Abstract
The tumor-suppressor protein p16 is paradoxically overexpressed in cervical cancer (CC). Despite its potential as a biomarker, its clinical value and the reasons for its failure in tumor suppression remain unclear. Our purpose was to determine p16 clinical and biological significance in CC. p16 expression pattern was examined by immunohistochemistry in 78 CC cases (high-grade squamous intraepithelial lesions (HSILs) and squamous cell carcinomas of the cervix –SCCCs). CC cell proliferation and invasion were monitored by real-time cell analysis and Transwell® invasion assay, respectively. Cytoplasmic p16 interactors were identified from immunoprecipitated extracts by liquid chromatography-tandem mass spectrometry, and colocalization was confirmed by double-immunofluorescence. We observed that SCCCs showed significantly more cytoplasmic than nuclear p16 expression than HSILs. Importantly, nuclear p16 absence significantly predicted poor outcome in SCCC patients irrespective of other clinical parameters. Moreover, we demonstrated that cytoplasmic p16 interacted with CDK4 and other unreported proteins, such as BANF1, AKAP8 and AGTRAP, which could sequester p16 to avoid nuclear translocation, and then, impair its anti-tumor function. Our results suggest that the absence of nuclear p16 could be a diagnostic biomarker between HSIL and SCCC, and an independent prognostic biomarker in SCCC; and explain why p16 overexpression fails to stop CC growth.
1. Introduction
Squamous cell carcinoma of the cervix (SCCC) accounts for 70–80% of all cervical cancer (CC) cases, which is the fourth most frequent cancer type in women worldwide [1], with a 5-year survival of 57–67% in Europe [2]. SCCC pathogenesis is a slow multi-stage process: when a high-risk human papillomavirus (HR-HPV) infection—present in 99.7% of cases [2,3]—persists in dividing cells, these can originate a high-grade squamous intraepithelial lesion (HSIL); if, additionally, some molecular alterations occur, tumor cells extend beyond the basement membrane of the epithelium, and then an invasive carcinoma appears [3,4]. Unfortunately, early SCCC is often asymptomatic and detected at an advanced stage when treatments are ineffective. Developments in early diagnosis are therefore much needed [1]. Currently, the standard method for SCCC diagnosis has many limitations because it is mainly based on subjective interpretation of histological changes in cervical tissue [2,4,5,6]. Intriguingly, as CC development seems to be a continuous process, it is quite difficult to morphologically distinguish an HSIL which has begun to invade and an SCCC which has just penetrated the basement membrane. However, these apparently similar tumors display very different clinical behaviors. Women with HSIL are treated with surgery that is usually successful, with more than 85% of patients being cured [7]; those with SCCC receive surgery plus chemoradiotherapy, but unfortunately, half of them die from the disease [2]. An accurate diagnosis is therefore crucial for the successful clinical management of these patients.
Recently, an HPV DNA test has been introduced as a screening tool; this has proved to be much more sensitive than traditional methods [2], indicating that molecular biomarkers are very informative and of great utility. Indeed, they are objective, quantitative and easily reproducible in all hospitals and laboratories, for many tumor types [8,9,10]. However, to date, no molecular panel has been available for SCCC, in contrast to other malignancies, such as breast [8,11], lung [12] and colorectal [13] cancers, so there is an urgent need for molecular biomarkers that will enable early diagnosis, prognosis and targeted therapy for SCCC [14]. A better understanding of the molecular mechanisms leading to SCCC could help us develop better clinical strategies to improve patients’ likelihood of survival. In fact, the European Society of Medical Oncology has recommended that more research be undertaken to identify molecular biomarkers in CC [2].
The cyclin-dependent kinase inhibitor 2A protein, also called p16, is a well-known tumor-suppressor protein which is down-regulated in many tumors, and is involved in regulating cell cycle, senescence, apoptosis, cell invasion and angiogenesis [3,5,15]. In spite of these well-recognized tumor-suppressor properties, p16 expression paradoxically increases with the severity of the cervical lesion [3,5,14,16], and has been then proposed as a useful marker for identifying such lesions [5,17,18,19,20]. In fact, its ancillary use in cervical cytology has improved the accuracy of histological diagnosis [5,21], mainly in premalignant lesions [4]. However, the specificity of p16 has been insufficiently studied in SCCC [18], and its prognostic value remains controversial [3,5,14].
As there is a lack of consensus and clear guidelines regarding the use of p16 expression in routine clinical management of SCCC [2,5,19], we aimed to shed some light on the clinical utility and biological significance of p16 overexpression during SCCC progression.
2. Results
2.1. Expression Pattern and Clinical Relevance of p16 in SCCC Patients
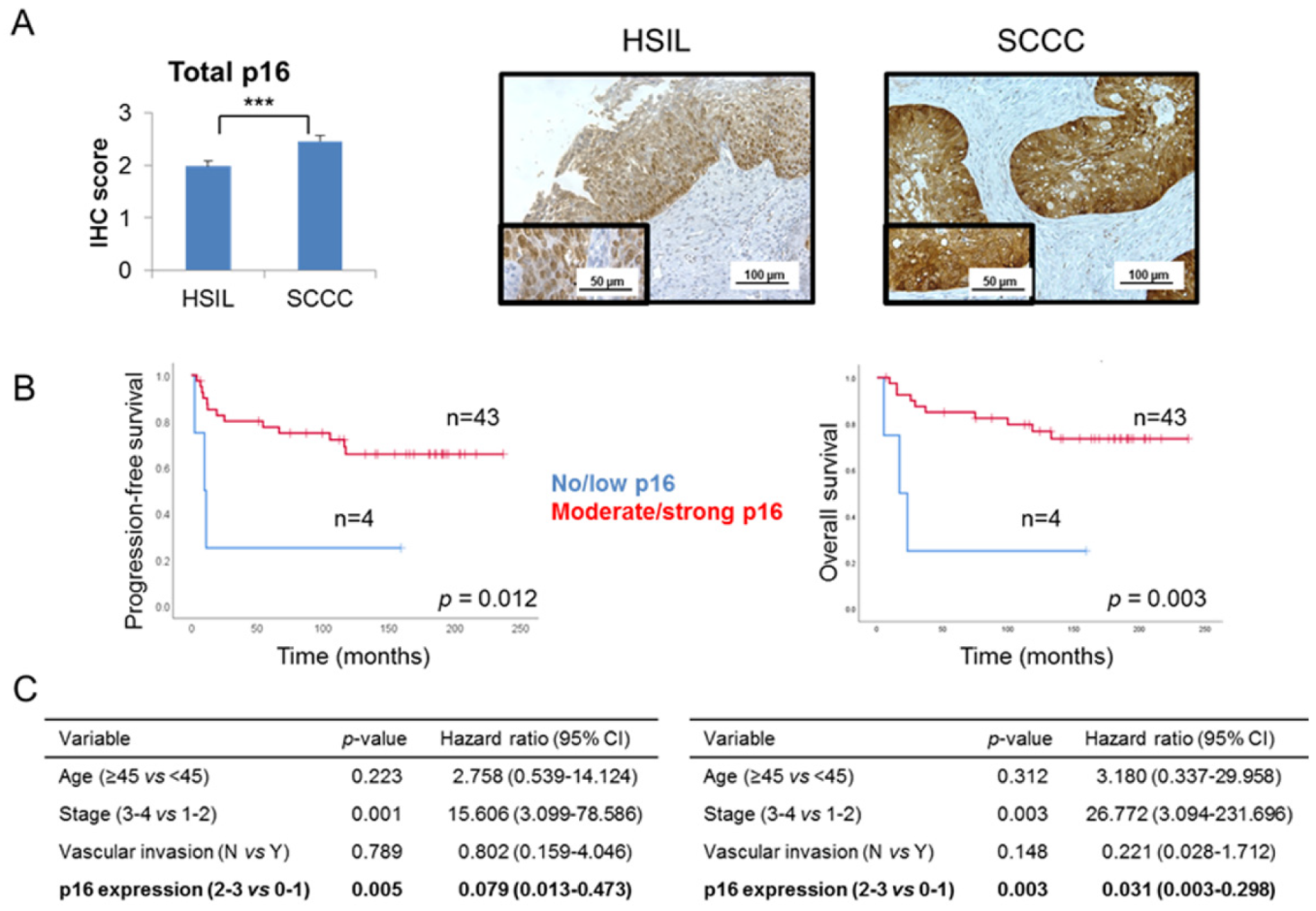
Global p16 expression was examined in our series of 29 HSILs and 49 SCCCs, taking into account the criteria which are routinely used in the clinical practice. We observed that SCCC tumors had a significantly higher level of p16 expression than did HSILs (p < 0.001) (Figure 1A and Table 1). No statistical association was found between global p16 expression and pathological variables (data not shown). Interestingly, significant associations between a high level of global p16 expression and good prognosis and outcome were found in SCCC patients (Figure 1B), independent of other clinical parameters crucial in SCCC prognosis, such as age, stage and vascular invasion (Figure 1C and Figure S1). Indeed, Cox analyses showed that strong expression of p16 had a protective effect against relapse and death (hazard ratios 0.079 and 0.031 for progression-free survival (PFS) and overall survival (OS), respectively) (Figure 1C). These results contribute to the paradox of p16 overexpression in SCCC, since a tumor-suppressor protein which significantly adds to a good outcome is overall overexpressed in this aggressive cancer type.
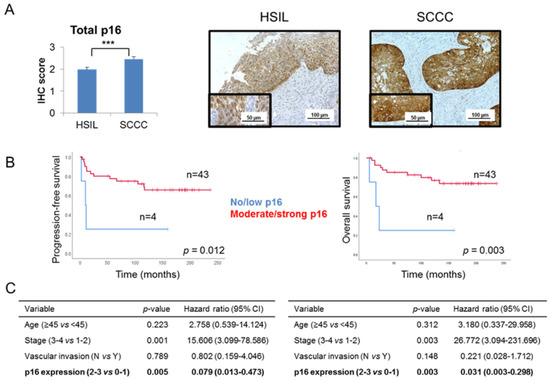
Figure 1.
Global p16 expression in cervical tumors. (A) Immunohistochemical expression of p16 was examined in a series of 29 high-grade squamous intraepithelial lesions (HSILs) and 49 squamous cell carcinomas of the cervix (SCCCs) (*** p < 0.001). Representative images are shown at 200× magnification, and details are highlighted at 400×. (B) Association between global p16 expression and progression-free survival (PFS) and overall survival (OS) in SCCC patients. (C) Multivariate analysis showing the independent association between p16 expression and PFS (left) or OS (right), regardless of vascular invasion, stage and age of SCCC patients. (CI, confidence interval).

Table 1.
Distribution of p16 expression and subcellular location in our series of 78 cervical tumors. Number and percentage of high-grade squamous intraepithelial lesions (HSILs) (n = 29) and squamous cell carcinomas of the cervix (SCCCs) (n = 49) patients with negative (0), weak (1), moderate (2) and strong (3) immunohistochemical expression of global, nuclear and cytoplasmic p16 protein levels. The cytoplasmic/nuclear p16 ratio was calculated by dividing cytoplasmic by nuclear p16 immunohistochemical score.
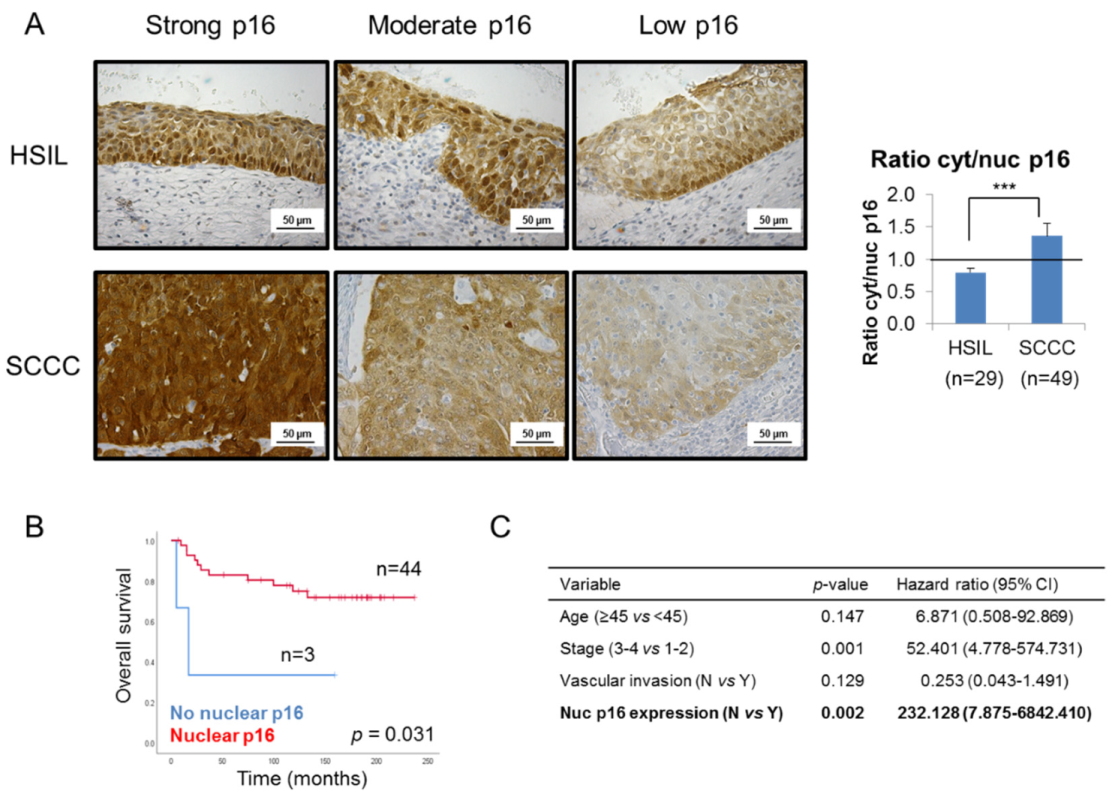
In order to shed some light on this controversy, we focused on p16 subcellular location, and observed that SCCC tumors showed stronger cytoplasmic than nuclear staining than the HSIL samples, regardless of the global expression of the protein (Figure 2A and Table 1). Indeed, SCCC samples displayed a significantly higher cytoplasmic/nuclear p16 ratio than HSIL (p < 0.001). This finding suggests that not only p16 expression, but also its subcellular location, could be diagnostic biomarkers which can distinguish between HSIL and SCCC.
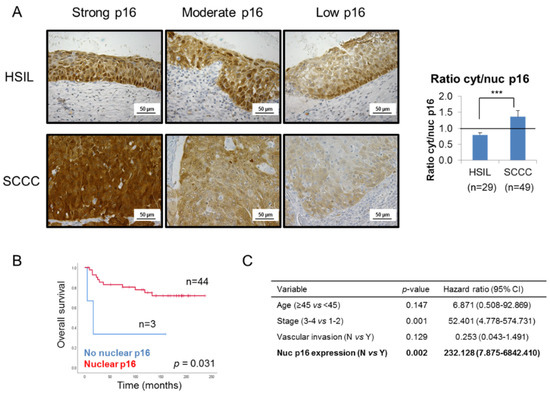
Figure 2.
Subcellular location of p16 in cervical tumors. (A) Representative images of p16 staining in 3 high-grade intraepithelial lesions (HSILs) and 3 squamous carcinomas of the cervix (SCCCs) at 400× magnification showing that SCCC samples (n = 49) showed stronger cytoplasmic than nuclear p16 expression than HSIL (n = 29), regardless of the degree of global p16 staining (strong, moderate or low). Note p16 negativity in the nuclei of the low p16-expressing SCCC sample. The ratio cytoplasmic to nuclear (cyt/nuc) p16 expression was calculated by dividing p16 immunohistochemical score in each subcellular compartment. The horizontal line in the histogram shows equal amounts of cytoplasmic and nuclear p16 (ratio cyt/nuc p16 = 1) (*** p < 0.001). (B) Association between p16 subcellular location and overall survival (OS) in SCCC patients. (C) Multivariate analysis revealed an independent association between p16 subcellular location and OS, regardless of vascular invasion, stage and age of SCCC patients. (CI, confidence interval).
Furthermore, although no significant association was found between nuclear or cytoplasmic p16 expression and pathological variables (data not shown), the absence of nuclear p16, observed in only 6% of SCCC patients though (Table 1), was significantly associated with a very poor outcome (p = 0.031) (Figure 2B and Table S1). Moreover, this association was significantly independent of clinical parameters like age, stage or vascular invasion (p = 0.002); and we found that the lack of nuclear p16 significantly increased the risk of death by more than 200-fold (Figure 2C).
These findings indicate that although p16 is globally overexpressed in SCCC, it is not mainly located in the nucleus, where it could exert its tumor-suppressor functions. Therefore, these results could explain the p16 paradox in SCCC.
2.2. p16 Subcellular Location in SCCC Cell Lines
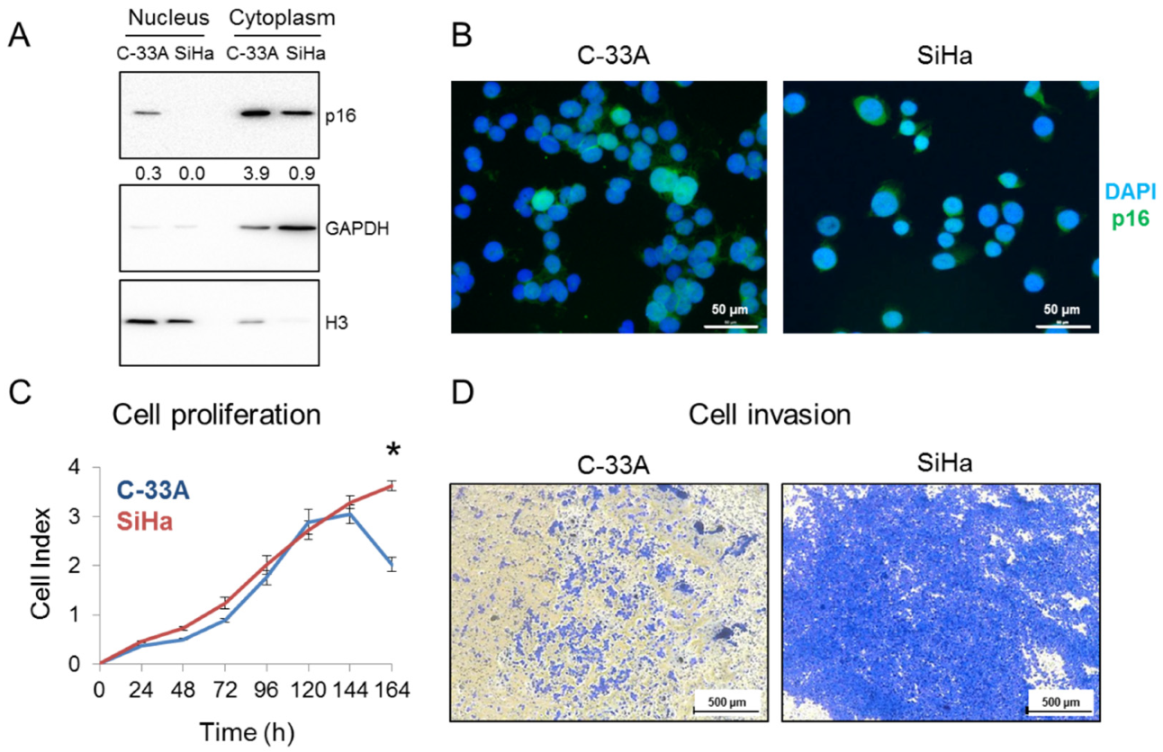
p16 expression pattern was also interrogated in SCCC cell lines. Western blot (Figure 3A) and immunofluorescence (Figure 3B) revealed that C-33A cells expressed p16 in both the nucleus and the cytoplasm, while SiHa cells only expressed it in the cytoplasm. Similar to what we observed in SCCC tumors, this differential subcellular location could affect cell aggressiveness, as SiHa had greater proliferative (Figure 3C) and invasive (Figure 3D) properties than did C-33A. These abilities were also examined in the HeLa cell line (Figure S2), which was derived from a cervical adenocarcinoma, with nearly identical clinical features to SCCC [22]. Interestingly, we found a very similar pattern of p16 expression and growth and invasive capacities in HeLa and SiHa cells, indicating that the absence of nuclear location of p16 is related to CC aggressiveness.
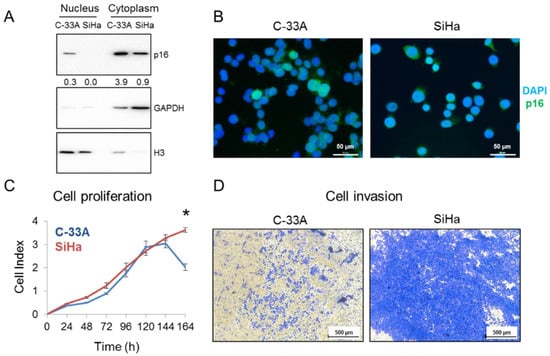
Figure 3.
p16 subcellular location in SCCC cell lines. (A) Nuclear and cytoplasmic protein fractions of C-33A and SiHa were separately subjected to western blot to check p16 expression. GAPDH and Histone H3 were used as loading controls of each subcellular fraction. Numbers indicate the ratio of p16 signal relative to that in the loading control, measured by densitometry. (B) p16 expression was examined by immunofluorescence in C-33A and SiHa cells. Images were acquired at 400× magnification. (C) Cell proliferation of C-33A and SiHa cell lines was measured by real-time cell analysis for 7 days (* p < 0.05). (D) Cell invasion of C-33A and SiHa cell lines was examined by their ability to penetrate a Matrigel® layer for 3 days. Images were acquired at 50× magnification.
2.3. Cytoplasmic p16 Interactome in SCCC
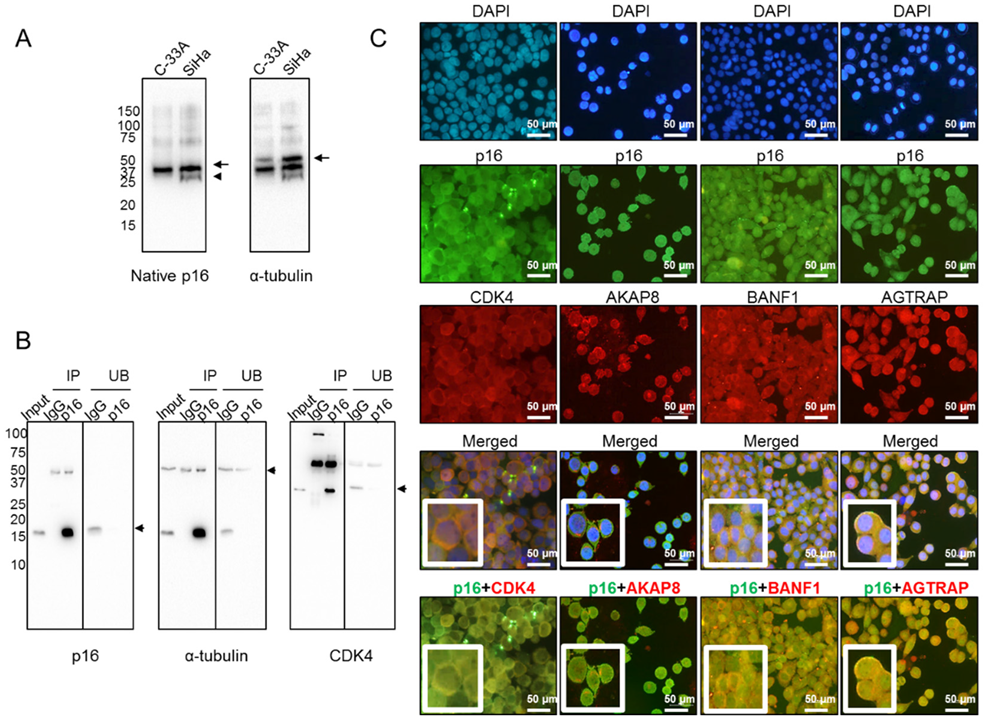
To understand why p16 is mainly confined to the cytoplasm in aggressive SCCC tumors, we hypothesized that it could be sequestered by other proteins to form large macromolecular complexes in order to avoid nuclear translocation, as has been described in other tumor types [15]. To test this hypothesis, we first examined the conformation of native p16 by western blot under non-denaturing conditions. We observed a band in SiHa, but not in C-33A, which may indicate a differential 3D conformation of native p16 (Figure 4A). This finding suggested that p16 could actually be bound to other proteins that sequester it in the cytoplasm. Therefore, cytoplasmic p16 was immunoprecipitated from SiHa cells, which lacked nuclear p16 (Figure 3A). Figure 4B shows immunoprecipitation efficiency regarding the pull-down of p16 and its interactors, as one of the well-known proteins that interacts with p16, the cyclin-dependent kinase 4 (CDK4) [15], was also detected in our immunoprecipitated extract (Figure 4B). To be confident in the identification of p16 interactors, those extracts were digested by taking a dual approach—in-gel and in-solution digestion—and then subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Table S2). Data mining of LC-MS/MS-generated proteomic data revealed that 16 proteins were simultaneously identified in both extracts pulled down with the anti-p16 antibody, but not with IgG (Table 2). They were proposed to be at least a part of the cytoplasmic p16 interactome in SiHa cells with a false-discovery rate (FDR) < 0.01. It is worth mentioning that no protein from HPV was found to be bound to cytoplasmic p16. Importantly, we observed that the vast majority of those human interacting proteins had not been previously reported in the BioGRID interaction repository, which currently contains 229 interactors for p16 (January 2020). The only two proteins found to be common to the BioGRID list and the immunoprecipitated extracts digested in-gel and in-solution were CDK4 and C1QBP (Figure S3). Our findings therefore include 14 new potentially interacting cytoplasmic p16 proteins.
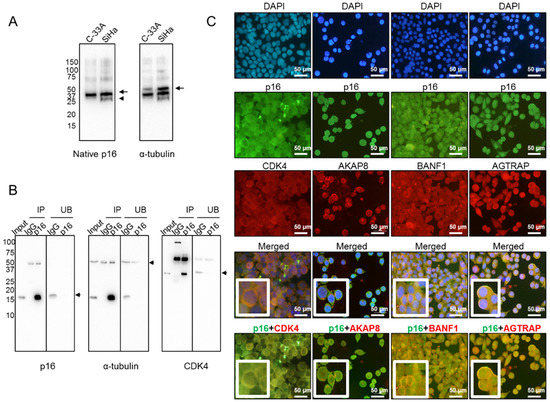
Figure 4.
Cytoplasmic p16 interactors in SiHa cells. (A) Native form of p16 protein was examined in both C-33A and SiHa cell lines by western blot under non-denaturing conditions. Arrows point to the proteins of interest, while the arrowhead indicates a different 3D conformation of native p16 in SiHa, as compared with C-33A. α-tubulin was used as a loading control. (B) Exclusively cytoplasmic p16 was immunoprecipitated from SiHa cells, and efficiency was checked by western blot. Immunoprecipitated (IP) and unbound (UB) fractions upon incubation with IgG and anti-p16 antibodies are shown. α-tubulin was used as a loading control. Immunoprecipitation of p16 interactors was checked by incubating the same membrane with an anti-CDK4 antibody, a very well-known p16-interacting protein. The arrows indicate the proteins of interest. (C) Colocalization in the cytoplasm of SiHa cells of p16 (in green) and four interactors (in red), identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS), was revealed by double immunofluorescence. Images were acquired at 400× magnification.

Table 2.
Cytoplasmic p16 interactome in SiHa cells. 16 proteins were simultaneously identified in two independent experiments through in-solution- and in-gel-digested extracts from immunoprecipitated p16 in the SiHa cell line (false-discovery rate (FDR) < 0.01).
2.4. Colocalization of Cytoplasmic p16 and Interactors in SiHa Cells
Some of the cytoplasmic p16 interactors identified by LC-MS/MS were selected for further examination by double immunofluorescence because of their involvement in other carcinogenic processes [23,24,25,26,27,28,29]. As seen in Figure 4C, we found that CDK4, the A-kinase anchor protein 8 (AKAP8), the barrier to autointegration factor (BANF1), and the type-1 angiotensin II receptor-associated protein (AGTRAP) colocalized with p16 in the cytoplasm of SiHa cells, confirming that these proteins interact with cytoplasmic p16 in SiHa cells.
3. Discussion
Although p16 overexpression is a potential biomarker for CC clinical management [5,17,18], there is a lack of consensus [5,19] and standard guidelines [2] concerning its routine use. Similarly, the biological significance of the paradoxical activation of this tumor-suppressor protein in CC is not yet well understood.
We confirmed p16 overexpression in our series of 78 cervical tumors, as extensively described [3,14,16,21], since strong p16 expression is a proven useful surrogate biomarker for tumors with transcriptionally active HR-HPV [5]. Importantly, we found that p16 expression—and especially the cytoplasmic/nuclear p16 ratio—significantly distinguished HSIL from SCCC. Our findings are therefore evidence of the usefulness of p16 as a diagnostic biomarker between HSIL and SCCC, as previously proposed [18,19]. In fact, it has facilitated inter-observer agreement and then has improved the accuracy of traditional diagnostic methods based on histological observation in lower grade lesions [4,5,6,21,30].
Notably, a high level of p16 expression was significantly associated with longer survival in our SCCC series, irrespective of other clinical parameters such as age, stage and vascular invasion. Although some reports have associated p16 overexpression with poor prognosis [3,14], our results are consistent with those of a meta-analysis involving 1633 CC patients [5]. It is thought that HR-HPV-related tumors are less genetically altered, and therefore tend to respond better to therapy and to have improved outcome [5,15]. Here, we noticed that 6% of SCCCs had null or low p16 levels. Although our sample number is limited, we have observed the same small proportion of p16 negative SCCC patients than a recent report where such patients negative for global p16, which represented only 4% of CC cases, had significantly worse OS [31].
It has been suggested that the lack of a standardized cut-off point for p16 overexpression could have masked its true clinical value [5]. However, as it is difficult to establish a cut-off value in subjectively evaluated immunohistochemical staining, the novelty of our study relies on the relevance of p16 subcellular location. To date, p16 expression pattern remains controversial, because there is no fundamental justification yet as to whether cytoplasmic/nuclear or only nuclear staining should be taken as positive [15]. As other authors have found [5,32], there was no association between p16 expression or location and pathological variables in our study, but we did find that the lack of nuclear p16 allowed SCCC to acquire aggressive features both in tumors and in vitro. This interesting finding goes some way to explaining the paradoxical p16 overexpression in SCCC. It is thought that retinoblastoma (Rb) protein inhibition by HPV E7 protein triggers a negative feedback that leads to p16 overexpression, in a failed attempt to stop cell proliferation [3,5,15,17]. Our results could explain this failure, since, although overexpressed, p16 would not be able to fulfil any anti-tumor function in the cytoplasm. Cytoplasmic levels of some typical nuclear cell cycle regulators, such as p16, p27 and PTEN, have been described and associated with tumor progression in uterine leiomyosarcomas, astrocytomas, gastrointestinal stromal tumors and breast and colorectal cancer [15,17]. However, cytoplasmic p16 has been subtly examined in CC, found to be not significantly associated with the histological grade of the lesion [17] and associated with a borderline-significantly worse two-year PFS in HSIL patients (p = 0.049) [33].
Cytoplasmic p16 has only recently been evaluated, having been considered a background factor in many studies, and so its significance is due for reinterpretation [15]. Some authors have characterized p16 expression in CC cell lines and found very opposite results in comparison with ours: exclusively cytoplasmic p16 in C-33A cells and nuclear/cytoplasmic p16 in SiHa and HeLa cells [18]. Nevertheless, it is important to mention that they described p16 subcellular location solely in cell lines, which are just models of disease with many limitations that can influence protein levels and location. In contrast, our conclusions are supported by observations in tissues. Based on this, our study demonstrates that cytoplasmic p16 is not an artefact but has important biological and clinical implications. It has been hypothesized that the binding of p16 to other cytoplasmic proteins could form large molecules that are unable to pass through the nuclear membrane pores. Among these proteins, AE1 is thought to be present in gastric and colon cancer cells [15]; while PCNA, MCM6, α/β/γ-actin and α/β-tubulin, among others, have been observed in a lymphoblastoid cell line [34]. Although it is conceivable that cytoplasmic p16 interactors are cellular-context-dependent, CDK4 has been proposed as a strong candidate for sequestering p16 in the cytoplasm [15]. In fact, the interaction between p16 and CDK4 has already been demonstrated via co-immunoprecipitation in SiHa cells [35]. Accordingly, we also found binding and colocalization of p16 and CDK4 in SiHa cell cytoplasm, which strengthened the validity of our results. We also detected another p16 interactor already reported in the BioGRID repository, the complement component 1 Q subcomponent-binding protein (C1QBP), which has been described as associating with the CDKN2A product to mediate apoptosis [36]. Apart from these, 14 new interactors of cytoplasmic p16 were also identified here. Importantly, we confirmed the colocalization of p16 with a protein subset previously involved in carcinogenesis with a specific role in nuclear shuttling. For instance, BANF1 has been reported to be involved in colony formation, migration and invasion in CC [37] and in the development of hepatocellular [23] and esophageal squamous cell [24] carcinomas and gastric cancer [26]. It is also known to interact with a protein which prevents nuclear accumulation of other proteins, such as β-catenin [38]. Another novel p16 interactor identified here, AKAP8, dynamically interacts with other proteins during cell cycle progression [39] and appears to play an important role in promoting lung [27], ovarian [28] and rectal [29] cancers. Moreover, it helps deliver cyclin D/E to CDK4 [40] and interacts with a kinase which phosphorylates p27 to prevent its translocation to the nucleus [41]. Finally, we identified AGTRAP, a HIF1α direct target related to reduced PFS in melanoma [25] and to tumor growth and angiogenesis in Lewis lung carcinoma [42].
Further evaluation is warranted to confirm these observations in larger cohorts of patients, and to answer many questions that have been raised. For instance, whether p16 sequestration in the cytoplasm per se is the reason for the failure of its tumor-suppressor function or whether it involves more complex mechanisms is still to be determined. Since p16 overexpression has been observed at the invasive front of endometrial, colorectal and basal cell carcinoma, it has been proposed that cytoplasmic p16 is involved in the dissociation of focal adhesions, and then, related to cell invasion [15], lymphangiogenesis and lymphatic metastasis [5].
The potential therapeutic value of p16 is also to be elucidated. On the one hand, depletion of p16 has been shown to promote chemo- and radioresistance in HeLa cells [32]. On the other hand, it has been suggested that p16 could be a therapeutic target in CC, since its knockdown inhibits cell proliferation, migration and invasion in SiHa and HeLa cell lines [43] and sensitizes SiHa cells to cisplatin [35]. However, given the importance of its tumor-suppressor properties in other cell types, it is not clear how p16 inhibition would affect organism homeostasis. Here, we draw attention to a potential therapeutic use for p16 that works not by targeting it directly, but by inhibiting the proteins which sequester it in the cytoplasm. This strategy would be expected to release and translocate p16 to the nucleus in order to exert its tumor-suppressor function.
Recently, novel proteins, such as PDL1 [44], survivin [45] and SIRT1 [46], have become of interest as diagnostic or prognostic biomarkers in cervical malignancies. Undoubtedly, these findings are very important for SCCC clinical management, but their implementation would have economic consequences. In contrast, our results imply that it is only necessary to evaluate the p16 immunohistochemical pattern accurately—and since this is already routinely performed—it would not incur any additional cost.
4. Materials and Methods
4.1. Patient Samples
We analyzed a series of 78 formalin-fixed, paraffin-embedded samples from women diagnosed with HSIL (n = 29) and SCCC (n = 49). All patients were diagnosed between 1995 and 2015 in the Pathology Department of the Complejo Hospitalario de Navarra (Pamplona, Spain), Hospital Universitari de Bellvitge, (L’Hospitalet de Llobregat, Spain), Complejo Asistencial Universitario (León, Spain), and Hospital Universitari Arnau de Vilanova (Lleida, Spain). Clinical characteristics of SCCC patients are summarized in Table S3. No clinical follow-up was available for HSIL patients, since they were all successfully cured by surgery. All tumors were surgically removed and staged according to their size, histological grade and lymph node involvement. All cases were ensured to harbor at least 70% tumor cells. None of the patients had received radiotherapy or chemotherapy before surgery. The study was approved by the Navarre Ethics Committee for Clinical Research (PI_2018/75) on September 30th 2019, procedures were in accordance with the Helsinki declaration, and informed consent was obtained according to the current Spanish legislation.
4.2. Immunohistochemistry
Sections (4-µm thick) of 29 HSILs and 49 SCCCs were placed on slides and deparaffinized. After performing antigen retrieval at 37 °C for 30 min with the cell conditioning 1 buffer (catalog number 950–124) at pH 8.4, samples were incubated with the anti-p16 antibody (catalog number 805–4713, clone E6H4) [31] at 1 µg/mL for 28 min at 37 °C in a Benchmark XT® Ventana immunostainer (all reagents and equipment from Roche, Basel, Switzerland). The pattern and intensity of expression were evaluated blind by two independent observers (T.Z. and E.M.-S.), and ascribed to one of four categories: 0, no expression; 1, weak expression; 2, moderate expression; and 3, strong expression. Images were acquired with a Leica DM4000B microscope (Leica, Wetzlar, Germany) and the NIS Elements v4.30 program (Nikon Instruments, Amsterdam, Netherlands).
4.3. Cell Lines
Two human SCCC cell lines were used in this study, C-33A (an HPV-negative carcinoma) and SiHa (an HPV16-positive SCCC), both of which were purchased from the American Type Cell Collection (ATCC, Rockville, MD, USA). Additionally, the HeLa cell line (HPV18-positive cervical adenocarcinoma) was also obtained from the ATCC. They were all grown in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (all from Life Technologies, Carlsbad, CA, USA), at 37 °C in a humidified atmosphere with 5% CO2. Experiments were performed with Mycoplasma-free and recently authenticated cell lines at low passage.
4.4. Western Blot
p16 expression levels were checked in both cytoplasmic and nuclear fractions in three CC cell lines by western blot. Briefly, for subcellular fractionation, C-33A, SiHa and HeLa cells were grown to 70–80% confluence and lysed with a phosphate-buffered saline (PBS)-based buffer containing 1% NP-40 (Thermo Fisher Scientific, Waltham, MA, USA) and protease inhibitors (Roche, Basel, Switzerland). Cells were then scraped and centrifuged at 13,000 rpm for 1 min at 4 °C. Cytoplasmic proteins in the supernatant were collected, while pellets were resuspended in 50 μL of lysis buffer (7M urea, 2M thiourea and 50 mM DTT), incubated on ice for 30 min and sonicated for two cycles of 20 s each in a Vibra-CellTM 75185 ultrasonic liquid processor (Bioblock Scientific, Illkirch, France). Finally, samples were centrifuged at 20,000× g for 20 min at 15 °C, and nuclear proteins were collected from the supernatant. Protein concentrations were quantified using the Protein DC kit (Bio-Rad, Hercules, CA, USA) in an Epoch plate reader (BioTek, Winooski, VT, USA), following the manufacturers’ recommendations. For western blot, 60 μg of proteins were resolved by SDS-PAGE in a 15% polyacrylamide gel and transferred onto a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA). The membrane was blocked with 5% non-fat milk and incubated with the anti-p16 antibody (805–4713, Roche, Basel, Switzerland) at 250 ng/mL overnight and at 4 °C. Then, it was incubated with the secondary anti-mouse antibody (Bio-Rad, Hercules, CA, USA) at 1:2000 for 1 h at room temperature. The signal was detected with the SuperSignal West Pico Chemiluminescent Substrate kit (Thermo Fisher Scientific, Waltham, MA, USA) in a ChemiDoc imaging system (Bio-Rad, Hercules, CA, USA) using ImageLab v5.2 software. The GAPDH (CB1001 from Calbiochem, Burlington, MA, USA) and Histone H3 (ab17684 from Abcam, Cambridge, UK) antibodies were employed as loading controls for the cytoplasmic and nuclear fractions, respectively. Finally, the intensity of bands was quantitated by densitometric analysis using the ImageJ v1.50i program.
4.5. Cell Proliferation and Invasion
To evaluate the proliferative ability of CC cell lines, C-33A, SiHa and HeLa cells were seeded (1 × 104 cells/well) into 400 µL of medium in an E-plate L8 device (iCELLigence system, ACEA Biosciences, San Diego, CA, USA), after measuring the background in 100 µL of medium. Two replicates for each condition were analyzed. Cell attachment, spreading and proliferation were monitored by real-time cell analysis for 7 days, on the basis of changes in cell-sensor impedance, as previously described [47]. To assess the invasive capability of CC cell lines, cells were starved overnight. Next day, extracellular matrix layer was prepared by adding 40 µL of Matrigel® (BD Biosciences, Franklin Lake, NJ, USA) into Transwell® inserts with 8-μm-pore membranes (Sarstedt, Nümbrecht, Germany). After gelling for 15–30 min at 37 °C, starved cells were seeded (1.25 × 105 cells/insert) on the gel layer in FBS-free medium, and the insert was placed in a well with FBS-containing medium. Cells were allowed to digest and penetrate the Matrigel® layer as far as the membrane for 72 h. Then, non-invading cells and the gel layer were removed, and membranes were fixed and stained with a crystal violet solution containing paraformaldehyde (Sigma-Aldrich, St Louis, MO, USA). Images were taken with a Leica DMi1 microscope and the Leica Application Suite v4.12 program (Leica, Wetzlar, Germany).
4.6. p16 Native form Identification
In order to explore whether p16 subcellular location depended on the 3D conformation of the p16+interactors complex, the p16 native form was interrogated in SCCC cell lines. To do this, C-33A and SiHa cells were scraped and centrifuged at 4000 rpm for 1 min. Pellets were resuspended in 50–200 µL of non-denaturing lysis buffer (20 mM Tris-HCl, 137 mM NaCl, 2 mM EDTA and protease inhibitors) and incubated on ice for 5 min. After centrifuging at 8000× g for 10 min at 4 °C, proteins contained in the supernatants were quantified, and 50 µg of native protein Were subjected to PAGE on an 8% polyacrylamide gel under non-denaturing conditions. Transfer, blocking, incubation with antibodies and revealing were performed as described above.
4.7. p16 Immunoprecipitation
To identify cytoplasmic p16 interactors, p16 was first immunoprecipitated from SiHa cells. Briefly, cells were grown to 70–80% confluence, lysed with a home-made IP buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% Triton and protease inhibitors) and centrifuged at maximum speed for 10 min. Proteins contained in the supernatants were quantified as described above, and three aliquots were separated: 50 µg Were denatured and stored to check the input by western blot; 5 mg Were incubated with 10 µg of anti-p16 antibody (ab108349, Abcam, Cambridge, UK); and 5 mg Were incubated with an irrelevant antibody (rabbit IgG, 10500C, Thermo Fisher Scientific, Waltham, MA, USA). Incubations were performed overnight at 4 °C in a rotating wheel. The next day, 30 µL of protein A/G-coated magnetic beads (Thermo Fisher Scientific, Waltham, MA, USA) Were added to each sample and incubated for 2 h at room temperature to allow them to bind to the antigen-antibody complex. Extracts were sequentially washed three times with IP buffer using a magnetic stand. Finally, 30 µL of IP buffer and 10 µL of Laemmli buffer 4× Were added, and samples were heated at 95 °C for 5 min to denature and unbind the magnetic beads. A magnetic stand was used to remove magnetic beads. A small 5-µL fraction of the eluted extract was separated to check immunoprecipitation efficiency by western blot, using the α-tubulin antibody (T-6074 from Sigma-Aldrich, St Louis, MO, USA) as a loading control, while 15 µL Were subjected to mass spectrometry.
4.8. Mass Spectrometry and Bioinformatic Analysis
SiHa protein extracts incubated with the anti-p16 and the IgG antibodies were analyzed by LC-MS/MS using two parallel strategies: i) extracts were subjected to SDS-PAGE in a Criterion™ TGX Stain-Free™ protein gel (Bio-Rad, Hercules, CA, USA), and bands were individually excised from the gel and then digested; ii) extracts were directly digested in solution. In both cases, protein extracts were trypsin-digested and cleaned with ZipTip. Resulting samples were examined in a Triple-TOF 5600+ (Sciex, Framingham, MA, USA) coupled to a NanoLC Ultra 1D Plus nanochromatograph (Eksigent, Dublin, CA, USA), as previously described [48]. MS/MS data were acquired using Analyst 1.7.1 (Sciex, Framingham, MA, USA) and spectra files were processed with Protein Pilot Software v.5.0 (Sciex, Framingham, MA, USA) using the Paragon™ algorithm for database searching, Progroup™ for data grouping, and searched against the concatenated target-decoy UniProt human proteome reference database (Proteome ID: UP000005640, 73045 proteins, April 2018) plus Human papillomavirus type 16 database (Proteome ID: UP000009251, 9 proteins, April 2018). The FDR was estimated using a non-linear fitting method and the results displayed were those for a global FDR of 1% or better. Proteins unequivocally detected in both workflows in the extract with the anti-p16 antibody but not present in the anti-IgG sample and with an FDR < 1% were considered to be potential p16 interactors. Their biological functions were interrogated using the UniprotKB/Swiss-Prot (https://www.uniprot.org/) database. Additionally, the p16 interactors identified here were compared with those available in the interaction repository BioGRID (Biological General Repository for Interaction Datasets, v3.5.173, https://thebiogrid.org/) [49].
4.9. Immunofluorescence
In order to confirm p16 expression pattern in cultured SCCC cells and to assess colocalization of cytoplasmic p16 and its potential interactors, cells were seeded on coverslips and allowed to attach overnight. Cells were then fixed with 4% paraformaldehyde, blocked with 5% FBS in PBS at room temperature for 1 h, and incubated at 4 °C overnight with primary antibodies: anti-p16 (805-4713, Roche, Basel, Switzerland) at 250 ng/mL, and anti-CDK4 (ab108357, Abcam, Cambridge, UK), anti-BANF1 (CSB-PA002550LA01HU), anti-AKAP8 (STJ91533) or anti-AGTRAP (CSB-PA744194LA01HU) (all from Antibodyplus, Inc., Brookline, MA, USA), at 1:150. All samples were simultaneously incubated with AlexaFluor 488 anti-mouse and AlexaFluor 568 anti-rabbit (1:200) (Life Technologies, Carlsbad, CA, USA) at room temperature for 1 h. Finally, samples were mounted on slides with DAPI to counterstain nuclei. Images were acquired with a Leica DM4000B microscope (Leica, Wetzlar, Germany), using the NIS Elements v4.30 program (Nikon Instruments, Amsterdam, Netherlands), and excitation/emission wavelengths of 340–380/425 nm, 450–490/525 nm, and 560/40–645/75 nm for blue, green and red, respectively.
4.10. Statistical Analysis
Demographic, clinical and pathological data were summarized as frequencies (and percentages) and means (and ranges) ± standard error of the mean, as appropriate. All statistical analyses were carried out using IBM SPSS Statistics v25. Immunohistochemical expression levels in HSIL and SCCC samples, as well as differences in cell line proliferation, were compared using the Mann-Whitney test. Association between clinical variables and p16 expression or location was tested using a χ2 contingency test. Kaplan-Meier plots and log-rank tests were used to examine the association of p16 expression and location, age, stage and vascular invasion with PFS and OS in SCCC patients. A multivariate Cox regression model was fitted to test the independent contribution of each variable to the patient’s outcome after adjusting. Hazard ratios and 95% confidence intervals were used to estimate the effect of each variable on the outcome.
5. Conclusions
In conclusion, we have gained further insight into the paradox of p16 overexpression in CC: it is expressed at high levels, but not located where it would act as a tumor-suppressor protein. We propose then that exploring p16 subcellular location can provide more useful information regarding anti-tumor activity than p16 overall expression examination. In particular, cytoplasmic p16, which is sequestered by novel proteins identified here for the first time, is more frequently observed in SCCC than HSIL and is associated with a worse outcome in SCCC. Therefore, the ratio of cytoplasmic to nuclear p16 and the absence of nuclear p16 could be independent biomarkers for SCCC diagnosis and prognosis, respectively.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/6/2125/s1.
Author Contributions
Conceptualization, E.M.-S.; methodology, E.M.-S.; software, J.F.-I., E.S. and E.M.-S.; validation, E.M.-S.; formal analysis, J.F.-I., E.S. and E.M.-S.; investigation, S.M., T.Z., K.A., M.J.D.d.C. and E.M.-S.; resources, J.F.-I., E.S., R.G., D.G.-S., A.V., J.S.-S., X.M.-G.; data curation, J.F.-I., E.S. and E.M.-S.; writing—original draft preparation, E.M.-S.; writing—review and editing, J.F.-I., E.S. and E.M.-S.; visualization, E.M.-S.; supervision, E.M.-S.; project administration, E.M.-S.; funding acquisition, D.G.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Health Department of the Navarre Government (Grant number 21/11), the Navarre Breast Cancer Patients’ Association (SARAY) and the “Proof of concept project on proteomic research” from Proteored (Grant number ProteoRed-0000029). The Proteomics Unit of Navarrabiomed is a member of Proteored, PRB3-ISCIII, and is supported by Grant PT17/0019/009, of the PE I + D + I 2013–2016 funded by ISCIII and ERDF. S.M. was a recipient of a predoctoral grant from the Public University of Navarre. E.M.-S. was a recipient of a fellowship from the Spanish Ministry of Science, Innovation and Universities (PTA2015-11895-I).
Acknowledgments
We thank the staff of the Biobank in Navarrabiomed and the Pathology Department of the Complejo Hospitalario de Navarra (both in Pamplona, Spain) for their excellent technical assistance with the histological and immunohistochemical techniques.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, the collection, analyses or interpretation of data, the writing of the manuscript or in the decision to publish the results.
Abbreviations
| AGTRAP | Type-1 angiotensin II receptor-associated protein |
| AKAP8 | A-kinase anchor protein 8 |
| ATCC | American Type Cell Collection |
| BANF1 | Barrier to autointegration factor |
| BioGRID | Biological General Repository for Interaction Datasets |
| C1QBP | Complement component 1 Q subcomponent-binding protein |
| CC | Cervical cancer |
| CDK4 | Cyclin-dependent kinase 4 |
| CI | Confidence interval |
| FBS | Fetal bovine serum |
| FDR | False-discovery rate |
| HR-HPV | High-risk human papillomavirus |
| HSIL | High-grade squamous intraepithelial lesion |
| LC-MS/MS | Liquid chromatography-tandem mass spectrometry |
| OS | Overall survival |
| p16 | Cyclin-dependent kinase inhibitor 2A protein |
| PBS | Phosphate-buffered saline |
| PFS | Progression-free survival |
| Rb | Retinoblastoma protein |
| SCCC | Squamous cell carcinoma of the cervix |
References
- World Health Organization. Available online: https://www.who.int/cancer/prevention/diagnosis-screening/cervical-cancer/en/ (accessed on 15 July 2019).
- Marth, C.; Landoni, F.; Mahner, S.; McCormack, M.; Gonzalez-Martin, A.; Colombo, N.; Committee, E.G. Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017, 28, iv72–iv83. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, K.P.; Brebi, P.M.; Roa, J.C. Epigenetic alterations in preneoplastic and neoplastic lesions of the cervix. Clin. Epigenet. 2012, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Reuschenbach, M.; Wentzensen, N.; Dijkstra, M.G.; von Knebel Doeberitz, M.; Arbyn, M. p16INK4a immunohistochemistry in cervical biopsy specimens: A systematic review and meta-analysis of the interobserver agreement. Am. J. Clin. Pathol. 2014, 142, 767–772. [Google Scholar] [CrossRef]
- Lin, J.; Albers, A.E.; Qin, J.; Kaufmann, A.M. Prognostic significance of overexpressed p16INK4a in patients with cervical cancer: A meta-analysis. PLoS ONE 2014, 9, e106384. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Shen, K.; Cao, D. Progress in immunocytochemical staining for cervical cancer screening. Cancer Manag. Res. 2019, 11, 1817–1827. [Google Scholar] [CrossRef] [PubMed]
- Kocken, M.; Helmerhorst, T.J.; Berkhof, J.; Louwers, J.A.; Nobbenhuis, M.A.; Bais, A.G.; Hogewoning, C.J.; Zaal, A.; Verheijen, R.H.; Snijders, P.J.; et al. Risk of recurrent high-grade cervical intraepithelial neoplasia after successful treatment: A long-term multi-cohort study. Lancet Oncol. 2011, 12, 441–450. [Google Scholar] [CrossRef]
- Coates, A.S.; Winer, E.P.; Goldhirsch, A.; Gelber, R.D.; Gnant, M.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.J.; Members, P. Tailoring therapies--improving the management of early breast cancer: St Gallen international expert consensus on the primary therapy of early breast cancer 2015. Ann. Oncol. 2015, 26, 1533–1546. [Google Scholar] [CrossRef]
- Bernicker, E.H.; Allen, T.C.; Cagle, P.T. Update on emerging biomarkers in lung cancer. J. Thorac. Dis. 2019, 11, S81–S88. [Google Scholar] [CrossRef]
- Wick, W.; Weller, M.; van den Bent, M.; Sanson, M.; Weiler, M.; von Deimling, A.; Plass, C.; Hegi, M.; Platten, M.; Reifenberger, G. MGMT testing--the challenges for biomarker-based glioma treatment. Nat. Rev. Neurol. 2014, 10, 372–385. [Google Scholar] [CrossRef]
- Harbeck, N.; Sotlar, K.; Wuerstlein, R.; Doisneau-Sixou, S. Molecular and protein markers for clinical decision making in breast cancer: Today and tomorrow. Cancer Treat. Rev. 2014, 40, 434–444. [Google Scholar] [CrossRef]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch. Pathol. Lab. Med. 2018, 142, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, A.R.; Hamilton, S.R.; Allegra, C.J.; Grody, W.; Cushman-Vokoun, A.M.; Funkhouser, W.K.; Kopetz, S.E.; Lieu, C.; Lindor, N.M.; Minsky, B.D.; et al. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J. Clin. Oncol. 2017, 35, 1453–1486. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, M.F.S.; Castelletti, C.H.M.; Lima-Filho, J.L.; Martins, D.B.G.; Teixeira, J.A.C. Putative biomarkers for cervical cancer: SNVs, methylation and expression profiles. Mutat. Res. 2017, 773, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Romagosa, C.; Simonetti, S.; López-Vicente, L.; Mazo, A.; Lleonart, M.E.; Castellvi, J.; Ramon y Cajal, S. p16(Ink4a) overexpression in cancer: A tumor suppressor gene associated with senescence and high-grade tumors. Oncogene 2011, 30, 2087–2097. [Google Scholar] [CrossRef]
- Sarma, U.; Biswas, I.; Das, A.; Das, G.C.; Saikia, C.; Sarma, B. p16INK4a expression in cervical lesions correlates with histologic grading—A tertiary level medical facility based retrospective study. Asian Pac. J. Cancer Prev. 2017, 18, 2643–2647. [Google Scholar] [CrossRef]
- Conesa-Zamora, P.; Doménech-Peris, A.; Orantes-Casado, F.J.; Ortiz-Reina, S.; Sahuquillo-Frías, L.; Acosta-Ortega, J.; García-Solano, J.; Pérez-Guillermo, M. Effect of human papillomavirus on cell cycle-related proteins p16, Ki-67, Cyclin D1, p53, and ProEx C in precursor lesions of cervical carcinoma: A tissue microarray study. Am. J. Clin. Pathol. 2009, 132, 378–390. [Google Scholar] [CrossRef]
- Vázquez-Vega, S.; Sánchez-Suárez, L.P.; Contreras-Paredes, A.; Castellanos-Juárez, E.; Peñarroja-Flores, R.; Lizano-Soberón, M.; Andrade-Cruz, R.; García-Carrancá, A.; Benítez-Bribiesca, L. Nuclear co-expression of p14ARF and p16INK4A in uterine cervical cancer-derived cell lines containing HPV. Cancer Biomark. 2010, 8, 341–350. [Google Scholar] [CrossRef]
- Pandey, A.; Chandra, S.; Nautiyal, R.; Shrivastav, V. Expression of p16INK4A and human papillomavirus 16 with associated risk factors in cervical premalignant and malignant lesions. South Asian J. Cancer 2018, 7, 236–239. [Google Scholar] [CrossRef]
- Volgareva, G.; Zavalishina, L.; Andreeva, Y.; Frank, G.; Krutikova, E.; Golovina, D.; Bliev, A.; Spitkovsky, D.; Ermilova, V.; Kisseljov, F. Protein p16 as a marker of dysplastic and neoplastic alterations in cervical epithelial cells. BMC Cancer 2004, 4, 58. [Google Scholar] [CrossRef]
- Wu, M.Z.; Wang, S.; Zheng, M.; Tian, L.X.; Wu, X.; Guo, K.J.; Zhang, Y.I.; Wu, G.P. The diagnostic utility of p16 immunostaining in differentiating cancer and HSIL from LSIL and benign in cervical cells. Cell Transpl. 2019, 28, 195–200. [Google Scholar] [CrossRef]
- Kurman, R.J.; Carcangiu, M.L.; Herrington, C.S.; Young, R.H. Tumours of the uterine cervix. In WHO Classification of Tumours of Female Reproductive Organs, 4th ed.; IARC Press: Lyon, France, 2014; Volume 6, pp. 169–207. [Google Scholar]
- Shen, Q.; Eun, J.W.; Lee, K.; Kim, H.S.; Yang, H.D.; Kim, S.Y.; Lee, E.K.; Kim, T.; Kang, K.; Kim, S.; et al. Barrier to autointegration factor 1, procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3, and splicing factor 3b subunit 4 as early-stage cancer decision markers and drivers of hepatocellular carcinoma. Hepatology 2018, 67, 1360–1377. [Google Scholar] [CrossRef]
- Li, J.; Wang, T.; Pei, L.; Jing, J.; Hu, W.; Sun, T.; Liu, H. Expression of VRK1 and the downstream gene BANF1 in esophageal cancer. Biomed. Pharmacother. 2017, 89, 1086–1091. [Google Scholar] [CrossRef]
- Loftus, S.K.; Baxter, L.L.; Cronin, J.C.; Fufa, T.D.; Pavan, W.J.; Program, N.C.S. Hypoxia-induced HIF1α targets in melanocytes reveal a molecular profile associated with poor melanoma prognosis. Pigment Cell Melanoma Res. 2017, 30, 339–352. [Google Scholar] [CrossRef]
- Li, J.; Hu, B.; Fang, L.; Gao, Y.; Shi, S.; He, H.; Liu, X.; Yuan, C. Barrier-to-autointegration factor 1: A novel biomarker for gastric cancer. Oncol. Lett. 2018, 16, 6488–6494. [Google Scholar] [CrossRef]
- Chen, Y.D.; Chen, X.X.; Shen, L.N.; Liang, F.C.; Ding, Y.; Yu, X.Y.; Xue, M.Q.; Zhang, Y.X. Expression of A-kinase anchor protein 95, cyclin E2, and connexin 43 in lung cancer tissue, clinical significance of their expression, and their expression correlation. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi 2012, 30, 725–729. [Google Scholar]
- Liu, W.; Hua, S.; Dai, Y.; Yuan, Y.; Yang, J.; Deng, J.; Huo, Y.; Chen, X.; Teng, B.; Yu, X.; et al. Roles of Cx43 and AKAP95 in ovarian cancer tissues in G1/S phase. Int. J. Clin. Exp. Pathol. 2015, 8, 14315–14324. [Google Scholar]
- Qi, F.; Yuan, Y.; Zhi, X.; Huang, Q.; Chen, Y.; Zhuang, W.; Zhang, D.; Teng, B.; Kong, X.; Zhang, Y. Synergistic effects of AKAP95, Cyclin D1, Cyclin E1, and Cx43 in the development of rectal cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 1666–1673. [Google Scholar]
- Van Zummeren, M.; Leeman, A.; Kremer, W.W.; Bleeker, M.C.G.; Jenkins, D.; van de Sandt, M.; Heideman, D.A.M.; Steenbergen, R.; Snijders, P.J.F.; Quint, W.G.V.; et al. Three-tiered score for Ki-67 and p16INK4A improves accuracy and reproducibility of grading CIN lesions. J. Clin. Pathol. 2018, 71, 981–988. [Google Scholar] [CrossRef]
- Nicolas, I.; Saco, A.; Barnadas, E.; Marimon, L.; Rakislova, N.; Fuste, P.; Rovirosa, A.; Gaba, L.; Bunesch, L.; Gil-Ibanez, B.; et al. Prognostic implications of genotyping and p16 immunostaining in HPV-positive tumors of the uterine cervix. Mod. Pathol. 2020, 33, 128–137. [Google Scholar] [CrossRef]
- Fu, H.C.; Chuang, I.C.; Yang, Y.C.; Chuang, P.C.; Lin, H.; Ou, Y.C.; Chang Chien, C.C.; Huang, H.S.; Kang, H.Y. Low P16INK4A expression associated with high expression of cancer stem cell markers predicts poor prognosis in cervical cancer after radiotherapy. Int. J. Mol. Sci. 2018, 19, 2541. [Google Scholar] [CrossRef]
- Andrade, C.E.; Scapulatempo-Neto, C.; Longatto-Filho, A.; Vieira, M.A.; Tsunoda, A.T.; Da Silva, I.D.; Fregnani, J.H. Prognostic scores after surgical treatment for cervical intraepithelial neoplasia: A proposed model and possible implications for post-operative follow-up. Acta Obstet. Gynecol. Scand. 2014, 93, 941–948. [Google Scholar] [CrossRef]
- Souza-Rodrígues, E.; Estanyol, J.M.; Friedrich-Heineken, E.; Olmedo, E.; Vera, J.; Canela, N.; Brun, S.; Agell, N.; Hübscher, U.; Bachs, O.; et al. Proteomic analysis of p16ink4a-binding proteins. Proteomics 2007, 7, 4102–4111. [Google Scholar] [CrossRef]
- Li, Y.; Xiao, S.; Dan, L.; Xue, M. P16INK4A is required for cisplatin resistance in cervical carcinoma SiHa cells. Oncol. Lett. 2015, 9, 1104–1108. [Google Scholar] [CrossRef]
- Itahana, K.; Clegg, H.V.; Zhang, Y. ARF in the mitochondria: The last frontier? Cell Cycle 2008, 7, 3641–3646. [Google Scholar] [CrossRef]
- Mao, L.; Zhang, Y.; Mo, W.; Yu, Y.; Lu, H. BANF1 is downregulated by IRF1-regulated microRNA-203 in cervical cancer. PLoS ONE 2015, 10, e0117035. [Google Scholar] [CrossRef]
- Markiewicz, E.; Tilgner, K.; Barker, N.; van de Wetering, M.; Clevers, H.; Dorobek, M.; Hausmanowa-Petrusewicz, I.; Ramaekers, F.C.; Broers, J.L.; Blankesteijn, W.M.; et al. The inner nuclear membrane protein emerin regulates beta-catenin activity by restricting its accumulation in the nucleus. EMBO J. 2006, 25, 3275–3285. [Google Scholar] [CrossRef]
- Chen, X.; Kong, X.; Zhuang, W.; Teng, B.; Yu, X.; Hua, S.; Wang, S.; Liang, F.; Ma, D.; Zhang, S.; et al. Dynamic changes in protein interaction between AKAP95 and Cx43 during cell cycle progression of A549 cells. Sci. Rep. 2016, 6, 21224. [Google Scholar] [CrossRef]
- Arsenijevic, T.; Degraef, C.; Dumont, J.E.; Roger, P.P.; Pirson, I. A novel partner for D-type cyclins: Protein kinase A-anchoring protein AKAP95. Biochem. J. 2004, 378, 673–679. [Google Scholar] [CrossRef]
- Larrea, M.D.; Hong, F.; Wander, S.A.; da Silva, T.G.; Helfman, D.; Lannigan, D.; Smith, J.A.; Slingerland, J.M. RSK1 drives p27Kip1 phosphorylation at T198 to promote RhoA inhibition and increase cell motility. Proc. Natl. Acad. Sci. USA 2009, 106, 9268–9273. [Google Scholar] [CrossRef]
- Imai, N.; Hashimoto, T.; Kihara, M.; Yoshida, S.; Kawana, I.; Yazawa, T.; Kitamura, H.; Umemura, S. Roles for host and tumor angiotensin II type 1 receptor in tumor growth and tumor-associated angiogenesis. Lab. Investig. 2007, 87, 189–198. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Bao, W.; Wang, L.H. Downregulation of p16(ink4a) inhibits cell proliferation and induces G1 cell cycle arrest in cervical cancer cells. Int. J. Mol. Med. 2014, 33, 1577–1585. [Google Scholar] [CrossRef]
- Nicol, A.F.; de Andrade, C.V.; Gomes, S.C.; Brusadelli, M.G.; Lodin, H.M.; Wells, S.I.; Nuovo, G.J. The distribution of novel biomarkers in carcinoma-in-situ, microinvasive, and squamous cell carcinoma of the uterine cervix. Ann. Diagn. Pathol. 2019, 38, 115–122. [Google Scholar] [CrossRef]
- Liu, H.Q.; Wang, Y.H.; Wang, L.L.; Hao, M. P16INK4A and survivin: Diagnostic and prognostic markers in cervical intraepithelial neoplasia and cervical squamous cell carcinoma. Exp. Mol. Pathol. 2015, 99, 44–49. [Google Scholar] [CrossRef]
- Velez-Perez, A.; Wang, X.I.; Li, M.; Zhang, S. SIRT1 overexpression in cervical squamous intraepithelial lesions and invasive squamous cell carcinoma. Hum. Pathol. 2017, 59, 102–107. [Google Scholar] [CrossRef]
- Martín-Sánchez, E.; Mendaza, S.; Ulazia-Garmendia, A.; Monreal-Santesteban, I.; Blanco-Luquin, I.; Córdoba, A.; Vicente-García, F.; Pérez-Janices, N.; Escors, D.; Megías, D.; et al. CHL1 hypermethylation as a potential biomarker of poor prognosis in breast cancer. Oncotarget 2017, 8, 15789–15801. [Google Scholar] [CrossRef]
- Lachén-Montes, M.; González-Morales, A.; Zelaya, M.V.; Pérez-Valderrama, E.; Ausín, K.; Ferrer, I.; Fernández-Irigoyen, J.; Santamaría, E. Olfactory bulb neuroproteomics reveals a chronological perturbation of survival routes and a disruption of prohibitin complex during Alzheimer’s disease progression. Sci. Rep. 2017, 7, 9115. [Google Scholar] [CrossRef]
- Oughtred, R.; Stark, C.; Breitkreutz, B.J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).