; Chen, W.; Wang, C.; Zhang, H.; Ge, G.; Shao, M.; You, D.; Fan, Z.; Xia, H.; et al. Bap1 promotes breast cancer cell proliferation and metastasis by deubiquitinating klf5. Nat. Commun. 2015, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Deng, T.; Ma, H.; Liu, Y.; Feng, P.; Wei, D.; Ling, N.; Li, L.; Qiu, S.; Zhang, L.; et al. Deubiquitinase dub3 regulates cell cycle progression via stabilizing cyclin a for proliferation of non-small cell lung cancer cells. Cells 2019, 8, 297. [Google Scholar] [CrossRef] [PubMed]
- Sobol, A.; Askonas, C.; Alani, S.; Weber, M.J.; Ananthanarayanan, V.; Osipo, C.; Bocchetta, M. Deubiquitinase otud6b isoforms are important regulators of growth and proliferation. Mol. Cancer Res. 2017, 15, 117–127. [Google Scholar] [CrossRef]
- Bonacci, T.; Suzuki, A.; Grant, G.D.; Stanley, N.; Cook, J.G.; Brown, N.G.; Emanuele, M.J. Cezanne/otud7b is a cell cycle-regulated deubiquitinase that antagonizes the degradation of apc/c substrates. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Bonacci, T.; Emanuele, M.J. Impressionist portraits of mitotic exit: Apc/c, k11-linked ubiquitin chains and cezanne. Cell Cycle 2019, 18, 652–660. [Google Scholar] [CrossRef]
- Moniz, S.; Bandarra, D.; Biddlestone, J.; Campbell, K.J.; Komander, D.; Bremm, A.; Rocha, S. Cezanne regulates e2f1-dependent hif2alpha expression. J. Cell Sci. 2015, 128, 3082–3093. [Google Scholar] [CrossRef]
- Wang, B.; Jie, Z.; Joo, D.; Ordureau, A.; Liu, P.; Gan, W.; Guo, J.; Zhang, J.; North, B.J.; Dai, X.; et al. Traf2 and otud7b govern a ubiquitin-dependent switch that regulates mtorc2 signalling. Nature 2017, 545, 365–369. [Google Scholar] [CrossRef]
- Liao, Y.; Liu, N.; Xia, X.; Guo, Z.; Li, Y.; Jiang, L.; Zhou, R.; Tang, D.; Huang, H.; Liu, J. Usp10 modulates the skp2/bcr-abl axis via stabilizing skp2 in chronic myeloid leukemia. Cell Discov. 2019, 5, 1–15. [Google Scholar] [CrossRef]
- Liao, Y.; Xia, X.; Liu, N.; Cai, J.; Guo, Z.; Li, Y.; Jiang, L.; Dou, Q.P.; Tang, D.; Huang, H.; et al. Growth arrest and apoptosis induction in androgen receptor-positive human breast cancer cells by inhibition of usp14-mediated androgen receptor deubiquitination. Oncogene 2018, 37, 1896–1910. [Google Scholar] [CrossRef]
- McFarlane, C.; Kelvin, A.A.; de la Vega, M.; Govender, U.; Scott, C.J.; Burrows, J.F.; Johnston, J.A. The deubiquitinating enzyme usp17 is highly expressed in tumor biopsies, is cell cycle regulated, and is required for g1-s progression. Cancer Res. 2010, 70, 3329–3339. [Google Scholar] [CrossRef] [PubMed]
- Ducker, C.; Chow, L.K.Y.; Saxton, J.; Handwerger, J.; McGregor, A.; Strahl, T.; Layfield, R.; Shaw, P.E. De-ubiquitination of elk-1 by usp17 potentiates mitogenic gene expression and cell proliferation. Nucleic Acids Res. 2019, 47, 4495–4508. [Google Scholar] [CrossRef] [PubMed]
- Fukuura, K.; Inoue, Y.; Miyajima, C.; Watanabe, S.; Tokugawa, M.; Morishita, D.; Ohoka, N.; Komada, M.; Hayashi, H. The ubiquitin-specific protease usp17 prevents cellular senescence by stabilizing the methyltransferase set8 and transcriptionally repressing p21. J. Biol. Chem. 2019, 294, 16429–16439. [Google Scholar] [CrossRef] [PubMed]
- Arceci, A.; Bonacci, T.; Wang, X.; Stewart, K.; Damrauer, J.S.; Hoadley, K.A.; Emanuele, M.J. Foxm1 deubiquitination by usp21 regulates cell cycle progression and paclitaxel sensitivity in basal-like breast cancer. Cell Rep. 2019, 26, 3076–3086. [Google Scholar] [CrossRef]
- Wu, Y.; Qin, J.; Li, F.; Yang, C.; Li, Z.; Zhou, Z.; Zhang, H.; Li, Y.; Wang, X.; Liu, R.; et al. Usp3 promotes breast cancer cell proliferation by deubiquitinating klf5. J. Biol. Chem. 2019, 294, 17837–17847. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, S.; Song, N.; Li, X.; Liu, L.; Yang, S.; Ding, X.; Shan, L.; Zhou, X.; Su, D.; et al. Stabilization of histone demethylase phf8 by usp7 promotes breast carcinogenesis. J. Clin. Investig. 2016, 126, 2205–2220. [Google Scholar] [CrossRef]
- Saldana, M.; VanderVorst, K.; Berg, A.L.; Lee, H.; Carraway, K.L. Otubain 1: A non-canonical deubiquitinase with an emerging role in cancer. Endocr. Relat. Cancer 2019, 26, R1–R14. [Google Scholar] [CrossRef]
- Piao, S.; Pei, H.Z.; Huang, B.; Baek, S.H. Ovarian tumor domain-containing protein 1 deubiquitinates and stabilizes p53. Cell. Signal. 2017, 33, 22–29. [Google Scholar] [CrossRef]
- Zhang, Z.; Fan, Y.; Xie, F.; Zhou, H.; Jin, K.; Shao, L.; Shi, W.; Fang, P.; Yang, B.; van Dam, H.; et al. Breast cancer metastasis suppressor otud1 deubiquitinates smad7. Nat. Commun. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Yuan, J.; Luo, K.; Zhang, L.; Cheville, J.C.; Lou, Z. Usp10 regulates p53 localization and stability by deubiquitinating p53. Cell 2010, 140, 384–396. [Google Scholar] [CrossRef]
- Liao, Y.; Liu, N.; Hua, X.; Cai, J.; Xia, X.; Wang, X.; Huang, H.; Liu, J. Proteasome-associated deubiquitinase ubiquitin-specific protease 14 regulates prostate cancer proliferation by deubiquitinating and stabilizing androgen receptor. Cell Death Dis. 2017, 8, e2585. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.; Jin, J.; Hu, H.; Li, H.S.; Romano, S.; Xiao, Y.; Nakaya, M.; Zhou, X.; Cheng, X.; Yang, P.; et al. Usp15 stabilizes mdm2 to mediate cancer-cell survival and inhibit antitumor t cell responses. Nat. Immunol. 2014, 15, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, P.J.; Rodon, L.; Gonzalez-Junca, A.; Dirac, A.; Gili, M.; Martinez-Saez, E.; Aura, C.; Barba, I.; Peg, V.; Prat, A.; et al. Usp15 stabilizes tgf-beta receptor i and promotes oncogenesis through the activation of tgf-beta signaling in glioblastoma. Nat. Med. 2012, 18, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, L.F.; Sparks, A.; Allende-Vega, N.; Xirodimas, D.P.; Lane, D.P.; Saville, M.K. The deubiquitinating enzyme usp2a regulates the p53 pathway by targeting mdm2. EMBO J. 2007, 26, 976–986. [Google Scholar] [CrossRef] [PubMed]
- Hock, A.K.; Vigneron, A.M.; Carter, S.; Ludwig, R.L.; Vousden, K.H. Regulation of p53 stability and function by the deubiquitinating enzyme usp42. EMBO J. 2011, 30, 4921–4930. [Google Scholar] [CrossRef]
- Cuella-Martin, R.; Oliveira, C.; Lockstone, H.E.; Snellenberg, S.; Grolmusova, N.; Chapman, J.R. 53bp1 integrates DNA repair and p53-dependent cell fate decisions via distinct mechanisms. Mol. Cell 2016, 64, 51–64. [Google Scholar] [CrossRef]
- Liu, J.; Chung, H.J.; Vogt, M.; Jin, Y.; Malide, D.; He, L.; Dundr, M.; Levens, D. Jtv1 co-activates fbp to induce usp29 transcription and stabilize p53 in response to oxidative stress. EMBO J. 2011, 30, 846–858. [Google Scholar] [CrossRef]
- Yun, S.I.; Kim, H.H.; Yoon, J.H.; Park, W.S.; Hahn, M.J.; Kim, H.C.; Chung, C.H.; Kim, K.K. Ubiquitin specific protease 4 positively regulates the wnt/beta-catenin signaling in colorectal cancer. Mol. Oncol. 2015, 9, 1834–1851. [Google Scholar] [CrossRef]
- Li, Z.; Hao, Q.; Luo, J.; Xiong, J.; Zhang, S.; Wang, T.; Bai, L.; Wang, W.; Chen, M.; Wang, W.; et al. Usp4 inhibits p53 and nf-κb through deubiquitinating and stabilizing hdac2. Oncogene 2016, 35, 2902–2912. [Google Scholar] [CrossRef]
- Zhang, X.; Berger, F.G.; Yang, J.; Lu, X. Usp4 inhibits p53 through deubiquitinating and stabilizing arf-bp1. EMBO J. 2011, 30, 2177–2189. [Google Scholar] [CrossRef]
- Luo, K.; Li, Y.; Yin, Y.; Li, L.; Wu, C.; Chen, Y.; Nowsheen, S.; Hu, Q.; Zhang, L.; Lou, Z.; et al. Usp49 negatively regulates tumorigenesis and chemoresistance through fkbp51-akt signaling. EMBO J. 2017, 36, 1434–1446. [Google Scholar] [CrossRef] [PubMed]
- Potu, H.; Peterson, L.F.; Pal, A.; Verhaegen, M.; Cao, J.; Talpaz, M.; Donato, N.J. Usp5 links suppression of p53 and fas levels in melanoma to the braf pathway. Oncotarget 2014, 5, 5559–5569. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; He, S.B.; Yao, Y.Z.; Qu, J.G.; Xie, R.; Ma, Y.Q.; Zong, M.H.; Chen, J.X. Tre2 (usp6nl) promotes colorectal cancer cell proliferation via wnt/beta-catenin pathway. Cancer Cell Int. 2019, 19, 102. [Google Scholar] [CrossRef] [PubMed]
- Cummins, J.M.; Vogelstein, B. Hausp is required for p53 destabilization. Cell Cycle 2004, 3, 689–692. [Google Scholar] [CrossRef]
- Li, M.; Brooks, C.L.; Kon, N.; Gu, W. A dynamic role of hausp in the p53-mdm2 pathway. Mol. Cell 2004, 13, 879–886. [Google Scholar] [CrossRef]
- Hu, M.; Gu, L.; Li, M.; Jeffrey, P.D.; Gu, W.; Shi, Y. Structural basis of competitive recognition of p53 and mdm2 by hausp/usp7: Implications for the regulation of the p53-mdm2 pathway. PLoS Biol. 2006, 4, e27. [Google Scholar] [CrossRef]
- Sheng, Y.; Saridakis, V.; Sarkari, F.; Duan, S.; Wu, T.; Arrowsmith, C.H.; Frappier, L. Molecular recognition of p53 and mdm2 by usp7/hausp. Nat. Struct. Mol. Biol. 2006, 13, 285–291. [Google Scholar] [CrossRef]
- Li, M.; Chen, D.; Shiloh, A.; Luo, J.; Nikolaev, A.Y.; Qin, J.; Gu, W. Deubiquitination of p53 by hausp is an important pathway for p53 stabilization. Nature 2002, 416, 648–653. [Google Scholar] [CrossRef]
- Yang, B.; Zhang, S.; Wang, Z.; Yang, C.; Ouyang, W.; Zhou, F.; Zhou, Y.; Xie, C. Deubiquitinase usp9x deubiquitinates beta-catenin and promotes high grade glioma cell growth. Oncotarget 2016, 7, 79515–79525. [Google Scholar] [CrossRef]
- Liu, H.; Chen, W.; Liang, C.; Chen, B.W.; Zhi, X.; Zhang, S.; Zheng, X.; Bai, X.; Liang, T. Wp1130 increases doxorubicin sensitivity in hepatocellular carcinoma cells through usp9x-dependent p53 degradation. Cancer Lett. 2015, 361, 218–225. [Google Scholar] [CrossRef]
- Liu, H.; Li, X.; Ning, G.; Zhu, S.; Ma, X.; Liu, X.; Liu, C.; Huang, M.; Schmitt, I.; Wullner, U.; et al. The machado-joseph disease deubiquitinase ataxin-3 regulates the stability and apoptotic function of p53. PLoS Biol. 2016, 14, e2000733. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Luo, Q.; Zhao, P.; Chang, W.; Wang, Y.; Shu, T.; Ding, F.; Li, B.; Liu, Z. Josd1 inhibits mitochondrial apoptotic signalling to drive acquired chemoresistance in gynaecological cancer by stabilizing mcl1. Cell Death Differ. 2020, 27, 55–70. [Google Scholar] [CrossRef]
- Potu, H.; Kandarpa, M.; Peterson, L.F.; Donato, N.J.; Talpaz, M. Tumor necrosis factor related apoptosis inducing ligand (trail) regulates deubiquitinase usp5 in tumor cells. Oncotarget 2019, 10, 5745–5754. [Google Scholar] [CrossRef][Green Version]
- Wang, S.; Juan, J.; Zhang, Z.; Du, Y.; Xu, Y.; Tong, J.; Cao, B.; Moran, M.F.; Zeng, Y.; Mao, X. Inhibition of the deubiquitinase usp5 leads to c-maf protein degradation and myeloma cell apoptosis. Cell Death Dis. 2017, 8, e3058. [Google Scholar] [CrossRef] [PubMed]
- Ismail, I.H.; Davidson, R.; Gagne, J.P.; Xu, Z.Z.; Poirier, G.G.; Hendzel, M.J. Germline mutations in bap1 impair its function in DNA double-strand break repair. Cancer Res. 2014, 74, 4282–4294. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Majada, V.; Welz, P.S.; Ermolaeva, M.A.; Schell, M.; Adam, A.; Dietlein, F.; Komander, D.; Buttner, R.; Thomas, R.K.; Schumacher, B.; et al. The tumour suppressor cyld regulates the p53 DNA damage response. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- de Vivo, A.; Sanchez, A.; Yegres, J.; Kim, J.; Emly, S.; Kee, Y. The otud5-ubr5 complex regulates fact-mediated transcription at damaged chromatin. Nucleic Acids Res. 2019, 47, 729–746. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, S.; Sagum, C.; Chen, J.; Singh, R.; Chaturvedi, A.; Horton, J.R.; Kashyap, T.R.; Fushman, D.; Cheng, X.; et al. Crosstalk between lys63- and lys11-polyubiquitin signaling at DNA damage sites is driven by cezanne. Genes Dev. 2019, 33, 1702–1717. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, V.; Vinas, L.; Gallego-Sanchez, A.; Andres, S.; Sacristan, M.P.; Bueno, A. Orderly progression through s-phase requires dynamic ubiquitylation and deubiquitylation of pcna. Sci. Rep. 2016, 6, 25513. [Google Scholar] [CrossRef]
- Nishi, R.; Wijnhoven, P.; le Sage, C.; Tjeertes, J.; Galanty, Y.; Forment, J.V.; Clague, M.J.; Urbe, S.; Jackson, S.P. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat. Cell Biol 2014, 16, 1016–1026, 1011–1018. [Google Scholar] [CrossRef]
- Whitehurst, C.B.; Vaziri, C.; Shackelford, J.; Pagano, J.S. Epstein-barr virus bplf1 deubiquitinates pcna and attenuates polymerase eta recruitment to DNA damage sites. J. Virol. 2012, 86, 8097–8106. [Google Scholar] [CrossRef] [PubMed]
- Castella, M.; Jacquemont, C.; Thompson, E.L.; Yeo, J.E.; Cheung, R.S.; Huang, J.W.; Sobeck, A.; Hendrickson, E.A.; Taniguchi, T. Fanci regulates recruitment of the fa core complex at sites of DNA damage independently of fancd2. PLoS Genet. 2015, 11, e1005563. [Google Scholar] [CrossRef] [PubMed]
- Nijman, S.M.; Huang, T.T.; Dirac, A.M.; Brummelkamp, T.R.; Kerkhoven, R.M.; D’Andrea, A.D.; Bernards, R. The deubiquitinating enzyme usp1 regulates the fanconi anemia pathway. Mol. Cell 2005, 17, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Olazabal-Herrero, A.; Garcia-Santisteban, I.; Rodriguez, J.A. Structure-function analysis of usp1: Insights into the role of ser313 phosphorylation site and the effect of cancer-associated mutations on autocleavage. Mol. Cancer 2015, 14, 33. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Orthwein, A.; Noordermeer, S.M.; Wilson, M.D.; Landry, S.; Enchev, R.I.; Sherker, A.; Munro, M.; Pinder, J.; Salsman, J.; Dellaire, G.; et al. A mechanism for the suppression of homologous recombination in g1 cells. Nature 2015, 528, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Zhu, Q.; Wani, G.; He, J.; Wang, Q.E.; Wani, A.A. Usp3 counteracts rnf168 via deubiquitinating h2a and gammah2ax at lysine 13 and 15. Cell Cycle 2014, 13, 106–114. [Google Scholar] [CrossRef]
- Uckelmann, M.; Densham, R.M.; Baas, R.; Winterwerp, H.H.K.; Fish, A.; Sixma, T.K.; Morris, J.R. Usp48 restrains resection by site-specific cleavage of the brca1 ubiquitin mark from h2a. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Zhu, Q.; Sharma, N.; He, J.; Wani, G.; Wani, A.A. Usp7 deubiquitinase promotes ubiquitin-dependent DNA damage signaling by stabilizing rnf168. Cell Cycle 2015, 14, 1413–1425. [Google Scholar] [CrossRef]
- McGarry, E.; Gaboriau, D.; Rainey, M.D.; Restuccia, U.; Bachi, A.; Santocanale, C. The deubiquitinase usp9x maintains DNA replication fork stability and DNA damage checkpoint responses by regulating claspin during s-phase. Cancer Res. 2016, 76, 2384–2393. [Google Scholar] [CrossRef]
- Kovalenko, A.; Chable-Bessia, C.; Cantarella, G.; Israel, A.; Wallach, D.; Courtois, G. The tumour suppressor cyld negatively regulates nf-kappa b signalling by deubiquitination. Nature 2003, 424, 801–805. [Google Scholar] [CrossRef]
- Trompouki, E.; Hatzivassiliou, E.; Tsichritzis, T.; Farmer, H.; Ashworth, A.; Mosialos, G. Cyld is a deubiquitinating enzyme that negatively regulates nf-kappa b activation by tnfr family members. Nature 2003, 424, 793–796. [Google Scholar] [CrossRef] [PubMed]
- Brummelkamp, T.R.; Nijman, S.M.; Dirac, A.M.; Bernards, R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating nf-kappab. Nature 2003, 424, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.C.; Lin, Y.C.; Liu, C.H.; Chung, H.C.; Wang, Y.T.; Lin, Y.W.; Ma, H.I.; Tu, P.H.; Lawler, S.E.; Chen, R.H. Usp11 regulates pml stability to control notch-induced malignancy in brain tumours. Nat. Commun. 2014, 5, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Zhang, R.; Su, M.; Liu, W. Usp13 serves as a tumor suppressor via the pten/akt pathway in oral squamous cell carcinoma. Cancer Manag. Res. 2019, 11, 9175–9183. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Stevens, P.D.; Yang, H.; Gulhati, P.; Wang, W.; Evers, B.M.; Gao, T. The deubiquitination enzyme usp46 functions as a tumor suppressor by controlling phlpp-dependent attenuation of akt signaling in colon cancer. Oncogene 2013, 32, 471–478. [Google Scholar] [CrossRef]
- Bott, M.; Brevet, M.; Taylor, B.S.; Shimizu, S.; Ito, T.; Wang, L.; Creaney, J.; Lake, R.A.; Zakowski, M.F.; Reva, B.; et al. The nuclear deubiquitinase bap1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat. Genet. 2011, 43, 668–672. [Google Scholar] [CrossRef]
- Kim, D.; Hong, A.; Park, H.I.; Shin, W.H.; Yoo, L.; Jeon, S.J.; Chung, K.C. Deubiquitinating enzyme usp22 positively regulates c-myc stability and tumorigenic activity in mammalian and breast cancer cells. J. Cell Physiol. 2017, 232, 3664–3676. [Google Scholar] [CrossRef]
- Diefenbacher, M.E.; Popov, N.; Blake, S.M.; Schulein-Volk, C.; Nye, E.; Spencer-Dene, B.; Jaenicke, L.A.; Eilers, M.; Behrens, A. The deubiquitinase usp28 controls intestinal homeostasis and promotes colorectal cancer. J. Clin. Inv. 2014, 124, 3407–3418. [Google Scholar] [CrossRef]
- Khan, O.M.; Carvalho, J.; Spencer-Dene, B.; Mitter, R.; Frith, D.; Snijders, A.P.; Wood, S.A.; Behrens, A. The deubiquitinase usp9x regulates fbw7 stability and suppresses colorectal cancer. J. Clin. Inv. 2018, 128, 1326–1337. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Lin, Y.; Liu, Y.; Wang, Y.; Jia, J.; Singh, P.; Chi, Y.I.; Wang, C.; Dong, C.; et al. Dub3 inhibition suppresses breast cancer invasion and metastasis by promoting snail1 degradation. Nat. Commun. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Lin, Y.; Wang, Y.; Shi, Q.; Yu, Q.; Liu, C.; Feng, J.; Deng, J.; Evers, B.M.; Zhou, B.P.; Wu, Y. Stabilization of the transcription factors slug and twist by the deubiquitinase dub3 is a key requirement for tumor metastasis. Oncotarget 2017, 8, 75127–75140. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, Y.; Zhu, R.; Ding, F.; Cao, X.; Lin, D.; Liu, Z. Otub1 promotes esophageal squamous cell carcinoma metastasis through modulating snail stability. Oncogene 2018, 37, 3356–3368. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhang, S.; Wu, H.; Lu, J.; Lu, Y.; Wang, F.; Zhao, W.; Zhan, P.; Lu, J.; Fang, Q.; et al. Deubiquitinase psmd14 enhances hepatocellular carcinoma growth and metastasis by stabilizing grb2. Cancer lett. 2020, 469, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Liu, W.; Li, J. Usp17 is upregulated in osteosarcoma and promotes cell proliferation, metastasis, and epithelial-mesenchymal transition through stabilizing smad4. Tumour Biol. 2017, 39, 1010428317717138. [Google Scholar] [CrossRef]
- Wu, X.; Liu, M.; Zhu, H.; Wang, J.; Dai, W.; Li, J.; Zhu, D.; Tang, W.; Xiao, Y.; Lin, J.; et al. Ubiquitin-specific protease 3 promotes cell migration and invasion by interacting with and deubiquitinating suz12 in gastric cancer. J. Exp. Clin. Cancer Res. 2019, 38, 277. [Google Scholar] [CrossRef]
- Sapmaz, A.; Berlin, I.; Bos, E.; Wijdeven, R.H.; Janssen, H.; Konietzny, R.; Akkermans, J.J.; Erson-Bensan, A.E.; Koning, R.I.; Kessler, B.M.; et al. Usp32 regulates late endosomal transport and recycling through deubiquitylation of rab7. Nat. Commun. 2019, 10, 1–18. [Google Scholar] [CrossRef]
- Kim, J.O.; Kim, S.R.; Lim, K.H.; Kim, J.H.; Ajjappala, B.; Lee, H.J.; Choi, J.I.; Baek, K.H. Deubiquitinating enzyme usp37 regulating oncogenic function of 14-3-3gamma. Oncotarget 2015, 6, 36551–36576. [Google Scholar] [CrossRef]
- Yuan, J.; Luo, K.; Deng, M.; Li, Y.; Yin, P.; Gao, B.; Fang, Y.; Wu, P.; Liu, T.; Lou, Z. Herc2-usp20 axis regulates DNA damage checkpoint through claspin. Nucleic Acids Res. 2014, 42, 13110–13121. [Google Scholar] [CrossRef]
- Jiang, Y.Z.; Ma, D.; Suo, C.; Shi, J.; Xue, M.; Hu, X.; Xiao, Y.; Yu, K.D.; Liu, Y.R.; Yu, Y.; et al. Genomic and transcriptomic landscape of triple-negative breast cancers: Subtypes and treatment strategies. Cancer Cell 2019, 35, 428–440. [Google Scholar] [CrossRef]
- Chiu, H.W.; Lin, H.Y.; Tseng, I.J.; Lin, Y.F. Otud7b upregulation predicts a poor response to paclitaxel in patients with triple-negative breast cancer. Oncotarget 2018, 9, 553–565. [Google Scholar] [CrossRef]
- Lin, D.D.; Shen, Y.; Qiao, S.; Liu, W.W.; Zheng, L.; Wang, Y.N.; Cui, N.; Wang, Y.F.; Zhao, S.; Shi, J.H. Upregulation of otud7b (cezanne) promotes tumor progression via akt/vegf pathway in lung squamous carcinoma and adenocarcinoma. Front. Oncol. 2019, 9, 862. [Google Scholar] [CrossRef] [PubMed]
- Anastas, J.N.; Zee, B.M.; Kalin, J.H.; Kim, M.; Guo, R.; Alexandrescu, S.; Blanco, M.A.; Giera, S.; Gillespie, S.M.; Das, J.; et al. Re-programing chromatin with a bifunctional lsd1/hdac inhibitor induces therapeutic differentiation in dipg. Cancer Cell 2019, 36, 528–544. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Kim, T.; Nguyen, T.; Hahn, M.J.; Yun, S.I.; Kim, K.K. A selective inhibitor of ubiquitin-specific protease 4 suppresses colorectal cancer progression by regulating beta-catenin signaling. Cell. Physiol. Biochem.: Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2019, 53, 157–171. [Google Scholar]
- Brooks, C.L.; Gu, W. P53 regulation by ubiquitin. FEBS Lett. 2011, 585, 2803–2809. [Google Scholar] [CrossRef]
- Brooks, C.L.; Li, M.; Hu, M.; Shi, Y.; Gu, W. The p53--mdm2--hausp complex is involved in p53 stabilization by hausp. Oncogene 2007, 26, 7262–7266. [Google Scholar] [CrossRef]
- Li, F.; Han, H.; Sun, Q.; Liu, K.; Lin, N.; Xu, C.; Zhao, Z.; Zhao, W. Usp28 regulates deubiquitination of histone h2a and cell proliferation. Exp. Cell Res. 2019, 379, 11–18. [Google Scholar] [CrossRef]
- Niendorf, S.; Oksche, A.; Kisser, A.; Lohler, J.; Prinz, M.; Schorle, H.; Feller, S.; Lewitzky, M.; Horak, I.; Knobeloch, K.P. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol. Cell Biol. 2007, 27, 5029–5039. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Fernald, K.; Kurokawa, M. Evading apoptosis in cancer. Trends Cell Biol. 2013, 23, 620–633. [Google Scholar] [CrossRef] [PubMed]
- Burrows, J.F.; McGrattan, M.J.; Rascle, A.; Humbert, M.; Baek, K.H.; Johnston, J.A. Dub-3, a cytokine-inducible deubiquitinating enzyme that blocks proliferation. J. Biol. Chem. 2004, 279, 13993–14000. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Takanashi, M.; Oikawa, K.; Tanaka, M.; Nishi, H.; Isaka, K.; Kudo, M.; Kuroda, M. Usp15 plays an essential role for caspase-3 activation during paclitaxel-induced apoptosis. Biochem Biophys Res. Commun 2009, 388, 366–371. [Google Scholar] [CrossRef]
- Jin, Z.; Li, Y.; Pitti, R.; Lawrence, D.; Pham, V.C.; Lill, J.R.; Ashkenazi, A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 2009, 137, 721–735. [Google Scholar] [CrossRef]
- Crook, N.E.; Clem, R.J.; Miller, L.K. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 1993, 67, 2168–2174. [Google Scholar] [CrossRef]
- Mei, Y.; Hahn, A.A.; Hu, S.; Yang, X. The usp19 deubiquitinase regulates the stability of c-iap1 and c-iap2. J. Biol. Chem. 2011, 286, 35380–35387. [Google Scholar] [CrossRef]
- Goncharov, T.; Niessen, K.; de Almagro, M.C.; Izrael-Tomasevic, A.; Fedorova, A.V.; Varfolomeev, E.; Arnott, D.; Deshayes, K.; Kirkpatrick, D.S.; Vucic, D. Otub1 modulates c-iap1 stability to regulate signalling pathways. EMBO J. 2013, 32, 1103–1114. [Google Scholar] [CrossRef]
- Engel, K.; Rudelius, M.; Slawska, J.; Jacobs, L.; Ahangarian Abhari, B.; Altmann, B.; Kurutz, J.; Rathakrishnan, A.; Fernandez-Saiz, V.; Brunner, A.; et al. Usp9x stabilizes xiap to regulate mitotic cell death and chemoresistance in aggressive b-cell lymphoma. EMBO Mol. Med. 2016, 8, 851–862. [Google Scholar] [CrossRef]
- Weber, A.; Heinlein, M.; Dengjel, J.; Alber, C.; Singh, P.K.; Hacker, G. The deubiquitinase usp27x stabilizes the bh3-only protein bim and enhances apoptosis. EMBO Rep. 2016, 17, 724–738. [Google Scholar] [CrossRef]
- Ghosal, G.; Chen, J. DNA damage tolerance: A double-edged sword guarding the genome. Transl. Cancer Res. 2013, 2, 107–129. [Google Scholar] [PubMed]
- Hoeijmakers, J.H. Genome maintenance mechanisms for preventing cancer. Nature 2001, 411, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Le, J.; Perez, E.; Nemzow, L.; Gong, F. Role of deubiquitinases in DNA damage response. DNA Repair 2019, 76, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef]
- Shanmugam, I.; Abbas, M.; Ayoub, F.; Mirabal, S.; Bsaili, M.; Caulder, E.K.; Weinstock, D.M.; Tomkinson, A.E.; Hromas, R.; Shaheen, M. Ubiquitin-specific peptidase 20 regulates rad17 stability, checkpoint kinase 1 phosphorylation and DNA repair by homologous recombination. J. Biol. Chem. 2014, 289, 22739–22748. [Google Scholar] [CrossRef]
- Yoshida, K.; Miki, Y. Role of brca1 and brca2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004, 95, 866–871. [Google Scholar] [CrossRef]
- Meyer, T.; Jahn, N.; Lindner, S.; Rohner, L.; Dolnik, A.; Weber, D.; Scheffold, A.; Kopff, S.; Paschka, P.; Gaidzik, V.I.; et al. Functional characterization of brcc3 mutations in acute myeloid leukemia with t(8;21)(q22;q22.1). Leukemia 2020, 34, 404–415. [Google Scholar] [CrossRef]
- Lancini, C.; van den Berk, P.C.; Vissers, J.H.; Gargiulo, G.; Song, J.Y.; Hulsman, D.; Serresi, M.; Tanger, E.; Blom, M.; Vens, C.; et al. Tight regulation of ubiquitin-mediated DNA damage response by usp3 preserves the functional integrity of hematopoietic stem cells. J. Exp. Med. 2014, 211, 1759–1777. [Google Scholar] [CrossRef]
- Bignell, G.R.; Warren, W.; Seal, S.; Takahashi, M.; Rapley, E.; Barfoot, R.; Green, H.; Brown, C.; Biggs, P.J.; Lakhani, S.R.; et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 2000, 25, 160–165. [Google Scholar] [CrossRef]
- Strobel, P.; Zettl, A.; Ren, Z.; Starostik, P.; Riedmiller, H.; Storkel, S.; Muller-Hermelink, H.K.; Marx, A. Spiradenocylindroma of the kidney: Clinical and genetic findings suggesting a role of somatic mutation of the cyld1 gene in the oncogenesis of an unusual renal neoplasm. Am. J. Surg. Pathol. 2002, 26, 119–124. [Google Scholar] [CrossRef]
- Hirai, Y.; Kawamata, Y.; Takeshima, N.; Furuta, R.; Kitagawa, T.; Kawaguchi, T.; Hasumi, K.; Sugai, S.; Noda, T. Conventional and array-based comparative genomic hybridization analyses of novel cell lines harboring hpv18 from glassy cell carcinoma of the uterine cervix. Int. J. Oncol. 2004, 24, 977–986. [Google Scholar] [CrossRef] [PubMed]
- Reiley, W.; Zhang, M.Y.; Sun, S.C. Negative regulation of jnk signaling by the tumor suppressor cyld. J. Biol. Chem. 2004, 279, 55161–55167. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, F.; Dikic, I. Cyld in ubiquitin signaling and tumor pathogenesis. Cell 2006, 125, 643–645. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Z.; Inuzuka, H.; Fukushima, H.; Wan, L.; Gao, D.; Shaik, S.; Sarkar, F.H.; Wei, W. Emerging roles of the fbw7 tumour suppressor in stem cell differentiation. EMBO Rep. 2011, 13, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. Emt: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Chen, Z.; Wu, X.; Cai, X.; Feng, S.; Lu, J.; Wang, H.; Liu, N. Ubiquitin-specific protease 3 promotes glioblastoma cell invasion and epithelial-mesenchymal transition via stabilizing snail. Mol. Cancer Res. MCR 2019, 17, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Hiraoka, E.; Mimae, T.; Ito, M.; Kadoya, T.; Miyata, Y.; Ito, A.; Okada, M. Correction to: Breast cancer cell motility is promoted by 14-3-3gamma. Breast Cancer 2019, 26, 594. [Google Scholar] [CrossRef]
- Raungrut, P.; Wongkotsila, A.; Champoochana, N.; Lirdprapamongkol, K.; Svasti, J.; Thongsuksai, P. Knockdown of 14-3-3gamma suppresses epithelial-mesenchymal transition and reduces metastatic potential of human non-small cell lung cancer cells. Anticancer Res. 2018, 38, 3507–3514. [Google Scholar] [CrossRef]
- Alonso-Curbelo, D.; Riveiro-Falkenbach, E.; Perez-Guijarro, E.; Cifdaloz, M.; Karras, P.; Osterloh, L.; Megias, D.; Canon, E.; Calvo, T.G.; Olmeda, D.; et al. Rab7 controls melanoma progression by exploiting a lineage-specific wiring of the endolysosomal pathway. Cancer Cell 2014, 26, 61–76. [Google Scholar] [CrossRef]
- Wang, T.; Ming, Z.; Xiaochun, W.; Hong, W. Rab7: Role of its protein interaction cascades in endo-lysosomal traffic. Cell. Sign. 2011, 23, 516–521. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, L.; Wang, S.; Wang, T. Rab7: Roles in membrane trafficking and disease. Biosci. Rep. 2009, 29, 193–209. [Google Scholar] [CrossRef] [PubMed]
- Richardson, P.G.; Barlogie, B.; Berenson, J.; Singhal, S.; Jagannath, S.; Irwin, D.; Rajkumar, S.V.; Srkalovic, G.; Alsina, M.; Alexanian, R.; et al. A phase 2 study of bortezomib in relapsed, refractory myeloma. N. Engl. J. Med. 2003, 348, 2609–2617. [Google Scholar] [CrossRef] [PubMed]
- Richardson, P.G.; Sonneveld, P.; Schuster, M.W.; Irwin, D.; Stadtmauer, E.A.; Facon, T.; Harousseau, J.L.; Ben-Yehuda, D.; Lonial, S.; Goldschmidt, H.; et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N. Engl. J. Med. 2005, 352, 2487–2498. [Google Scholar] [CrossRef] [PubMed]
- Cacan, E.; Ozmen, Z.C. Regulation of fas in response to bortezomib and epirubicin in colorectal cancer cells. J. Chemother. 2020, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Okazuka, K.; Ishida, T. Proteasome inhibitors for multiple myelomaJap. J. Clin. Oncol. 2018, 48, 785–793. [Google Scholar]
- Kuhn, D.J.; Chen, Q.; Voorhees, P.M.; Strader, J.S.; Shenk, K.D.; Sun, C.M.; Demo, S.D.; Bennett, M.K.; van Leeuwen, F.W.; Chanan-Khan, A.A.; et al. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood 2007, 110, 3281–3290. [Google Scholar] [CrossRef] [PubMed]
- Jakubowiak, A.J.; Dytfeld, D.; Griffith, K.A.; Lebovic, D.; Vesole, D.H.; Jagannath, S.; Al-Zoubi, A.; Anderson, T.; Nordgren, B.; Detweiler-Short, K.; et al. A phase 1/2 study of carfilzomib in combination with lenalidomide and low-dose dexamethasone as a frontline treatment for multiple myeloma. Blood 2012, 120, 1801–1809. [Google Scholar] [CrossRef]
- Richardson, P.G.; Zweegman, S.; O’Donnell, E.K.; Laubach, J.P.; Raje, N.; Voorhees, P.; Ferrari, R.H.; Skacel, T.; Kumar, S.K.; Lonial, S. Ixazomib for the treatment of multiple myeloma. Expert Opin. Pharmacother. 2018, 19, 1949–1968. [Google Scholar] [CrossRef]
- Moreau, P.; Masszi, T.; Grzasko, N.; Bahlis, N.J.; Hansson, M.; Pour, L.; Sandhu, I.; Ganly, P.; Baker, B.W.; Jackson, S.R.; et al. Oral ixazomib, lenalidomide, and dexamethasone for multiple myeloma. N. Engl. J. Med. 2016, 374, 1621–1634. [Google Scholar] [CrossRef]
- Petroski, M.D. The ubiquitin system, disease, and drug discovery. BMC Biochem. 2008, 9, S7. [Google Scholar] [CrossRef]
- Cheon, K.W.; Baek, K.H. Hausp as a therapeutic target for hematopoietic tumors (review). Int. J. Oncol. 2006, 28, 1209–1215. [Google Scholar] [CrossRef] [PubMed]
- Issaeva, N.; Bozko, P.; Enge, M.; Protopopova, M.; Verhoef, L.G.; Masucci, M.; Pramanik, A.; Selivanova, G. Small molecule rita binds to p53, blocks p53-hdm-2 interaction and activates p53 function in tumors. Nature Med. 2004, 10, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In vivo activation of the p53 pathway by small-molecule antagonists of mdm2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef] [PubMed]
- Tovar, C.; Rosinski, J.; Filipovic, Z.; Higgins, B.; Kolinsky, K.; Hilton, H.; Zhao, X.; Vu, B.T.; Qing, W.; Packman, K.; et al. Small-molecule mdm2 antagonists reveal aberrant p53 signaling in cancer: Implications for therapy. Proc. Natl. Acad. Sci. USA 2006, 103, 1888–1893. [Google Scholar] [CrossRef]
- Stuhmer, T.; Chatterjee, M.; Hildebrandt, M.; Herrmann, P.; Gollasch, H.; Gerecke, C.; Theurich, S.; Cigliano, L.; Manz, R.A.; Daniel, P.T.; et al. Nongenotoxic activation of the p53 pathway as a therapeutic strategy for multiple myeloma. Blood 2005, 106, 3609–3617. [Google Scholar] [CrossRef]
- Colland, F.; Formstecher, E.; Jacq, X.; Reverdy, C.; Planquette, C.; Conrath, S.; Trouplin, V.; Bianchi, J.; Aushev, V.N.; Camonis, J.; et al. Small-molecule inhibitor of usp7/hausp ubiquitin protease stabilizes and activates p53 in cells. Mol. Cancer Ther. 2009, 8, 2286–2295. [Google Scholar] [CrossRef]
- Chitta, K.; Paulus, A.; Akhtar, S.; Blake, M.K.; Caulfield, T.R.; Novak, A.J.; Ansell, S.M.; Advani, P.; Ailawadhi, S.; Sher, T.; et al. Targeted inhibition of the deubiquitinating enzymes, usp14 and uchl5, induces proteotoxic stress and apoptosis in waldenstrom macroglobulinaemia tumour cells. Br. J. Haematol. 2015, 169, 377–390. [Google Scholar] [CrossRef]
- Vogel, R.I.; Pulver, T.; Heilmann, W.; Mooneyham, A.; Mullany, S.; Zhao, X.; Shahi, M.; Richter, J.; Klein, M.; Chen, L.; et al. Usp14 is a predictor of recurrence in endometrial cancer and a molecular target for endometrial cancer treatment. Oncotarget 2016, 7, 30962–30976. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, X.; Lin, P.; He, Y.; Zhang, Y.; Cao, B.; Zhang, Z.; Sethi, G.; Liu, J.; Zhou, X.; et al. Inhibition of the deubiquitinase usp9x induces pre-b cell homeobox 1 (pbx1) degradation and thereby stimulates prostate cancer cell apoptosis. J. Biol. Chem. 2019, 294, 4572–4582. [Google Scholar] [CrossRef]
- Byun, S.; Lee, S.Y.; Lee, J.; Jeong, C.H.; Farrand, L.; Lim, S.; Reddy, K.; Kim, J.Y.; Lee, M.H.; Lee, H.J.; et al. Usp8 is a novel target for overcoming gefitinib resistance in lung cancer. Clin. Cancer Res. 2013, 19, 3894–3904. [Google Scholar] [CrossRef]
- Liu, Y.; Lashuel, H.A.; Choi, S.; Xing, X.; Case, A.; Ni, J.; Yeh, L.A.; Cuny, G.D.; Stein, R.L.; Lansbury, P.T., Jr. Discovery of inhibitors that elucidate the role of uch-l1 activity in the h1299 lung cancer cell line. Chem. Biol. 2003, 10, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; D’Arcy, P.; Wang, X.; Ray, A.; Tai, Y.T.; Hu, Y.; Carrasco, R.D.; Richardson, P.; Linder, S.; Chauhan, D.; et al. A novel small molecule inhibitor of deubiquitylating enzyme usp14 and uchl5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood 2014, 123, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Mistry, H.; Hsieh, G.; Buhrlage, S.J.; Huang, M.; Park, E.; Cuny, G.D.; Galinsky, I.; Stone, R.M.; Gray, N.S.; D’Andrea, A.D.; et al. Small-molecule inhibitors of usp1 target id1 degradation in leukemic cells. Mol. Cancer Ther. 2013, 12, 2651–2662. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Dexheimer, T.S.; Zhang, P.; Rosenthal, A.S.; Villamil, M.A.; You, C.; Zhang, Q.; Chen, J.; Ott, C.A.; Sun, H.; et al. A selective usp1-uaf1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 2014, 10, 298–304. [Google Scholar] [CrossRef]
- Chen, J.; Dexheimer, T.S.; Ai, Y.; Liang, Q.; Villamil, M.A.; Inglese, J.; Maloney, D.J.; Jadhav, A.; Simeonov, A.; Zhuang, Z. Selective and cell-active inhibitors of the usp1/uaf1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem. Biol. 2011, 18, 1390–1400. [Google Scholar] [CrossRef]
- Davis, M.I.; Pragani, R.; Fox, J.T.; Shen, M.; Parmar, K.; Gaudiano, E.F.; Liu, L.; Tanega, C.; McGee, L.; Hall, M.D.; et al. Small molecule inhibition of the ubiquitin-specific protease usp2 accelerates cyclin d1 degradation and leads to cell cycle arrest in colorectal cancer and mantle cell lymphoma models. J. Biol. Chem. 2016, 291, 24628–24640. [Google Scholar] [CrossRef]
- Issaenko, O.A.; Amerik, A.Y. Chalcone-based small-molecule inhibitors attenuate malignant phenotype via targeting deubiquitinating enzymes. Cell Cycle 2012, 11, 1804–1817. [Google Scholar] [CrossRef]
- Okada, K.; Ye, Y.Q.; Taniguchi, K.; Yoshida, A.; Akiyama, T.; Yoshioka, Y.; Onose, J.; Koshino, H.; Takahashi, S.; Yajima, A.; et al. Vialinin a is a ubiquitin-specific peptidase inhibitor. Bioorg. Med. Chem. Lett. 2013, 23, 4328–4331. [Google Scholar] [CrossRef]
- Chauhan, D.; Tian, Z.; Nicholson, B.; Kumar, K.G.; Zhou, B.; Carrasco, R.; McDermott, J.L.; Leach, C.A.; Fulcinniti, M.; Kodrasov, M.P.; et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell 2012, 22, 345–358. [Google Scholar] [CrossRef]
- Weinstock, J.; Wu, J.; Cao, P.; Kingsbury, W.D.; McDermott, J.L.; Kodrasov, M.P.; McKelvey, D.M.; Suresh Kumar, K.G.; Goldenberg, S.J.; Mattern, M.R.; et al. Selective dual inhibitors of the cancer-related deubiquitylating proteases usp7 and usp47. ACS Med. Chem. Lett. 2012, 3, 789–792. [Google Scholar] [CrossRef]
- Pal, A.; Young, M.A.; Donato, N.J. Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Res. 2014, 74, 4955–4966. [Google Scholar] [CrossRef] [PubMed]
- Kapuria, V.; Peterson, L.F.; Fang, D.; Bornmann, W.G.; Talpaz, M.; Donato, N.J. Deubiquitinase inhibition by small-molecule wp1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010, 70, 9265–9276. [Google Scholar] [CrossRef]
- Zhou, B.; Zuo, Y.; Li, B.; Wang, H.; Liu, H.; Wang, X.; Qiu, X.; Hu, Y.; Wen, S.; Du, J.; et al. Deubiquitinase inhibition of 19s regulatory particles by 4-arylidene curcumin analog ac17 causes nf-kappab inhibition and p53 reactivation in human lung cancer cells. Mol. Cancer Ther. 2013, 12, 1381–1392. [Google Scholar] [CrossRef] [PubMed]
- D’Arcy, P.; Linder, S. Proteasome deubiquitinases as novel targets for cancer therapy. Int. J. Biochem. Cell Biol. 2012, 44, 1729–1738. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; D’Arcy, P.; Caulfield, T.R.; Paulus, A.; Chitta, K.; Mohanty, C.; Gullbo, J.; Chanan-Khan, A.; Linder, S. Synthesis and evaluation of derivatives of the proteasome deubiquitinase inhibitor b-ap15. Chem. Biol. Drug Des. 2015, 86, 1036–1048. [Google Scholar] [CrossRef] [PubMed]

| Organelle | DUBs |
|---|---|
| Nucleolus | USP36, USP39 |
| Nucleus | BAP1, MYSM1, USP1, USP11, USP22, USP26, USP28, USP29, USP3, USP42, USP44, USP49, USP51, USP7, USPL1, ZUP1 |
| Golgi | USP32, USP33 |
| Endoplasmic reticulum | ATXN3, USP13, USP19, USP33, YOD1 |
| Microtubules | CYLD, USP21 |
| Centriole | USP21, USP33, USP9X |
| Early endosome and multivesicular body | AMSH, AMSH-LP, USP2a, USP8 |
| Lipid droplet | USP35 |
| Peroxisome and mitochondrion | USP30 |
| Cajal body | USPL1 |
| Stress granule | USP10, USP13, USP5 |
| Plasma membrane | JOSD1, USP6 |
| Cytoplasm | A20, CYLD, PSMD14, UCHL5, USP14 |
| Functions | DUBs | Targets | References |
|---|---|---|---|
| Cell cycle control | BAP1 | KLF5 | [22] |
| DUB3 | cyclin A | [23] | |
| OTUD6B-2 | cyclin D1 and c-Myc | [24] | |
| OTUD7B | APC/C, GβL, HIF2α and E2F1 | [25,26,27,28] | |
| USP10 | SKP2, Bcr-Abl | [29] | |
| USP14 | AR | [30] | |
| USP17 | p21, ELK-1, Su(var)3-9, Enhancer-of-zeste, and Trithorax domain-containing protein 8 | [31,32,33] | |
| USP21 | FOXM1 | [34] | |
| USP3 | KLF5 | [35] | |
| USP7 | PHF8 | [36] | |
| Cell proliferation | OTUB1 | p53 | [37] |
| OTUD1 | p53, SMAD7 | [38,39] | |
| USP10 | p53 | [40] | |
| USP14 | AR | [41] | |
| USP15 | MDM2, TGF-β receptor | [42,43] | |
| USP2 | MDM2 | [44] | |
| USP28 | p53, p21, and p16INK4a | [45,46] | |
| USP29 | p53 | [47] | |
| USP4 | β-catenin, p53 and NF-κB | [48,49,50] | |
| USP42 | P53 | [45] | |
| USP49 | FKBP51 | [51] | |
| USP5 | P53 | [52] | |
| USP6NL | β-catenin | [53] | |
| USP7 | MDM2 | [54,55,56,57,58] | |
| USP9X | β-catenin, p53 | [59,60] | |
| Cell apoptosis | ATXN3 | p53 | [61] |
| JOSD1 | MCL1 | [62] | |
| USP5 | p53, MAF bZIP | [63,64] | |
| DNA damage repair | BAP1 | PR-DUB | [65] |
| CYLD | p53 | [66] | |
| OTUD5 | SPT16 | [67] | |
| OTUD7A | Rap80/BRCA1-A complex | [68] | |
| OTUD7B | Rap80/BRCA1-A complex | [68] | |
| UBP12 | PCNA | [69] | |
| UBP2 | PCNA | [69] | |
| UCHL5 | NFRKB | [70] | |
| USP1 | PCNA | [71,72,73,74] | |
| USP11 | BRCA2 | [75] | |
| UBP15 | PCNA | [69] | |
| USP3 | γH2AX and H2A | [76] | |
| USP48 | BRCA1 | [77] | |
| USP7 | PHF8, pBmi1, Bmi1, RNF168, and BRCA1 | [36,78] | |
| USP9X | claspin | [79] | |
| Tumor suppression | CYLD | tumor necrosis factor receptor-associated factor 2, IKKγ | [80,81,82] |
| USP11 | PML | [83] | |
| USP13 | PTEN | [84] | |
| USP46 | PHLPP | [85] | |
| Oncogene | BAP1 | ASXL1 | [86] |
| USP22 | c-Myc | [87] | |
| USP28 | MYC | [88] | |
| USP9X | FBW7 | [89] | |
| Metastasis | DUB3 | Snail, Slug and Twist | [90,91] |
| OTUB1 | Snail | [92] | |
| PSMD14 | GRB2 | [93] | |
| USP17 | SMAD4 | [94] | |
| USP3 | SUZ12 | [95] | |
| USP32 | RAB7 | [96] | |
| USP37 | 14-3-3γ | [97] |
| DUBs | DUBs Inhibitors | Therapeutic Targets | Functional Effects | References |
|---|---|---|---|---|
| USP8 | 9-Ethyloxyimino-9H-indeno[1,2-b]pyrazine-2,3-dicarbonitrile | Non-small cell lung cancer | Downregulation of receptor tyrosine kinases including EGFR, ERBB2, ERBB3, and MET | [160] |
| UCHL1 | LDN-57444 | Lung cancer cell line | Inhibit proliferation | [161] |
| UCHL1, UCHL3 | TCID | Multiple myeloma | Induce apoptosis | [162] |
| USP1 | Pimozide | Leukemic cell lines | Promoted the degradation of ID1 | [163] |
| USP1-UAF1 | ML323 | Non-small cell lung cancer and osteosarcoma cells | Induced DNA damage | [164] |
| USP1-UAF1 | Pimozide and GW7647 | Non–small cell lung cancer | Inhibit cell proliferation | [165] |
| USP2 | ML346 | Colorectal cancer nad mantle cell lymphoma | Accelerate cyclin D1 degradation, cell cycle arrest | [166] |
| USP2a/USP2b/USP5/USP8 | AM146, RA-9 and RA-14 | Breast, ovarian and cervical cancer cell lines | Downregulation cell-cycle promoter, and upregulation of tumor suppressor | [167] |
| USP5/IsoT, USP4 | Vialinin A | Basophilic leukemia cells | Inhibit the release of TNFα | [168] |
| USP7 | HBX 41,108 | Colorectal carcinoma | Induced p53-dependent apoptosis | [156] |
| USP7/USP47 | P5091 and Compound 1 | Multiple myeloma | Induce apoptosis, inhibit tumor growth | [169,170] |
| USP9X/USP5/USP24 | WP1130 | Mantle cell lymphoma | Downregulation of antiapoptotic and upregulation of proapoptotic proteins, such as MCL-1 and p53 | [171,172] |
| USP14/ UCHL5 | AC17 | Human lung cancer cells | Inhibit NFκB pathway and reactive p53 | [173] |
| USP14/UCHL5 | b-AP15 (WO2013058691) | Multiple myeloma/ colorectal carcinoma | Downregulation of CDC25C, CDC2, and cyclin B1/ overexpression of the anti-apoptotic mediator Bcl-2 and anti-tumor activity | [162,174] |
| USP14/UCHL5 | VLX1570 | Colon carcinoma cell | Inhibit proteasome DUB activity | [175] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, K.P.; Chen, J.; Tse, W.K.F. Role of Deubiquitinases in Human Cancers: Potential Targeted Therapy. Int. J. Mol. Sci. 2020, 21, 2548. https://doi.org/10.3390/ijms21072548
Lai KP, Chen J, Tse WKF. Role of Deubiquitinases in Human Cancers: Potential Targeted Therapy. International Journal of Molecular Sciences. 2020; 21(7):2548. https://doi.org/10.3390/ijms21072548
Chicago/Turabian StyleLai, Keng Po, Jian Chen, and William Ka Fai Tse. 2020. "Role of Deubiquitinases in Human Cancers: Potential Targeted Therapy" International Journal of Molecular Sciences 21, no. 7: 2548. https://doi.org/10.3390/ijms21072548
APA StyleLai, K. P., Chen, J., & Tse, W. K. F. (2020). Role of Deubiquitinases in Human Cancers: Potential Targeted Therapy. International Journal of Molecular Sciences, 21(7), 2548. https://doi.org/10.3390/ijms21072548





