CDK6 Inhibition: A Novel Approach in AML Management
Abstract
1. Introduction
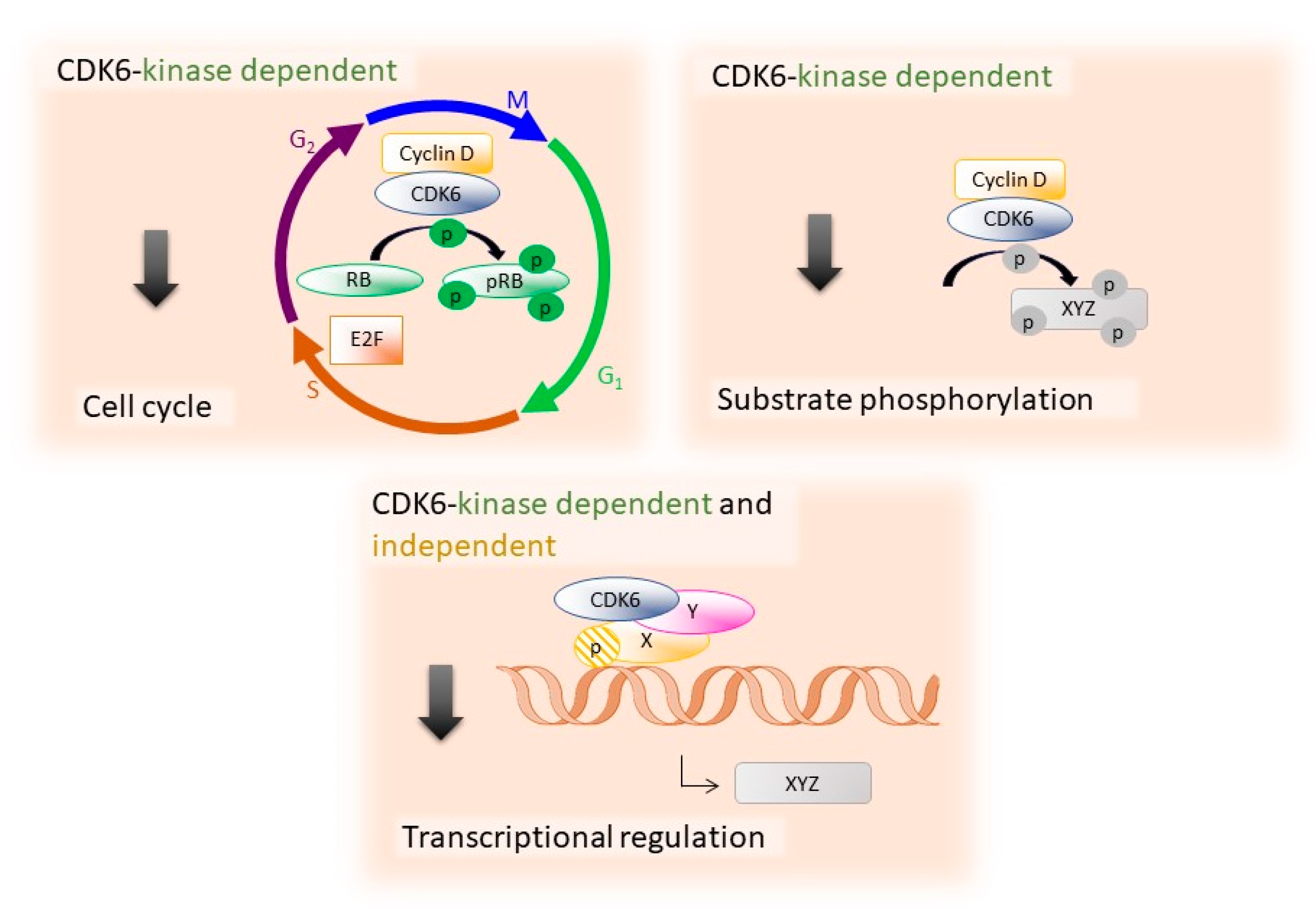
2. The Specific Functions of CDK6
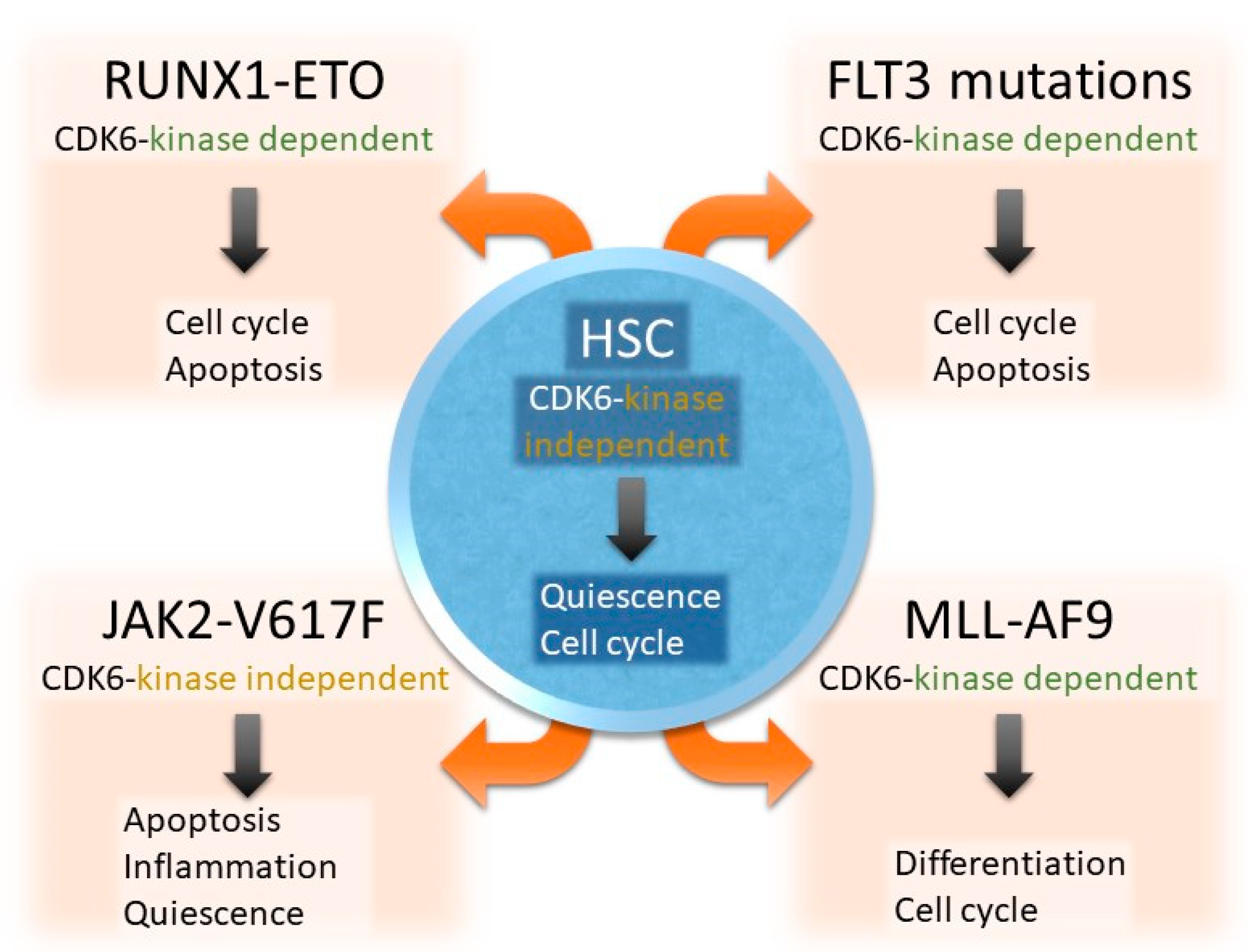
2.1. CDK6 in Hematopoietic Stem Cells
2.2. CDK6 Acts Largely Kinase-Independent in JAK2-V617F+ HSCs
3. The Role of CDK6 in AML
3.1. CDK6 as Driver and Therapeutic Target in MLL Rearrangements
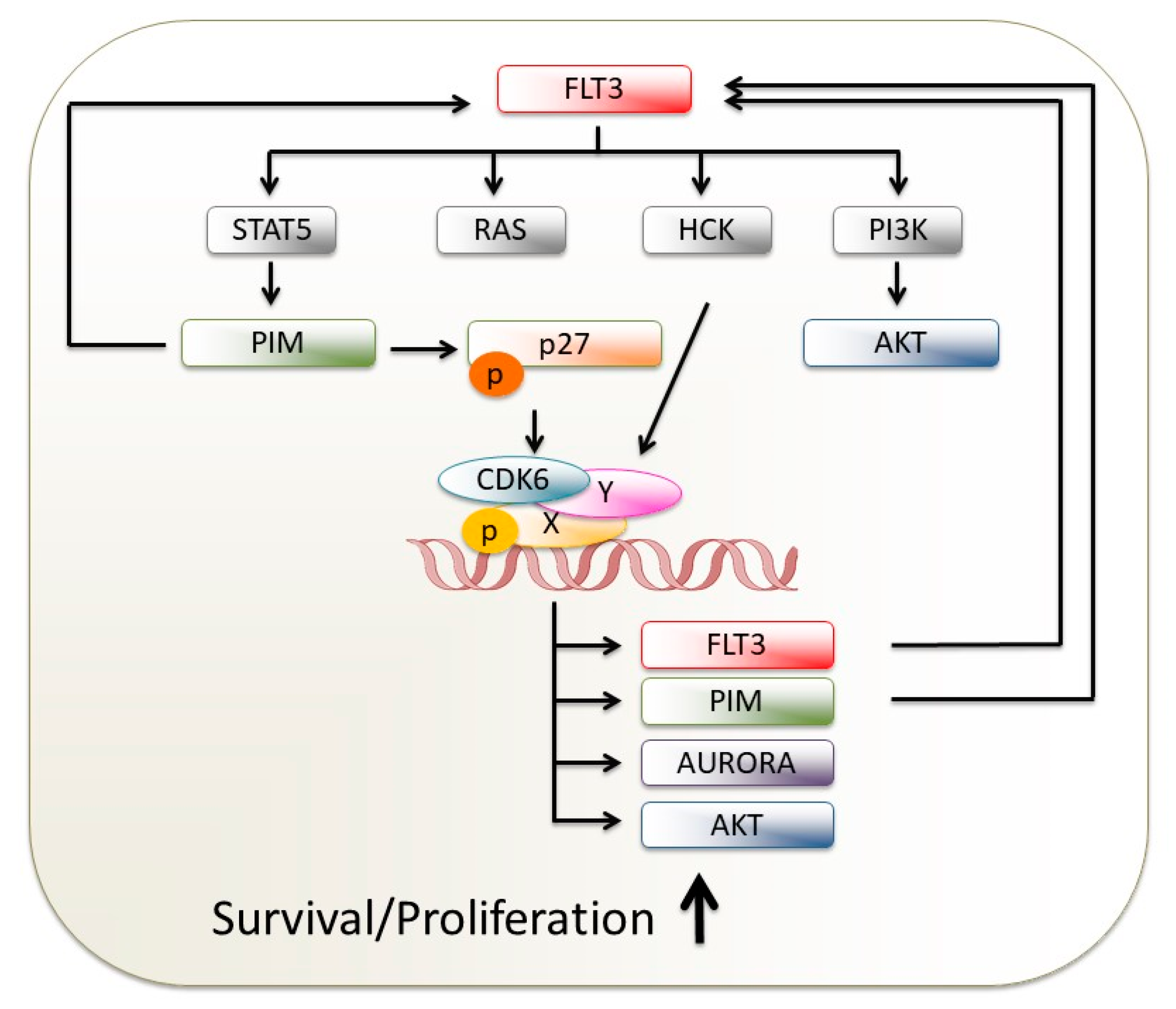
3.2. CDK6 Blockage Attacks FLT3-Driven AML via Several Roads
3.3. CDK6 Kinase Inhibition Targets RUNX1/ETO-Driven Disease Formation
4. CDK6 Protein Degradation Versus CDK6 Kinase Inhibition
5. CDK6 Inhibitors in Clinical Trials for AML
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AF4 | ALL1-fused gene from chromosome 4 protein |
| AF9 | ALL1-fused gene from chromosome 9 protein |
| ALL | Acute lymphoid leukemia |
| AML | Acute myeloid leukemia |
| AP-1 | Activator protein 1 |
| Ara-C | Cytarabine |
| Atf3 | Activating transcription factor 3 |
| AURK | Aurora kinase |
| BCR-ABL | Breakpoint cluster region - abelson 1 |
| Btg2 | B-cell translocation gene 2 |
| CDK | Cyclin-dependent kinase |
| dHSC | Dormant hematopoietic stem cell |
| EGR-1 | Early growth response protein 1 |
| ER | Estrogen receptor |
| ERK | Extracellular signal-regulated kinase |
| ET | Essential thrombocythaemia |
| FDA | U.S. Food and Drug Administration |
| FLT3 | FMS-like tyrosine kinase 3 |
| HCK | Hemopoietic cell kinase |
| HER2 | Human epidermal growth factor receptor 2 |
| HSC | Hematopoietic stem cell |
| HSPC | Hematopoietic stem/progenitor cell |
| IL | Interleukin |
| ITD | Internal tandem duplications |
| JAK2 | Janus kinase 2 |
| Klf6 | Krueppel-like factor 6 |
| KMT2A | Lysine-specific MethylTransferase 2A |
| LSC | Leukemic stem cell |
| LSK | Purified Lineage− Sca1+cKit+ cell |
| MDS | Myelodysplastic syndrome |
| MLL | Mixed-lineage leukemia |
| MLLr | MLL-rearranged |
| MPN | Myeloproliferative neoplasm |
| MTG8 | Myeloid translocation gene on 8 |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NFY | Nuclear transcription factor Y |
| NFκBiz | NF-κB inhibitor zeta |
| OS | Overall survival |
| PDGFR | Platelet-derived growth factor receptor |
| PIM1 | Proviral integration site for Moloney murine leukemia virus 1 |
| Pmaip1 | Phorbol-12-myristate-13-acetate-induced protein 1 |
| PMF | Primary myelofibrosis |
| Poly (I:C) | Polyinosinic:polycytidylic acid |
| Protein kinase B | PKB |
| PV | Polycythaemia vera |
| R/R | Refractory/recurrent |
| RAS | Rat sarcoma |
| RB | Retinoblastoma |
| RNA-Seq | RNA sequencing |
| RUNX1 | Runt-related transcription factor 1 |
| shRNA | Small hairpin RNA |
| Socs3 | Suppressor of cytokine signaling 3 |
| SP1 | Specific protein 1 |
| SRC | v-Src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| STAT | Signal transducer and activator of transcription |
| TKD | Tyrosine kinase domain |
| TKI | Tyrosine kinase inhibitor |
| VEGF-A | Vascular endothelial growth factor A |
References
- Visser, O.; Trama, A.; Maynadié, M.; Stiller, C.; Marcos-Gragera, R.; De Angelis, R.; Mallone, S.; Tereanu, C.; Allemani, C.; Ricardi, U.; et al. Incidence, survival and prevalence of myeloid malignancies in Europe. Eur. J. Cancer 2012, 48, 3257–3266. [Google Scholar] [CrossRef] [PubMed]
- Mehdipour, P.; Santoro, F.; Minucci, S. Epigenetic alterations in acute myeloid leukemias. FEBS J. 2015, 282, 1786–1800. [Google Scholar] [CrossRef] [PubMed]
- Tenen, D.G. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer 2003, 3, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Kayser, S.; Levis, M.J. Advances in targeted therapy for acute myeloid leukaemia. Br. J. Haematol. 2018, 180, 484–500. [Google Scholar] [CrossRef] [PubMed]
- Aleem, E.; Arceci, R.J. Targeting cell cycle regulators in hematologic malignancies. Front. Cell Dev. Biol. 2015, 3, 16. [Google Scholar] [CrossRef] [PubMed]
- Kozar, K.; Sicinski, P. Cell cycle progression without cyclin D-CDK4 and cyclin D-CDK6 complexes. Cell Cycle 2005, 4, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Sotillo, R.; Santamaría, D.; Galán, J.; Cerezo, A.; Ortega, S.; Dubus, P.; Barbacid, M. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell 2004, 118, 493–504. [Google Scholar] [CrossRef]
- Uras, I.Z.; Scheicher, R.M.; Kollmann, K.; Glösmann, M.; Prchal-Murphy, M.; Tigan, A.S.; Fux, D.A.; Altamura, S.; Neves, J.; Muckenthaler, M.U.; et al. Cdk6 contributes to cytoskeletal stability in erythroid cells. Haematologica 2017, 102, 995–1005. [Google Scholar] [CrossRef]
- Anders, L.; Ke, N.; Hydbring, P.; Choi, Y.J.; Widlund, H.R.; Chick, J.M.; Zhai, H.; Vidal, M.; Gygi, S.P.; Braun, P.; et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell 2011, 20, 620–634. [Google Scholar] [CrossRef]
- Chilosi, M.; Doglioni, C.; Yan, Z.; Lestani, M.; Menestrina, F.; Sorio, C.; Benedetti, A.; Vinante, F.; Pizzolo, G.; Inghirami, G. Differential Expression of Cyclin-Dependent Kinase 6 in Cortical Thymocytes and T-Cell Lymphoblastic Lymphoma/Leukemia. Am. J. Pathol. 1998, 152, 209–217. [Google Scholar]
- Lien, H.C.; Lin, C.W.; Huang, P.H.; Chang, M.L.; Hsu, S.M. Expression of cyclin-dependent kinase 6 (cdk6) and frequent loss of CD44 in nasal-nasopharyngeal NK/T-cell lymphomas: Comparison with CD56-negative peripheral T-cell lymphomas. Lab. Investig. 2000, 80, 893–900. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schwartz, R.; Engel, I.; Fallahi-Sichani, M.; Petrie, H.T.; Murre, C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc. Natl. Acad. Sci. USA 2006, 103, 9976–9981. [Google Scholar] [CrossRef] [PubMed]
- Nagel, S.; Leich, E.; Quentmeier, H.; Meyer, C.; Kaufmann, M.; Drexler, H.G.; Zettl, A.; Rosenwald, A.; MacLeodet, R.A.F. Amplification at 7q22 targets cyclin-dependent kinase 6 in T-cell lymphoma. Leukemia 2008, 22, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Giacomini, C.P.; Sun, S.; Varma, S.; Hunter Shain, A.; Giacomini, M.M.; Balagtas, J.; Sweeney, R.T.; Lai, E.; Del Vecchio, C.A.; Forster, A.D.; et al. Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types. PLoS Genet. 2013, 9, e1003464. [Google Scholar] [CrossRef] [PubMed]
- Hayette, S.; Tigaud, I.; Callet-Bauchu, E.; Ffrench, M.; Gazzo, S.; Wahbi, K.; Callanan, M.; Felman, P.; Dumontet, C.; Magaud, J.P.; et al. In B-cell chronic lymphocytic leukemias, 7q21 translocations lead to overexpression of the CDK6 gene. Blood 2003, 102, 1549–1550. [Google Scholar] [CrossRef] [PubMed]
- Su, X.Y.; Busson, M.; Della Valle, V.; Ballerini, P.; Dastugue, N.; Talmant, P.; Ferrando, A.A.; Baudry-Bluteau, D.; Romana, S.; Berger, R.; et al. Various types of rearrangements target TLX3 locus in T-cell acute lymphoblastic leukemia. Genes. Chromosomes Cancer 2004, 41, 243–249. [Google Scholar] [CrossRef]
- Corcoran, M.M.; Mould, S.J.; Orchard, J.A.; Ibbotson, R.E.; Chapman, R.M.; Boright, A.P.; Platt, C.; Tsui, L.C.; Scherer, S.W.; Oscier, D.G. Dysregulation of cyclin dependent kinase 6 expression in splenic marginal zone lymphoma through chromosome 7q translocations. Oncogene 1999, 18, 6271–6277. [Google Scholar] [CrossRef]
- Cavazzini, F.; Hernandez, J.A.; Gozzetti, A.; Russo Rossi, A.; De Angeli, C.; Tiseo, R.; Bardi, A.; Tammiso, E.; Crupi, R.; Lenoci, M.P.; et al. Chromosome 14q32 translocations involving the immunoglobulin heavy chain locus in chronic lymphocytic leukaemia identify a disease subset with poor prognosis. Br. J. Haematol. 2008, 142, 529–537. [Google Scholar] [CrossRef]
- Brito-Babapulle, V.; Gruszka-Westwood, A.M.; Platt, G.; Andersen, C.L.; Elnenaei, M.O.; Matutes, E.; Wotherspoon, A.C.; Weston-Smith, S.G.; Catovsky, D. Translocation t(2;7)(p12;q21-22) with dysregulation of the CDK6 gene mapping to 7q21-22 in a non-Hodgkin’s lymphoma with leukemia. Mol. Hematol. 2002, 87, 357–362. [Google Scholar]
- Chen, D.; Law, M.E.; Theis, J.D.; Gamez, J.D.; Caron, L.B.; Vrana, J.A.; Dogan, A. Clinicopathologic features of CDK6 translocation-associated B-cell lymphoproliferative disorders. Am. J. Surg. Pathol. 2009, 33, 720–729. [Google Scholar] [CrossRef]
- Haferlach, C.; Bacher, U.; Schnittger, S.; Alpermann, T.; Zenger, M.; Kern, W.; Haferlach, T. ETV6 rearrangements are recurrent in myeloid malignancies and are frequently associated with other genetic events. Genes. Chromosomes Cancer 2012, 51, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Kollmann, K.; Heller, G.; Schneckenleithner, C.; Warsch, W.; Scheicher, R.; Ott, R.G.; Schäfer, M.; Fajmann, S.; Schlederer, M.; Schiefer, A.I.; et al. A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell 2013, 24, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Handschick, K.; Beuerlein, K.; Jurida, L.; Bartkuhn, M.; Müller, H.; Soelch, J.; Weber, A.; Dittrich-Breiholz, O.; Schneider, H.; Scharfe, M.; et al. Cyclin-dependent kinase 6 is a chromatin-bound cofactor for NF-κB-dependent gene expression. Mol. Cell 2014, 53, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Scheicher, R.; Hoelbl-kovacic, A.; Bellutti, F.; Tigan, A.S.; Prchal-Murphy, M.; Heller, G.; Schneckenleithner, C.; Salazar-Roa, M.; Zöchbauer-Müller, S.; Zuber, J.; et al. CDK6 as a key regulator of hematopoietic and leukemic stem cell activation. Blood 2015, 125, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Uras, I.Z.; Maurer, B.; Nivarthi, H.; Jodl, P.; Kollmann, K.; Prchal-Murphy, M.; Milosevic Feenstra, J.D.; Zojer, M.; Lagger, S.; Grausenburger, R.; et al. CDK6 coordinates JAK2V617F mutant MPN via NF-κB and apoptotic networks. Blood 2019, 133, 1677–1690. [Google Scholar] [CrossRef]
- Uras, I.Z.; Walter, G.J.; Scheicher, R.; Bellutti, F.; Prchal-Murphy, M.; Tigan, A.S.; Valent, P.; Heidel, F.H.; Kubicek, S.; Scholl, C.; et al. Palbociclib treatment of FLT3-ITD+AML cells uncovers a kinase-dependent transcriptional regulation of FLT3 and PIM1 by CDK6. Blood 2016, 127, 2890–2902. [Google Scholar] [CrossRef]
- Uras, I.; Maurer, B.; Nebenfuehr, S.; Zojer, M.; Valent, P.; Sexl, V. Therapeutic Vulnerabilities in FLT3-Mutant AML Unmasked by Palbociclib. Int. J. Mol. Sci. 2018, 19, 3987. [Google Scholar] [CrossRef]
- Kollmann, K.; Sexl, V. CDK6 and p16INK4A in lymphoid malignancies. Oncotarget 2013, 4, 1858–1859. [Google Scholar] [CrossRef]
- Otto, T.; Sicinski, P. The kinase-independent, second life of CDK6 in transcription. Cancer Cell 2013, 24, 141–143. [Google Scholar] [CrossRef]
- Bellutti, F.; Tigan, A.-S.; Nebenfuehr, S.; Dolezal, M.; Zojer, M.; Grausenburger, R.; Hartenberger, S.; Kollmann, S.; Doma, E.; Prchal-Murphy, M.; et al. CDK6 antagonizes P53-induced responses during tumorigenesis. Cancer Discov. 2018, 8, 884–897. [Google Scholar] [CrossRef]
- Goel, S.; DeCristo, M.J.; McAllister, S.S.; Zhao, J.J. CDK4/6 Inhibition in Cancer: Beyond Cell Cycle Arrest. Trends Cell Biol. 2018, 28, 911–925. [Google Scholar] [CrossRef] [PubMed]
- Thill, M.; Schmidt, M. Management of adverse events during cyclin-dependent kinase 4/6 (CDK4/6) inhibitor-based treatment in breast cancer. Ther. Adv. Med. Oncol. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Martin, M.; Rugo, H.S.; Jones, S.; Im, S.A.; Gelmon, K.; Harbeck, N.; Lipatov, O.N.; Walshe, J.M.; Moulder, S.; et al. Palbociclib and Letrozole in Advanced Breast Cancer. N. Engl. J. Med. 2016, 375, 1925–1936. [Google Scholar] [CrossRef] [PubMed]
- Hortobagyi, G.N.; Stemmer, S.M.; Burris, H.A.; Yap, Y.S.; Sonke, G.S.; Paluch-Shimon, S.; Campone, M.; Blackwell, K.L.; André, F.; Winer, E.P.; et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. N. Engl. J. Med. 2016, 375, 1738–1748. [Google Scholar] [CrossRef]
- Goetz, M.P.; Toi, M.; Campone, M.; Sohn, J.; Paluch-Shimon, S.; Huober, J.; Park, I.H.; Trédan, O.; Chen, S.C.; Manso, L.; et al. MONARCH 3: Abemaciclib As Initial Therapy for Advanced Breast Cancer. J. Clin. Oncol. 2017, 35, 3638–3646. [Google Scholar] [CrossRef]
- Dickler, M.N.; Tolaney, S.M.; Rugo, H.S.; Cortés, J.; Diéras, V.; Patt, D.; Wildiers, H.; Hudis, C.A.; O’Shaughnessy, J.; Zamora, E.; et al. MONARCH 1, A Phase II Study of Abemaciclib, a CDK4 and CDK6 Inhibitor, as a Single Agent, in Patients with Refractory HR+/HER2- Metastatic Breast Cancer. Clin. Cancer Res. 2017, 23, 5218–5224. [Google Scholar] [CrossRef]
- Leonard, J.P.; LaCasce, A.S.; Smith, M.R.; Noy, A.; Chirieac, L.R.; Rodig, S.J.; Yu, J.Q.; Vallabhajosula, S.; Schoder, H.; English, P.; et al. Selective CDK4/6 inhibition with tumor responses by PD0332991 in patients with mantle cell lymphoma. Blood 2012, 119, 4597–4607. [Google Scholar] [CrossRef]
- Dickson, M.A.; Schwartz, G.K.; Keohan, M.L.; D’Angelo, S.P.; Gounder, M.M.; Chi, P.; Antonescu, C.R.; Landa, J.; Qin, L.X.; Crago, A.M.; et al. Progression-Free Survival Among Patients With Well-Differentiated or Dedifferentiated Liposarcoma Treated With CDK4 Inhibitor Palbociclib. JAMA Oncol. 2016, 2, 937–940. [Google Scholar] [CrossRef]
- Patnaik, A.; Rosen, L.S.; Tolaney, S.M.; Tolcher, A.W.; Goldman, J.W.; Gandhi, L.; Papadopoulos, K.P.; Beeram, M.; Rasco, D.W.; Hilton, J.F.; et al. Efficacy and Safety of Abemaciclib, an Inhibitor of CDK4 and CDK6, for Patients with Breast Cancer, Non-Small Cell Lung Cancer, and Other Solid Tumors. Cancer Discov. 2016, 6, 740–753. [Google Scholar] [CrossRef]
- Geoerger, B.; Bourdeaut, F.; DuBois, S.G.; Fischer, M.; Geller, J.I.; Gottardo, N.G.; Marabelle, A.; Pearson, A.D.J.; Modak, S.; Cash, T.; et al. A Phase I Study of the CDK4/6 Inhibitor Ribociclib (LEE011) in Pediatric Patients with Malignant Rhabdoid Tumors, Neuroblastoma, and Other Solid Tumors. Clin. Cancer Res. 2017, 23, 2433–2441. [Google Scholar] [CrossRef]
- Placke, T.; Faber, K.; Nonami, A.; Putwain, S.L.; Salih, H.R.; Heidel, F.H.; Krämer, A.; Root, D.E.; Barbie, A.D.; Krivtsov, A.V.; et al. Requirement for CDK6 in MLL-rearranged acute myeloid leukemia. Blood 2014, 124, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Laurenti, E.; Frelin, C.; Xie, S.; Ferrari, R.; Dunant, C.F.; Zandi, S.; Neumann, A.; Plumb, I.; Doulatov, S.; Chen, J.; et al. CDK6 Levels Regulate Quiescence Exit in Human Hematopoietic Stem Cells. Cell Stem Cell 2015, 16, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Laurenti, E.; Oser, G.; van der Wath, R.C.; Blanco-Bose, W.; Jaworski, M.; Offner, S.; Dunant, C.F.; Eshkind, L.; Bockamp, E.; et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 2008, 135, 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Min, I.M.; Pietramaggiori, G.; Kim, F.S.; Passegué, E.; Stevenson, K.E.; Wagers, A.J. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell 2008, 2, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Volkert, S.; Kohlmann, A.; Schnittger, S.; Kern, W.; Haferlach, T.; Haferlach, C. Association of the type of 5q loss with complex karyotype, clonal evolution, TP53 mutation status, and prognosis in acute myeloid leukemia and myelodysplastic syndrome. Genes. Chromosomes Cancer 2014, 53, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Joslin, J.M.; Fernald, A.A.; Tennant, T.R.; Davis, E.M.; Kogan, S.C.; Anastasi, J.; Crispino, J.D.; Le Beau, M.M. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood 2007, 110, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Calogero, A.; Arcella, A.; De Gregorio, G.; Porcellini, A.; Mercola, D.; Liu, C.; Lombari, V.; Zani, M.; Giannini, G.; Gagliardi, F.M.; et al. The early growth response gene EGR-1 behaves as a suppressor gene that is down-regulated independent of ARF/Mdm2 but not p53 alterations in fresh human gliomas. Clin. Cancer Res. 2001, 7, 2788–2796, published september 2001. [Google Scholar]
- Huang, R.-P.; Fan, Y.; De Belle, I.; Niemeyer, C.; Gottardis, M.M.; Mercola, D.; Adamson, E.D. Decreased Egr-1 expression in human, mouse and rat mammary cells and tissues correlates with tumor formation. Int. J. Cancer 1997, 72, 102–109. [Google Scholar] [CrossRef]
- Stoddart, A.; Fernald, A.A.; Wang, J.; Davis, E.M.; Karrison, T.; Anastasi, J.; Le Beau, M.M. Haploinsufficiency of del(5q) genes, Egr1 and Apc, cooperate with Tp53 loss to induce acute myeloid leukemia in mice. Blood 2014, 123, 1069–1078. [Google Scholar] [CrossRef]
- Tanaka, S.; Miyagi, S.; Sashida, G.; Chiba, T.; Yuan, J.; Mochizuki-Kashio, M.; Suzuki, Y.; Sugana, S.; Nakaseko, C.; Yokote, K.; et al. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood 2012, 120, 1107–1117. [Google Scholar] [CrossRef]
- Kharbanda, S.; Nakamura, T.; Stone, R.; Hass, R.; Bernstein, S.; Datta, R.; Sukhatme, V.P.; Kufe, D. Expression of the early growth response 1 and 2 zinc finger genes during induction of monocytic differentiation. J. Clin. Investig. 1991, 88, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.Q.; Hoffman-Liebermann, B.; Liebermann, D.A. The zinc finger transcription factor Egr-1 is essential for and restricts differentiation along the macrophage lineage. Cell 1993, 72, 197–209. [Google Scholar] [CrossRef]
- Krishnaraju, K.; Hoffman, B.; Liebermann, D.A. The zinc finger transcription factor Egr-1 activates macrophage differentiation in M1 myeloblastic leukemia cells. Blood 1998, 92, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.E.; Wang, L.; Kropf, P.L.; Duan, R.; Johnson, D.E. Src family kinase gene targets during myeloid differentiation: Identification of the EGR-1 gene as a direct target. Leukemia 2009, 23, 1933–1935. [Google Scholar] [CrossRef][Green Version]
- Shafarenko, M.; Liebermann, D.A.; Hoffman, B. Egr-1 abrogates the block imparted by c-Myc on terminal M1 myeloid differentiation. Blood 2005, 106, 871–878. [Google Scholar] [CrossRef][Green Version]
- Gibbs, J.D.; Liebermann, D.A.; Hoffman, B. Leukemia suppressor function of Egr-1 is dependent on transforming oncogene. Leukemia 2008, 22, 1909–1916. [Google Scholar] [CrossRef]
- Baxter, E.J.; Scott, L.M.; Campbell, P.J.; East, C.; Fourouclas, N.; Swanton, S.; Vassiliou, G.S.; Bench, A.J.; Boyd, E.M.; Curtin, N.; et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet (Lond. Engl.) 2005, 365, 1054–1061. [Google Scholar] [CrossRef]
- Kralovics, R.; Passamonti, F.; Buser, A.S.; Teo, S.S.; Tiedt, R.; Passweg, J.R.; Tichelli, A.; Cazzola, M.; Skoda, R.C. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 2005, 352, 1779–1790. [Google Scholar] [CrossRef]
- Levine, R.L.; Wadleigh, M.; Cools, J.; Ebert, B.L.; Wernig, G.; Huntly, B.J.; Boggon, T.J.; Wlodarska, I.; Clark, J.J.; Moore, S.; et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 2005, 7, 387–397. [Google Scholar] [CrossRef]
- Jamieson, C.H.M.; Gotlib, J.; Durocher, J.A.; Chao, M.P.; Mariappan, M.R.; Lay, M.; Jones, C.; Zehnder, J.L.; Lilleberg, S.L.; Weissman, I.L. The JAK2 V617F mutation occurs in hematopoietic stem cells in polycythemia vera and predisposes toward erythroid differentiation. Proc. Natl. Acad. Sci. USA 2006, 103, 6224–6229. [Google Scholar] [CrossRef]
- Kirschner, K.; Samarajiwa, S.A.; Cairns, J.M.; Menon, S.; Pérez-Mancera, P.A.; Tomimatsu, K.; Bermejo-Rodriguez, C.; Ito, Y.; Chandra, T.; Narita, M.; et al. Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53. PLoS Genet. 2015, 11, e1005053. [Google Scholar] [CrossRef] [PubMed]
- Oda, E.; Ohki, R.; Murasawa, H.; Nemoto, J.; Shibue, T.; Yamashita, T.; Tokino, T.; Taniguchi, T.; Tanaka, N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 2000, 288, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Benzeno, S.; Narla, G.; Allina, J.; Cheng, G.Z.; Reeves, H.L.; Banck, M.S.; Odin, J.A.; Diehl, J.A.; Germain, D.; Friedman, S.L. Cyclin-Dependent Kinase Inhibition by the KLF6 Tumor Suppressor Protein through Interaction with Cyclin D1. Cancer Res. 2004, 64, 3885–3891. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Li, X.; Guo, B. KLF6 Induces Apoptosis in Prostate Cancer Cells through Up-regulation of ATF3. J. Biol. Chem. 2008, 283, 29795–29801. [Google Scholar] [CrossRef]
- Sato, A.; Nakama, K.; Watanabe, H.; Satake, A.; Yamamoto, A.; Omi, T.; Hiramoto, A.; Masutani, M.; Wataya, Y.; Kim, H.S. Role of activating transcription factor 3 protein ATF3 in necrosis and apoptosis induced by 5-fluoro-2′-deoxyuridine. FEBS J. 2014, 281, 1892–1900. [Google Scholar] [CrossRef]
- Carow, B.; Rottenberg, M.E. SOCS3, a Major Regulator of Infection and Inflammation. Front. Immunol. 2014, 5, 58. [Google Scholar] [CrossRef]
- Molavi, O.; Wang, P.; Zak, Z.; Gelebart, P.; Belch, A.; Lai, R. Gene methylation and silencing of SOCS3 in mantle cell lymphoma. Br. J. Haematol. 2013, 161, 348–356. [Google Scholar] [CrossRef]
- Frobøse, H.; Groth Rønn, S.; Heding, P.E.; Mendoza, H.; Cohen, P.; Mandrup-Poulsen, T.; Billestrup, N. Suppressor of Cytokine Signaling-3 Inhibits Interleukin-1 Signaling by Targeting the TRAF-6/TAK1 Complex. Mol. Endocrinol. 2006, 20, 1587–1596. [Google Scholar] [CrossRef]
- Nair, S.; Pandey, A.D.; Mukhopadhyay, S. The PPE18 Protein of Mycobacterium tuberculosis Inhibits NF-κB/rel–Mediated Proinflammatory Cytokine Production by Upregulating and Phosphorylating Suppressor of Cytokine Signaling 3 Protein. J. Immunol. 2011, 186, 5413–5424. [Google Scholar] [CrossRef]
- Reynaud, D.; Pietras, E.; Barry-Holson, K.; Mir, A.; Binnewies, M.; Jeanne, M.; Sala-Torra, O.; Radich, J.P.; Passegué, E. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell 2011, 20, 661–773. [Google Scholar] [CrossRef]
- Schürch, C.M.; Riether, C.; Ochsenbein, A.F. Cytotoxic CD8+ T Cells Stimulate Hematopoietic Progenitors by Promoting Cytokine Release from Bone Marrow Mesenchymal Stromal Cells. Cell Stem Cell 2014, 14, 460–472. [Google Scholar] [CrossRef] [PubMed]
- Nagareddy, P.R.; Kraakman, M.; Masters, S.L.; Stirzaker, R.A.; Gorman, D.J.; Grant, R.W.; Dragoljevic, D.; Hong, E.S.; Abdel-Latif, A.; Smyth, S.S.; et al. Adipose tissue macrophages promote myelopoiesis and monocytosis in obesity. Cell Metab. 2014, 19, 821–835. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, Y.; Cheng, H.; Liu, G.; Cheng, T.; Liu, Y.; Liu, L. System modeling reveals the molecular mechanisms of HSC cell cycle alteration mediated by Maff and Egr3 under leukemia. BMC Syst. Biol. 2017, 11 (Suppl. S5), 91. [Google Scholar] [CrossRef][Green Version]
- Cheng, H.; Hao, S.; Liu, Y.; Pang, Y.; Ma, S.; Dong, F.; Xu, J.; Zheng, G.; Li, S.; Yuan, W.; et al. Leukemic marrow infiltration reveals a novel role for Egr3 as a potent inhibitor of normal hematopoietic stem cell proliferation. Blood 2015, 126, 1302–1313. [Google Scholar] [CrossRef] [PubMed]
- Liebermann, D.A.; Gregory, B.; Hoffman, B. AP-1 (Fos/Jun) transcription factors in hematopoietic differentiation and apoptosis. Int. J. Oncol. 1998, 12, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Santaguida, M.; Schepers, K.; King, B.; Sabnis, A.J.; Forsberg, E.C.; Attema, J.L.; Braun, B.S.; Passagué, E. JunB protects against myeloid malignancies by limiting hematopoietic stem cell proliferation and differentiation without affecting self-renewal. Cancer Cell 2009, 15, 341–352. [Google Scholar] [CrossRef]
- Alder, J.K.; Georgantas, R.W.; Hildreth, R.L.; Civin, C.I. 348. Kruppel-Like Factor 4 Regulates Proliferation of Human and Mouse Hematopoietic Stem-Progenitor Cells, but Is Not Essential for Mouse Hematopoietic Repopulation. Mol. Ther. 2006, 13, S132–S133. [Google Scholar] [CrossRef]
- Vanegas, N.-D.P.; Vernot, J.-P. Loss of quiescence and self-renewal capacity of hematopoietic stem cell in an in vitro leukemic niche. Exp. Hematol. Oncol. 2017, 6, 2. [Google Scholar] [CrossRef]
- Ueharu, H.; Higuchi, M.; Nishimura, N.; Yoshida, S.; Shibuya, S.; Sensui, K.; Kato, T.; Kato, Y. Expression of Krüppel-like factor 6, KLF6, in rat pituitary stem/progenitor cells and its regulation of the PRRX2 gene. J. Reprod. Dev. 2014, 60, 304–311. [Google Scholar] [CrossRef]
- Land, R.H.; Rayne, A.K.; Vanderbeck, A.N.; Barlowe, T.S.; Manjunath, S.; Gross, M.; Eiger, S.; Klein, P.S.; Cunningham, N.R.; Huang, J.; et al. The Orphan Nuclear Receptor NR4A1 Specifies a Distinct Subpopulation of Quiescent Myeloid-Biased Long-Term HSCs. Stem Cells 2015, 33, 278–288. [Google Scholar] [CrossRef]
- Sirin, O.; Lukov, G.L.; Mao, R.; Conneely, O.M.; Goodell, M.A. The orphan nuclear receptor Nurr1 restricts the proliferation of haematopoietic stem cells. Nat. Cell Biol. 2010, 12, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Lorenzana, D.; Avilés-Vazquez, S.; Sandoval Esquivel, M.A.; Alvarado-Moreno, A.; Ortiz-Navarrete, V.; Torres-Martínez, H.; Ayala-Sánchez, M.; Mayani, H.; Chavez-Gonzalez, A. CDKIs p18(INK4c) and p57(Kip2) are involved in quiescence of CML leukemic stem cells after treatment with TKI. Cell Cycle 2016, 15, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Shea, K.L.; Xiang, W.; LaPorta, V.S.; Licht, J.D.; Keller, C.; Basson, M.A.; Brack, A.S. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell 2010, 6, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Bigot, A.; Duddy, W.J.; Ouandaogo, Z.G.; Negroni, E.; Mariot, V.; Ghimbovschi, S.; Harmon, B.; Wielgosik, A.; Loiseau, C.; Devaney, J.; et al. Age-Associated Methylation Suppresses SPRY1, Leading to a Failure of Re-quiescence and Loss of the Reserve Stem Cell Pool in Elderly Muscle. Cell Rep. 2015, 13, 1172–1182. [Google Scholar] [CrossRef]
- Meyer, C.; Burmeister, T.; Gröger, D.; Tsaur, G.; Fechina, L.; Renneville, A.; Sutton, R.; Venn, N.C.; Emerenciano, M.; Pombo-de-Oliveira, M.S.; et al. The MLL recombinome of acute leukemias in 2017. Leukemia 2018, 32, 273–284. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; Feng, Z.; Lemieux, M.E.; Faber, J.; Vempati, S.; Sinha, A.U.; Xia, X.; Jesneck, J.; Bracken, A.P.; Silverman, L.B.; et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 2008, 14, 355–368. [Google Scholar] [CrossRef]
- Van der Linden, M.; Willekes, M.; van Roon, E.; Seslija, L.; Schneider, P.; Pieters, R.; Stam, R.W. MLL fusion-driven activation of CDK6 potentiates proliferation in MLL- rearranged infant ALL. Cell Cycle 2014, 13, 834–844. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, Y.; Wang, Y.; Huang, J. Molecular dynamics simulations and statistical coupling analysis reveal functional coevolution network of oncogenic mutations in the CDKN2A-CDK6 complex. FEBS Lett. 2013, 587, 136–141. [Google Scholar] [CrossRef]
- Wang, L.; Wang, J.; Blaser, B.W.; Duchemin, A.M.; Kusewitt, D.F.; Liu, T.; Caligiuri, M.A.; Briesewitz, R. Pharmacologic inhibition of CDK4/6: Mechanistic evidence for selective activity or acquired resistance in acute myeloid leukemia. Blood 2007, 110, 2075–2083. [Google Scholar] [CrossRef]
- Keegan, K.; Li, C.; Li, Z.; Ma, J.; Ragains, M.; Coberly, S.; Hollenback, D.; Eksterowicz, J.; Liang, L.; Weidner, M.; et al. Preclinical evaluation of AMG 925, a FLT3/CDK4 dual kinase inhibitor for treating acute myeloid leukemia. Mol. Cancer Ther. 2014, 13, 880–889. [Google Scholar] [CrossRef]
- Li, Z.; Wang, X.; Eksterowicz, J.; Gribble, M.W., Jr.; Alba, G.Q.; Ayres, M.; Carlson, T.J.; Chen, A.; Chen, X.; Cho, R.; et al. Discovery of AMG 925, a FLT3 and CDK4 Dual Kinase Inhibitor with Preferential Affinity for the Activated State of FLT3. J. Med. Chem. 2014, 57, 3430–3449. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, L.; Liang, L.; Xia, Z.; Li, Z.; Wang, X.; McGee, L.R.; Newhall, K.; Sinclair, A.; Kamb, A.; et al. AMG 925 Is a Dual FLT3/CDK4 Inhibitor with the Potential to Overcome FLT3 Inhibitor Resistance in Acute Myeloid Leukemia. Mol. Cancer Ther. 2015, 14, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Lopez, S.; Voisset, E.; Tisserand, J.C.; Mosca, C.; Prebet, T.; Santamaria, D.; Dubreuil, P.; De Sepulveda, P. An essential pathway links FLT3-ITD, HCK and CDK6 in acute myeloid leukemia. Oncotarget 2016, 7, 51163–51173. [Google Scholar] [CrossRef] [PubMed]
- Poh, A.R.; Love, C.G.; Masson, F.; Preaudet, A.; Tsui, C.; Whitehead, L.; Monard, S.; Khakham, Y.; Burstroem, L.; Lessene, G.; et al. Inhibition of Hematopoietic Cell Kinase Activity Suppresses Myeloid Cell-Mediated Colon Cancer Progression. Cancer Cell 2017, 31, 563–575.e5. [Google Scholar] [CrossRef]
- Zhang, Y.; Hsu, C.-P.; Lu, J.F.; Kuchimanchi, M.; Sun, Y.N.; Ma, J.; Xu, G.; Zhang, Y.; Xu, Y.; Weidner, M.; et al. FLT3 and CDK4/6 inhibitors: Signaling mechanisms and tumor burden in subcutaneous and orthotopic mouse models of acute myeloid leukemia. J. Pharmacokinet. Pharmacodyn. 2014, 41, 675–691. [Google Scholar] [CrossRef]
- Natarajan, K.; Xie, Y.; Burcu, M.; Linn, D.E.; Qiu, Y.; Baer, M.R. Pim-1 kinase phosphorylates and stabilizes 130 kDa FLT3 and promotes aberrant STAT5 signaling in acute myeloid leukemia with FLT3 internal tandem duplication. PLoS ONE 2013, 8, e74653. [Google Scholar] [CrossRef]
- Peschel, I.; Podmirseg, S.R.; Taschler, M.; Duyster, J.; Götze, K.S.; Sill, H.; Nachbaur, D.; Jäkel, H.; Hengst, L. FLT3 and FLT3-ITD phosphorylate and inactivate the cyclin-dependent kinase inhibitor p27Kip1 in acute myeloid leukemia. Haematologica 2017, 102, 1378–1389. [Google Scholar] [CrossRef]
- Uras, I.Z.; Bellutti, F.; Sexl, V. p27 in FLT3-driven acute myeloid leukemia: Many roads lead to ruin. Haematologica 2017, 102, 1299–1301. [Google Scholar] [CrossRef]
- Winer, E.S.; Stone, R.M. Novel therapy in Acute myeloid leukemia (AML): Moving toward targeted approaches. Ther. Adv. Hematol. 2019, 10. [Google Scholar] [CrossRef]
- Yuan, T.; Qi, B.; Jiang, Z.; Dong, W.; Zhong, L.; Bai, L.; Tong, R.; Yu, J.; Shi, J. Dual FLT3 inhibitors: Against the drug resistance of acute myeloid leukemia in recent decade. Eur. J. Med. Chem. 2019, 178, 468–483. [Google Scholar] [CrossRef]
- Martinez-Soria, N.; McKenzie, L.; Draper, J.; Ptasinska, A.; Issa, H.; Potluri, S.; Blair, H.J.; Pickin, A.; Isa, A.; Chin, P.S.; et al. The Oncogenic Transcription Factor RUNX1/ETO Corrupts Cell Cycle Regulation to Drive Leukemic Transformation. Cancer Cell 2018, 34, 626–642.e8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Y.; Zhou, G.-B.; Yin, T.; Chen, B.; Shi, J.Y.; Liang, W.X.; Jin, X.L.; You, J.H.; Yang, G.; Shen, Z.X.; et al. AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: Implication in stepwise leukemogenesis and response to Gleevec. Proc. Natl. Acad. Sci. USA 2005, 102, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.; Jiang, B.; Bauer, S.; Donovan, K.A.; Liang, Y.; Wang, E.S.; Nowal, R.P.; Yuan, J.C.; Zhang, T.; Kwiatkowski, N.; et al. Homolog-Selective Degradation as a Strategy to Probe the Function of CDK6 in AML. Cell Chem. Biol. 2019, 26, 300–306.e9. [Google Scholar] [CrossRef] [PubMed]
- De Dominici, M.; Porazzi, P.; Xiao, Y.; Chao, A.; Tang, H.Y.; Kumar, G.; Fortina, P.; Spinelli, O.; Rambaldi, A.; Peterson, L.F.; et al. Selective inhibition of Ph-positive ALL cell growth through kinase-dependent and independent effects by CDK6-specific PROTACs. Blood 2020. [Google Scholar] [CrossRef]
- Kadia, T.M.; Konopleva, M.Y.; Garcia-Manero, G.; Benton, C.B.; Wierda, W.G.; Bose, P.; Yilmaz, M.E.; Jabbour, E.J.; Kornblau, S.M.; Bhalla, K.N.; et al. Phase I Study of Palbociclib Alone and in Combination in Patients with Relapsed and Refractory (R/R) Leukemias. Blood 2018, 132 (Suppl. S1), 4057. [Google Scholar] [CrossRef]
- Fröhling, S.; Agrawal, M.; Jahn, N.; Fransecky, L.R.; Baldus, C.D.; Wäsch, R.; Lübbert, M.; Walter, G.; Jensen, P.; Scholl, C.; et al. CDK4/6 Inhibitor Palbociclib for Treatment of KMT2A-Rearranged Acute Myeloid Leukemia: Interim Analysis of the AMLSG 23-14 Trial. Blood 2016, 128, 1608. [Google Scholar] [CrossRef]
- Daver, N.; Pollyea, D.A.; Rizzieri, D.A.; Palmer, J.; Rampal, R.K.; Dinner, S.; Bixby, D.L.; Percival, M.-E.M.; Kovacsovics, T.; Begna, K.H.; et al. A Phase I Study of FLX925, a Dual FLT3 and CDK4/6 Inhibitor in Patients with Relapsed or Refractory Acute Myeloid Leukemia (AML). Blood 2017, 130 (Suppl. S1), 1343. [Google Scholar] [CrossRef]
- Yang, C.; Boyson, C.A.; Liberto, M.D.; Huang, X.; Hannah, J.; Dorn, D.C.; Moore, M.A.; Chen-Klang, S.; Zhou, P. CDK4/6 Inhibitor PD 0332991 Sensitizes Acute Myeloid Leukemia to Cytarabine-Mediated Cytotoxicity. Cancer Res. 2015, 75, 1838–1846. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uras, I.Z.; Sexl, V.; Kollmann, K. CDK6 Inhibition: A Novel Approach in AML Management. Int. J. Mol. Sci. 2020, 21, 2528. https://doi.org/10.3390/ijms21072528
Uras IZ, Sexl V, Kollmann K. CDK6 Inhibition: A Novel Approach in AML Management. International Journal of Molecular Sciences. 2020; 21(7):2528. https://doi.org/10.3390/ijms21072528
Chicago/Turabian StyleUras, Iris Z., Veronika Sexl, and Karoline Kollmann. 2020. "CDK6 Inhibition: A Novel Approach in AML Management" International Journal of Molecular Sciences 21, no. 7: 2528. https://doi.org/10.3390/ijms21072528
APA StyleUras, I. Z., Sexl, V., & Kollmann, K. (2020). CDK6 Inhibition: A Novel Approach in AML Management. International Journal of Molecular Sciences, 21(7), 2528. https://doi.org/10.3390/ijms21072528




