Towards a New Understanding of Decision-Making by Hematopoietic Stem Cells
Abstract
1. Introduction
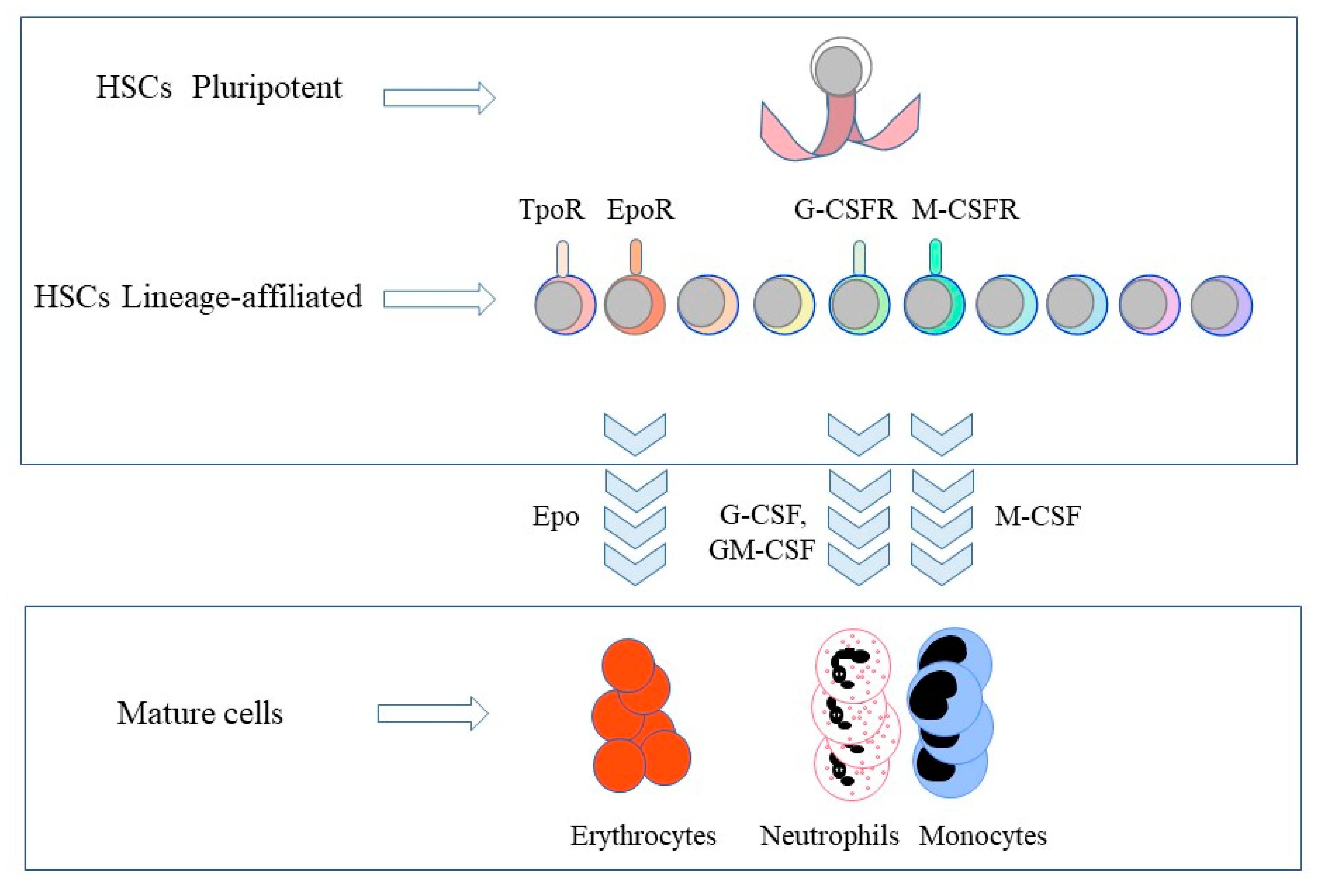
2. The Selective Expression of Cytokine Receptors by Hematopoietic Stem Cells
3. The Instructive Action of Hematopoietic Cytokines
4. HSC Commitment to Differentiation and Finding a New Niche
5. Natural Variation within Hematopoietic Stem Cells
6. A Natural Selection Model of Hematopoiesis
7. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Schofield, R. The relationship between the spleen colony-forming cell and the haematopoietic stem cell. Blood Cells 1978, 41, 7–25. [Google Scholar]
- Pinho, S.; Frenette, P.S. Haematopoietic stem cell activity and interactions with the niche. Nat. Rev. Mol. Cell Biol. 2019, 20, 303–320. [Google Scholar] [CrossRef] [PubMed]
- Okumura, N.; Ebihara, Y.; Tanaka, I.; Koike, K.; Komiyama, A.; Nakahata, T. Chemotactic and chemokinetic activities of stem cell factor on murine hematopoietic progenitor cells. Blood 1996, 87, 4100–4108. [Google Scholar] [CrossRef] [PubMed]
- Driessen, R.L.; Johnson, H.M.; Nilsson, S.K. Membrane-bound stem cell factor is a key regulator in the initial lodgement of stem cells within the endosteal marrow region. Exp. Hematol. 2003, 31, 1284–1291. [Google Scholar] [CrossRef] [PubMed]
- Ashman, L.K. The biology of stem cell factor and its receptor C-kit. Int. J. Biochem. Cell. Biol. 1999, 31, 1037–1051. [Google Scholar] [CrossRef]
- Sharma, S.; Gurudutta, G.U.; Satija, N.K.; Pati, S.; Gupta, P.; Verma, Y.K.; Singh, V.K.; Tripathi, R.P. Stem cell c-KIT and HOXB4 genes: Critical roles and mechanisms in self-renewal, proliferation, and differentiation. Stem Cells Dev. 2006, 15, 755–778. [Google Scholar] [CrossRef]
- Ceredig, R.; Rolink, A.G.; Brown, G. Models of haematopoiesis: Seeing the wood for the trees. Nat. Rev. Immunol. 2009, 9, 293–300. [Google Scholar] [CrossRef]
- Brown, G.; Ceredig, R. Modelling the hematopoietic landscape. Front. Cell Dev. Biol. 2019, 7, 104. [Google Scholar] [CrossRef]
- Hume, D.A.; MacDonald, K.P. Therapeutic applications of macrophage colony-stimulating factor (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood 2012, 119, 1810–1811, 1820. [Google Scholar] [CrossRef]
- Koury, M.J.; Bondurant, M.C. Erythropoietin retards DNA breakdown and prevents programmed death in erythroid progenitor cells. Science 1990, 248, 378–381. [Google Scholar] [CrossRef]
- Demetri, G.D.; Griffin, J.D. Granulocyte colony-stimulating factor and its receptor. Blood 1991, 78, 2791–2808. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, D. Hematopoietic regulators: Redundancy or subtlety? Blood 1993, 82, 3515–3523. [Google Scholar] [CrossRef] [PubMed]
- Gasson, J.C. Molecular physiology of granulocyte-macrophage colony stimulating factor. Blood 1991, 77, 1131–1145. [Google Scholar] [CrossRef] [PubMed]
- Kaushansky, K. Thrombopoietin: The primary regulator of megakaryocyte and platelet production. Thromb. Haemost. 1995, 74, 521–525. [Google Scholar] [CrossRef]
- Mooney, C.J.; Cunningham, A.; Tsapogas, P.; Toellner, K.-M.; Brown, G. Selective expression of Flt3 within the mouse hematopoietic stem cell compartment. Int. J. Mol. Sci. 2017, 18, 1037. [Google Scholar] [CrossRef]
- Osawa, M.; Hanada, K.; Hamada, H.; Nakauchi, H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 1996, 273, 242–245. [Google Scholar] [CrossRef]
- Shinjo, K.; Takeshita, A.; Higuchi, M.; Ohnishi, K.; Ohno, R. Erythropoietin receptor expression on human bone marrow erythroid precursor cells by a newly-devised quantitative flow-cytometric assay. Br. J. Haematol. 1997, 96, 551–558. [Google Scholar] [CrossRef]
- Zriwil, A.; Bolers, C.; Wittmann, L.; Green, J.C.A.; Woll, P.S.; Jacobsen, S.E.W.; Sitnicka, E. Macrophage colony-stimulating factor receptor marks and regulates a fetal myeloid-primed B-cell progenitor in mice. Blood 2016, 128, 217–226. [Google Scholar] [CrossRef]
- Mossadegh-Keller, N.; Sarrazin, S.; Kandalla, P.K.; Espinosa, L.; Stanley, E.R.; Nutt, S.L.; Moore, J.; Sieweke, M.H. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature 2013, 497, 239–243. [Google Scholar] [CrossRef]
- Stoffel, R.; Ziegler, S.; Ghilhardi, N.; Ledermann, B.; de Sauvage, F.J.; Skoda, R.C. Permissive role of thrombopoietin and granulocyte colony-stimulating factor receptors in hematopoietic cell fate decisions in vivo. Proc. Natl. Acad Sci. USA 1999, 96, 698–702. [Google Scholar] [CrossRef]
- Mansson, R.; Hultquist, A.; Luc, S.; Yang, L.; Anderson, K.; Kharazi, S.; Al-Hashmi, S.; Liuba, K.; Thoren, L.; Adolffsson, J.; et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity 2007, 26, 407–419. [Google Scholar] [CrossRef] [PubMed]
- De Graaf, C.; Metcalf, D. Thrombopoietin and hematopoietic stem cells. Cell Cycle 2011, 10, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, H.; Arai, F.; Hosokawa, K.; Hagiwara, T.; Takubo, K.; Nakamura, Y.; Gomei, Y.; Iwasaki, H.; Matsuoka, S.; Miyanoto, K.; et al. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell 2007, 1, 685–697.22. [Google Scholar] [CrossRef] [PubMed]
- Morrison, S.J.; Wright, D.E.; Weissman, I.L. Cyclophosphamide/granulocyte colony-stimulating factor induces hematopoietic stem cells to proliferate prior to mobilization. Proc. Natl. Acad. Sci. USA 1997, 94, 1908–1913. [Google Scholar] [CrossRef]
- Kondo, M.; Scherer, D.C.; Miyamoto, T.; King, A.G.; Akashi, K.; Sugamura, K.; Weissman, I.L. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature 2000, 407, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Tsapogas, P.; Mooney, C.J.; Brown, G.; Rolink, A. The cytokine Flt3-ligand in normal and malignant hematopoiesis. Int. J. Mol. Sci. 2017, 18, 1115. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, R.; Morita, Y.; Ooehara, J.; Hamanaka, S.; Onodera, M.; Rudolph, K.L.; Ema, H.; Nakauchi, H. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell 2013, 154, 1112–1126. [Google Scholar] [CrossRef]
- Shin, J.Y.; Hu, W.; Naramura, M.; Park, C.Y. High c-Kit expression identifies hematopoietic stem cells with impaired self-renewal and megakaryocytic bias. J. Exp. Med. 2014, 211, 217–231. [Google Scholar] [CrossRef]
- Sanjuan-Pla, A.; Macaulay, I.C.; Jensen, C.T.; Woll, P.S.; Luis, T.C.; Mead, A.; Moore, S.; Carella, C.; Bouriez Jones, T.; Chowdhury, O.; et al. Platelet-biased stem cells reside at the apex of the hematopoietic stem-cell hierarchy. Nature 2013, 502, 232–236. [Google Scholar] [CrossRef]
- Grover, A.; Mancini, E.; Moore, S.; Mead, A.J.; Atkinson, D.; Rasmussen, K.D.; O’Carroll, D.; Jacobsen, S.E.; Nerlove, C. Erythropoietin guides multipotent hematopoietic progenitor cells towards an erythroid fate. J. Exp. Med. 2014, 211, 181–188. [Google Scholar] [CrossRef]
- Metcalf, D.; Burgess, A.W. Clonal analysis of progenitor cell commitment of granulocyte or macrophage production. J. Cell Physiol. 1982, 111, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Rieger, M.A.; Hoppe, P.S.; Smejkal, B.M.; Eitelhuber, A.C.; Schroeder, T. Hematopoietic cytokines can instruct lineage choice. Science 2009, 325, 217–218. [Google Scholar] [CrossRef] [PubMed]
- Tsapogas, P.; Swee, L.K.; Nusser, A.; Nuber, N.; Kreuzaler, M.; Capoferri, G.; Rolink, H.; Ceredig, R.; Rolink, A. In vivo evidence for an instructive role of fms-like tyrosine kinase-3 (FLT3) ligand in hematopoietic development. Haematologica 2014, 99, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Colmore, A.; Amorim, M.; Pontier, A.L.; Wang, S.; Jablonski, E.; Sipkins, D.A. Leukaemia cells create bone marrow niches that disrupt the behaviour of normal hematopoietic progenitor cells. Science 2008, 322, 1861–1865. [Google Scholar] [CrossRef]
- Chasis, J.A.; Mohandas, N. Erythroblastic islands: Niches for erythropoiesis. Blood 2008, 112, 470–478. [Google Scholar] [CrossRef]
- Comazzetto, S.; Murphy, M.M.; Berto, S.; Jeffery, E.; Zhao, Z.; Morrison, S.J. Restricted hematopoietic progenitors and reythropoiesis require SCF from leptin receptor+ niche cells in bone marrow. Cell Stem Cell 2019, 245, 477–486. [Google Scholar] [CrossRef]
- Zwezdaryk, K.J.; Coffelt, S.B.; Figueroa, Y.G.; Liu, J.; Phinney, D.G.; LaMarca, H.L.; Florez, L.; Morris, C.B.; Hoyle, G.W.; Scandurro, A.B. Erythropoietin, a hypoxia-regulated factor, elicits a pro-angiogenic program in human mesenchymal stem cells. Exp. Hemato. 2007, 35, 640–652. [Google Scholar] [CrossRef]
- Anagnostou, A.; Lee, E.S.; Keissiman, N.; Levinson, R.; Steiner, M. Erytrhopoietin has a mitogenic and postive chemotactic effect on endothelial cells. Proc. Natl. Acad. Sci. USA 1990, 87, 5978–5982. [Google Scholar] [CrossRef]
- Poniewierska-Baran, A.; Rajewska, J.R.; Ratajczak, M.Z. Erythropoietin enhances migration of human nuroblastoma cells: In vitro studies and potential therapeutic implication. J. Cancer Stem Cell Res. 2017, 5, e1003. [Google Scholar]
- Gomez-Cambronero, J.; Horn, J.; Paul, C.C.; Baumann, M.A. Granulocyte-macrophage colony-stimulating factor is a chemoattractant cytokine for human neutrophils: Involvement of the ribosomal p70 S6 kinase signalling pathway. J. Immunol. 2003, 171, 6846–6855. [Google Scholar] [CrossRef]
- Bussolino, F.; Wang, J.M.; Defilippi, P.; Turrini, F.; Sanavio, F.; Edgell, J.; Aglietta, M.; Arese, P.; Mantovani, A. Granulocyte- and granulocyte-macrophage-colony stimulating factors induce human endothelial cells to migrateand proliferate. Nature 1989, 337, 471. [Google Scholar] [CrossRef] [PubMed]
- Vaillant, P.; Muller, V.; Martinet, Y.; Martinet, N. Human granulocyte and granulocyte-macrophage-colony stimulating factors are chemotactic and “competence” growth factors for human mesenchymal cells. Biochem. Biophys. Res. Commun. 1993, 192, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Fuller, K.; Owens, J.M.; Jagger, A.; Wilson, A.; Moss, R.; Chambers, T.J. Macrophage colony-stimulating factors stimulates survival and chemotactic behaviour in isolated osteoclasts. J. Exp. Med. 1993, 178, 1733–1744. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.M.; Griffin, J.D.; Rambaldi, A.; Chen, Z.D.; Mantovani, A. Induction of monocyte migration by recombinant macrophage colony-stimulating factor. J. Immunol. 1988, 141, 575–579. [Google Scholar]
- Vedham, V.; Phee, H.; Coggeshall, K.M. Vav activation and function as a Rac guanine nucleotide exchange factor in macrophage colony-stimulating factor-induced macrophage chemotaxis. Mol Cell Biol. 2005, 25, 4211–4220. [Google Scholar] [CrossRef][Green Version]
- Pierce, J.H.; Di Marco, E.; Cox, G.W.; Lombardi, D.; Ruggiero, M.; Varesio, L.; Wang, L.M.; Choudhury, G.C.; Sakaguchi, A.Y.; Di Fiore, P.P. Macrophage-colony-stimulating factor (CSF-1) induces proliferation, chemotaxis, and reversible monocytic differentiation in myeloid progenitor cells transfected with human c-fms/CSF-1 receptor cDNA. Proc. Natl. Acad. Sci. USA 1990, 87, 5613–5617. [Google Scholar] [CrossRef]
- Pollard, J.W.; Bartocci, A.; Arceci, R.; Orlofsky, A.; Ladner, M.B.; Stanley, E.R. Apparent role of the macrophage growth factor, CSF-1, in placental development. Nature 1987, 330, 484–486. [Google Scholar] [CrossRef]
- Filderman, A.E.; Bruckner, A.; Kacinski, B.M.; Deng, N.; Remold, H.G. Macrophage colony-stimulating factor (CSF-1) enhances invasiveness in CSF-1 receptor-positive carcinoma cell lines. Cancer Res. 1992, 52, 3661–3666. [Google Scholar]
- Alberti-Servera, L.; von Muenchow, L.; Tsapogas, P.; Capoferri, G.; Eschbach, K.; Beisel, C. Single-cell RNA sequencing reveals developmental heterogeneity among early lymphoid progenitors. EMBO J. 2017, 36, 3619–3633. [Google Scholar] [CrossRef]
- Weinreb, C.; Rodriguez-Fraticelli, A.R.; Camargo, F.; Klein, A.M. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science 2020, 367, pii: eaaw3381. [Google Scholar] [CrossRef]
- Nestorowa, S.; Hamey, F.K.; Pijuan Sala, B.; Diamanti, E.; Shepherd, M.; Laurenti, E.; Wilson, N.K.; Kent, D.G.; Gottens, B. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 2016, 128, e20–e31. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, C.; Kaneko, K. A dynamical-systems view of stem cell biology. Science 2012, 338, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Raser, J.M.; O’Shea, E.K. Noise in gene expression: Origins, consequences, and control. Science 2005, 309, 2010–2013. [Google Scholar] [CrossRef] [PubMed]
- Lund, R.J.; Narva, E.; Lahesmaa, R. Genetic and epigenetic stability of human pluripotent stem cells. Nat. Rev. Genet. 2012, 13, 732–744. [Google Scholar] [CrossRef]
- Barroso, G.V.; Puzovic, N.; Dutheil, J.Y. The evolution of gene-specific transcriptional noise is driven by selection at the pathway level. Genetics 2018, 208, 1173–1189. [Google Scholar] [CrossRef]
- Doi, Y.; Yokota, Y.; Satoh, Y.; Okuzaki, D.; Tokunaga, M.; Ishibashi, T.; Sudo, T.; Ueda, T.; Shingai, Y.; Ichi, M. Variable SATB1 levels regulate hematopoietic stem cell heterogeneity with distinct lineage fate. Cell Rep. 2018, 23, 3223–3235. [Google Scholar] [CrossRef]
- Chang, H.H.; Hemberg, M.; Barahana, M.; Ingber, D.E.; Huang, S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 2008, 453, 544–547. [Google Scholar] [CrossRef]
- Yokota, T. “Hierarchy” and “Holoaciary”: A paradigm of the hematopoietic system. Cells 2019, 8, pii E1138. [Google Scholar] [CrossRef]
- Lloyd, A.M.; Schena, M.; Walbot, V.; Davis, R.W. Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science 1994, 266, 436–439. [Google Scholar] [CrossRef]
- Yu, Q.; Li, P.; Liang, N.; Wang, H.; Xu, M.; Wu, S. Cell-fate specification in Arabidopsis roots requires coordinative action of lineage instruction and positional reprogramming. Plant Physiol. 2017, 175, 816–827. [Google Scholar] [CrossRef]
- Szyf, M.; McGowan, P.M.; Meaney, M.J. The social environment and the epigenome. Environ. Mol. Mutagenesis 2008, 49, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Bloom, A.B.; Zaman, M.H. Influence of the microenvironment on cell fate determination and migration. Physiol. Genomics 2014, 46, 309–314. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brown, G. Towards a New Understanding of Decision-Making by Hematopoietic Stem Cells. Int. J. Mol. Sci. 2020, 21, 2362. https://doi.org/10.3390/ijms21072362
Brown G. Towards a New Understanding of Decision-Making by Hematopoietic Stem Cells. International Journal of Molecular Sciences. 2020; 21(7):2362. https://doi.org/10.3390/ijms21072362
Chicago/Turabian StyleBrown, Geoffrey. 2020. "Towards a New Understanding of Decision-Making by Hematopoietic Stem Cells" International Journal of Molecular Sciences 21, no. 7: 2362. https://doi.org/10.3390/ijms21072362
APA StyleBrown, G. (2020). Towards a New Understanding of Decision-Making by Hematopoietic Stem Cells. International Journal of Molecular Sciences, 21(7), 2362. https://doi.org/10.3390/ijms21072362



