Effect of Arginine on Chaperone-Like Activity of HspB6 and Monomeric 14-3-3ζ
Abstract
1. Introduction
2. Results
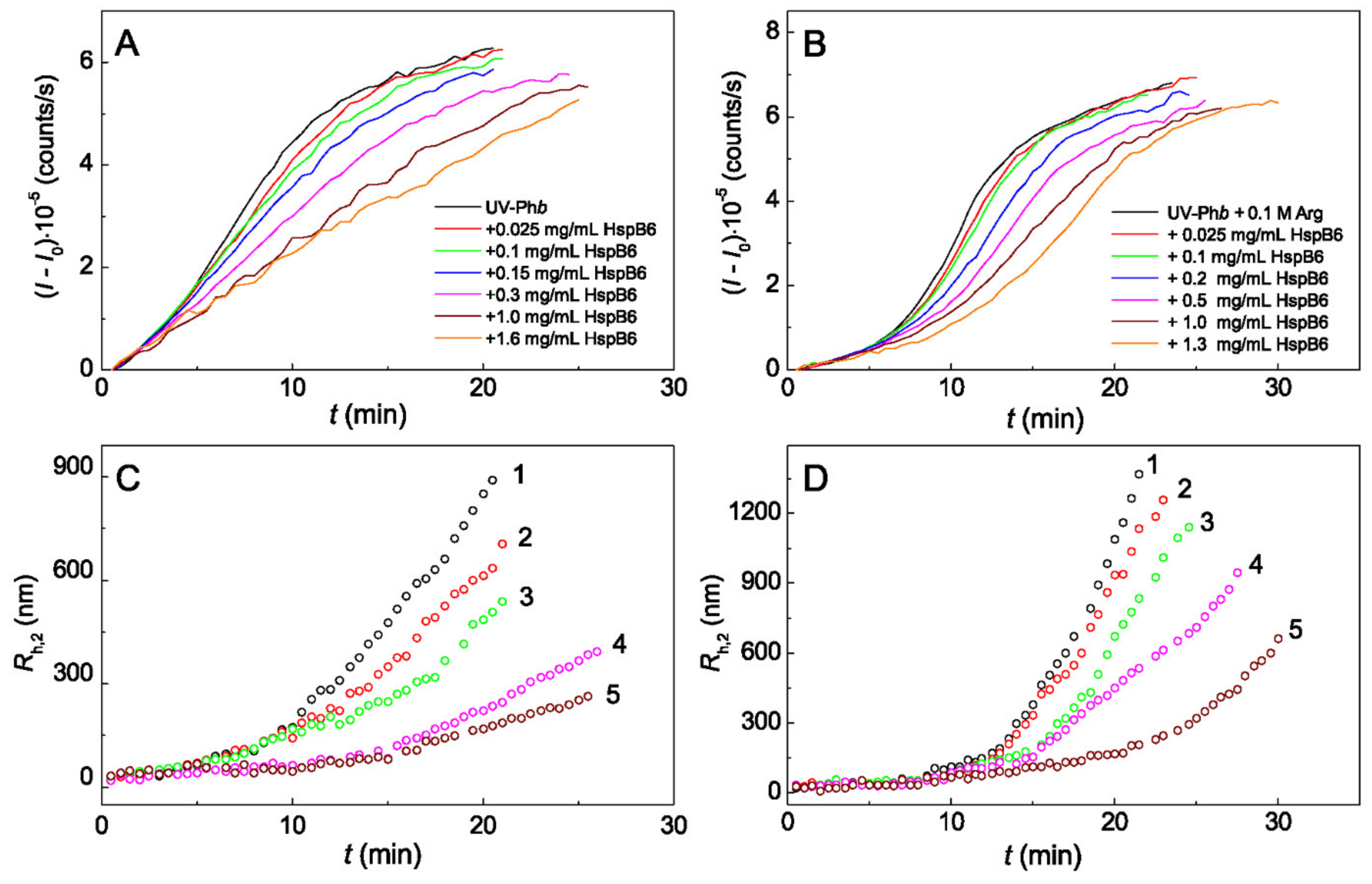
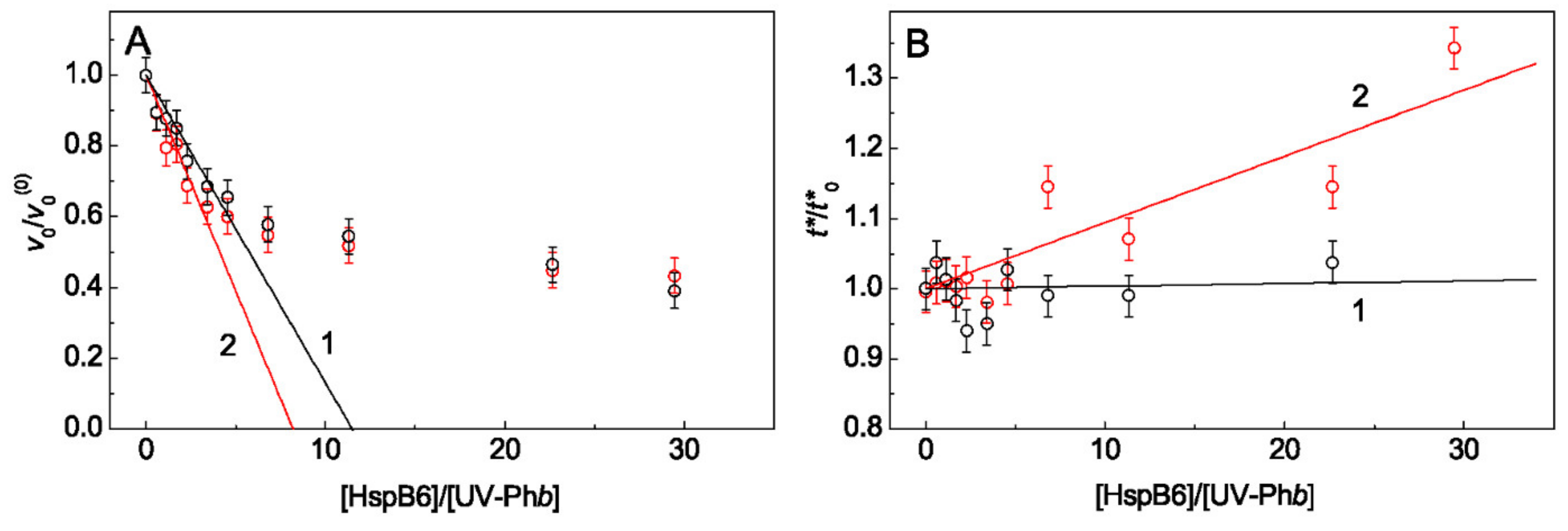
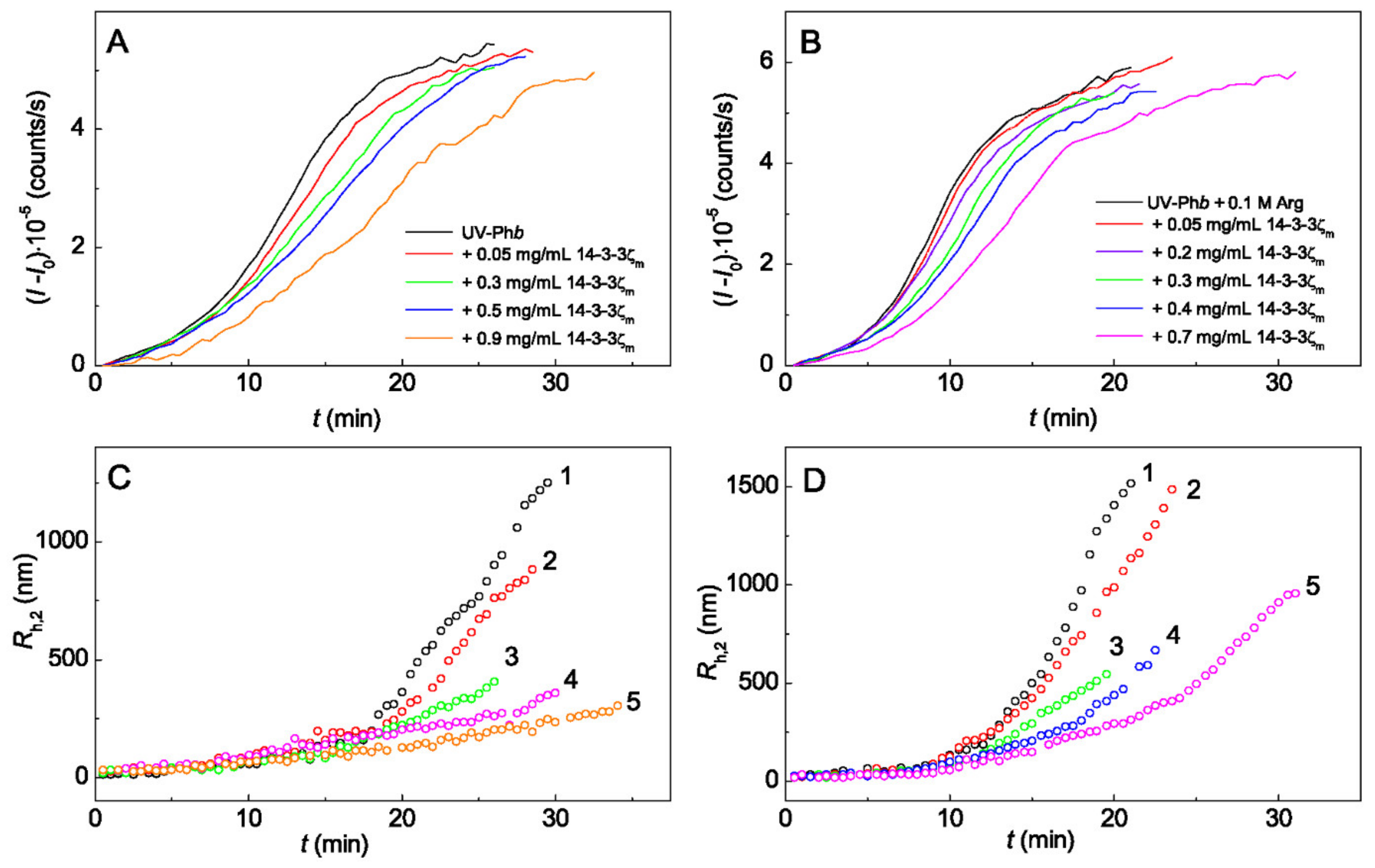
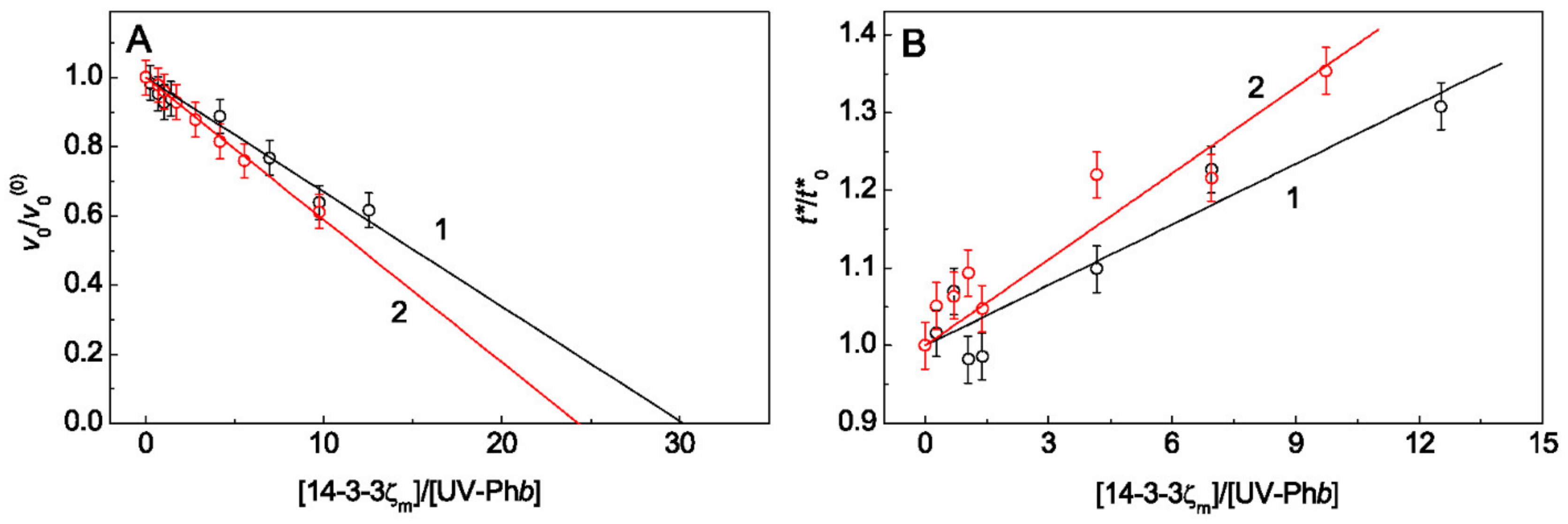
2.1. The Effect of Arg on the Chaperone-Like Activity of HspB6 and Monomeric 14-3-3ζ
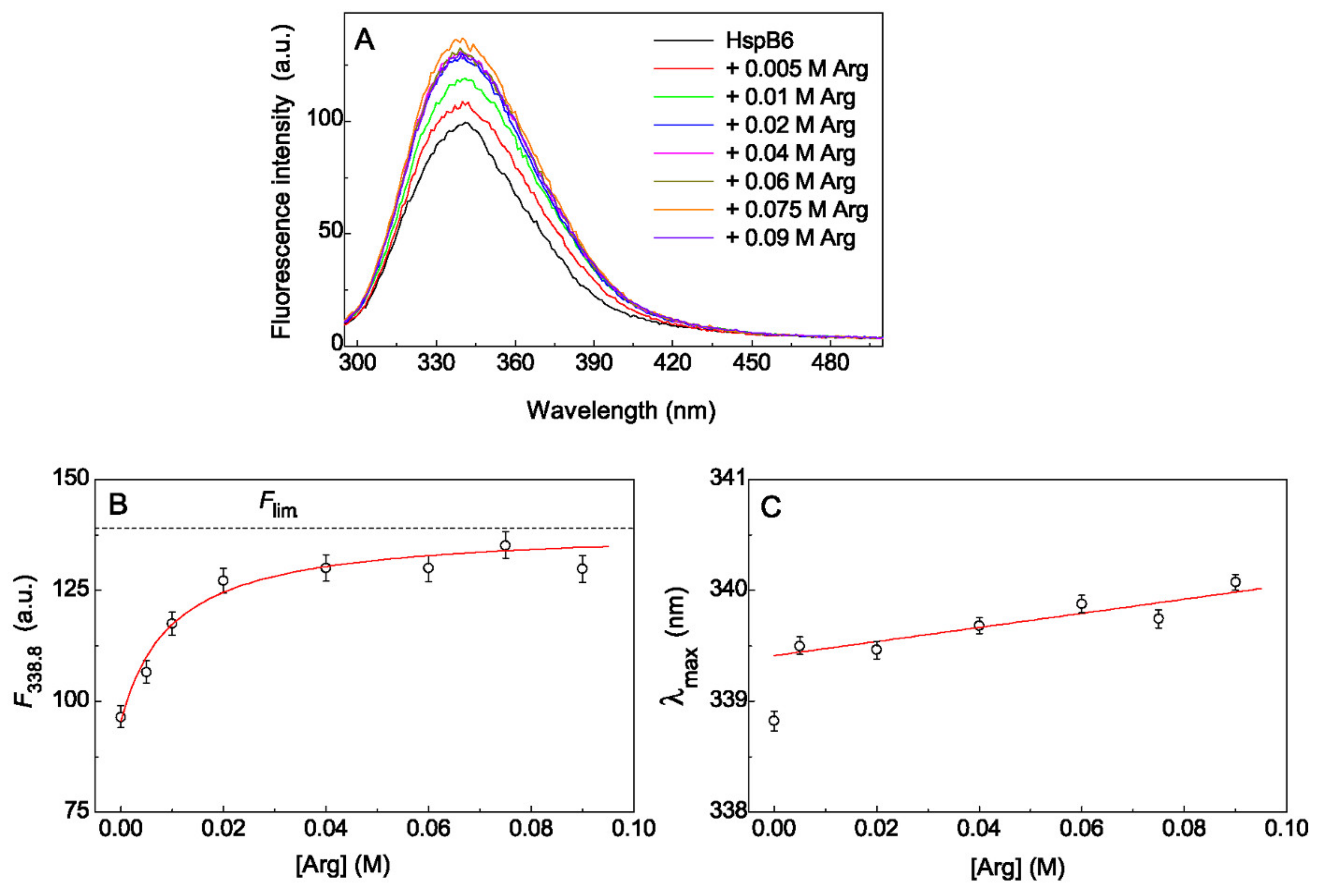
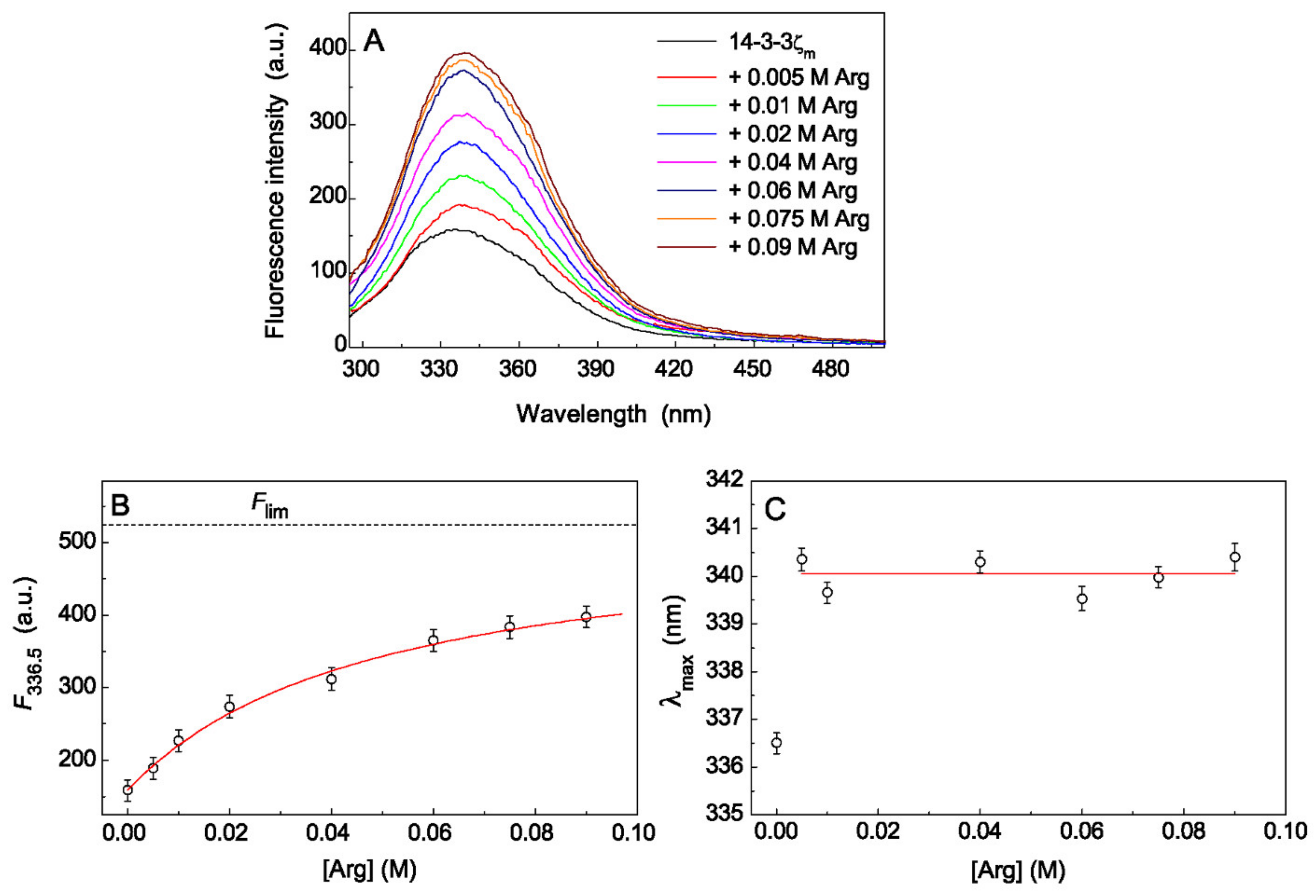
2.2. The Effect of Arg on Tryptophan Fluorescence Spectra of Chaperones
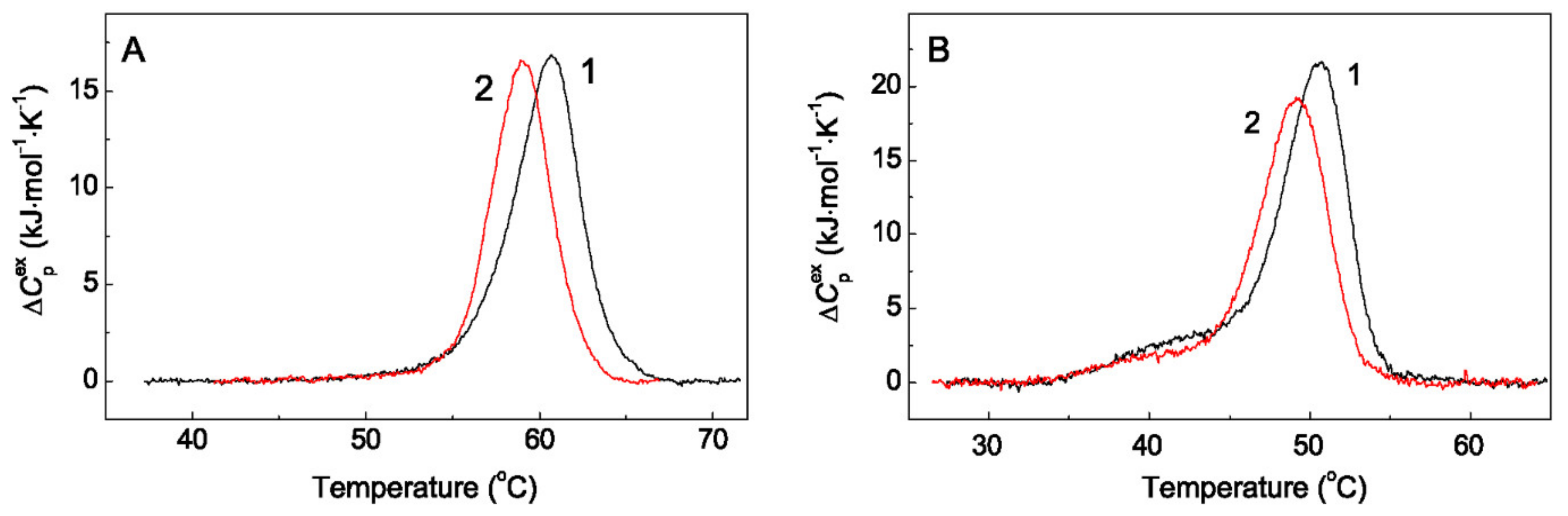
2.3. The Effect of Arg on Thermal Stability of Chaperones
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Isolation of Chaperones
4.3. Light Scattering Intensity Measurements
4.4. Tryptophan Fluorescence of Chaperones
4.5. Thermal Denaturation of Chaperones
Author Contributions
Funding
Conflicts of Interest
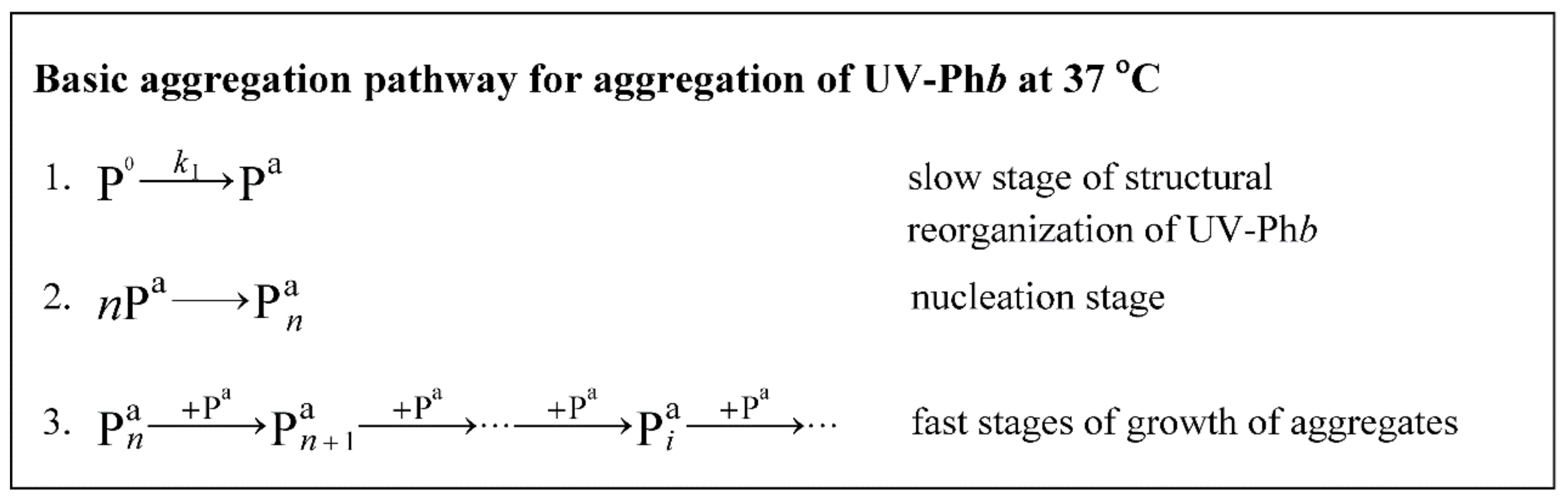
Appendix A. Calculation of the Initial Aggregation Rate for the Stage of Aggregate Growth
References
- Stefani, M.; Dobson, C.M. Protein aggregation and aggregate toxicity: New insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. (Berl.) 2003, 81, 678–699. [Google Scholar] [CrossRef] [PubMed]
- Markossian, K.A.; Kurganov, B.I. Protein folding, misfolding, and aggregation. Formation of inclusion bodies and aggresomes. Biochemistry 2004, 69, 971–984. [Google Scholar] [CrossRef] [PubMed]
- Mahler, H.C.; Friess, W.; Grauschopf, U.; Kiese, S. Protein aggregation: Pathways, induction factors and analysis. J. Pharm. Sci. 2009, 98, 2909–2934. [Google Scholar] [CrossRef] [PubMed]
- Panchal, J.; Kotarek, J.; Marszal, E.; Topp, E.M. Analyzing subvisible particles in protein drug products: A comparison of dynamic light scattering (DLS) and resonant mass measurement (RMM). AAPS J. 2014, 16, 440–451. [Google Scholar] [CrossRef]
- Siddiqi, M.K.; Alam, P.; Chaturvedi, S.K.; Shahein, Y.E.; Khan, R.H. Mechanisms of protein aggregation and inhibition. Front. Biosci. 2017, 9, 1–20. [Google Scholar] [CrossRef]
- Wang, W.; Roberts, C.J. Protein aggregation—Mechanisms, detection, and control. Int. J. Pharm. 2018, 550, 251–268. [Google Scholar] [CrossRef]
- Tyedmers, J.; Mogk, A.; Bukau, B. Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 2010, 11, 777–788. [Google Scholar] [CrossRef]
- Chen, B.; Retzlaff, M.; Roos, T.; Frydman, J. Cellular strategies of protein quality control. Cold Spring Harb. Perspect. Biol. 2011, 3, a004374. [Google Scholar] [CrossRef]
- Mymrikov, E.V.; Seit-Nebi, A.S.; Gusev, N.B. Large potentials of small heat shock proteins. Physiol. Rev. 2011, 91, 1123–1159. [Google Scholar] [CrossRef]
- Haslbeck, M.; Vierling, E.A. First line of stress defense: Small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 2015, 427, 1537–1548. [Google Scholar] [CrossRef]
- Webster, J.M.; Darling, A.L.; Uversky, V.N.; Blair, L.J. Small heat shock proteins, big impact on protein aggregation in neurodegenerative disease. Front. Pharmacol. 2019, 10, 1047. [Google Scholar] [CrossRef]
- Sluchanko, N.N.; Chebotareva, N.A.; Gusev, N.B. Quaternary structure of human small heat shock protein HSPB6 (Hsp20) in crowded media modeled by trimethylamine N-oxide (TMAO): Effect of protein phosphorylation. Biochimie 2015, 108, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Hochberg, G.K.; Benesch, J.L. Dynamical structure of αB-crystallin. Prog. Biophys. Mol. Biol. 2014, 115, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Hochberg, G.K.; Ecroyd, H.; Liu, C.; Cox, D.; Cascio, D.; Sawaya, M.R.; Collier, M.P.; Stroud, J.; Carver, J.A.; Baldwin, A.J.; et al. The structured core domain of alphaB-crystallin can prevent amyloid fibrillation and associated toxicity. Proc. Natl. Acad. Sci. USA 2014, 111, E1562–E1570. [Google Scholar] [CrossRef]
- Delbecq, S.P.; Klevit, R.E. One size does not fit all: The oligomeric states of alphaB crystallin. FEBS Lett. 2013, 587, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Bukach, O.V.; Seit-Nebi, A.S.; Marston, S.B.; Gusev, N.B. Some properties of human small heat shock protein Hsp20 (HspB6). Eur. J. Biochem. 2004, 271, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Karolczak-Bayatti, M.; Sweeney, M.; Treumann, A.; Morrissey, K.; Uirich, S.M.; Europe-Finner, G.N.; Taggart, M.J. Lysine deacetylase inhibition promotes relaxation of arterial tone and C-terminal acetylation of HSPB6 (Hsp20) in vascular smooth muscle cells. Physiol. Rep. 2013, 1, e00127. [Google Scholar] [CrossRef]
- Nagasawa, T.; Matsushima-Nishiwaki, R.; Toyoda, H.; Matsuura, J.; Kumada, T.; Kozawa, O. Heat shock protein 20 (HSPB6) regulates apoptosis in human hepatocellular carcinoma cells: Direct association with Bax. Oncol. Rep. 2014, 32, 1291–1295. [Google Scholar] [CrossRef]
- Matsushima-Nishiwaki, R.; Toyoda, H.; Nagasawa, T.; Yasuda, E.; Chiba, N.; Okuda, S.; Maeda, A.; Kaneoka, Y.; Kumada, T.; Kozawa, O. Phosphorylated heat shock protein 20 (HSPB6) regulates transforming growth factor-α-induced migration and invasion of hepatocellular carcinoma cells. PLoS ONE 2016, 1, e0151907. [Google Scholar] [CrossRef]
- Lu, T.; Zou, Y.; Zhou, X.; Peng, W.; Hu, Z. The mechanism on phosphorylation of Hsp20 Ser16 inhibit GA stress and ER stress during OGD/R. PLoS ONE 2019, 14, e0213410. [Google Scholar] [CrossRef]
- Mackintosh, C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 2004, 381, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Sluchanko, N.N.; Gusev, N.B. Oligomeric structure of 14-3-3 protein: What we know about monomers? FEBS Lett. 2012, 586, 4249–4256. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lee, W.H.; Sobott, F.; Papagrigoriou, E.; Robinson, C.V.; Grossmann, J.; Sundström, M.; Doyle, D.A.; Elkins, J.M. Structural basis for protein–protein interactions in the 14-3-3 protein family. Proc. Natl. Acad. Sci. USA 2006, 103, 17237–17242. [Google Scholar] [CrossRef] [PubMed]
- Sluchanko, N.N.; Artemova, N.V.; Sudnitsyna, M.V.; Safenkova, I.V.; Antson, A.A.; Levitsky, D.I.; Gusev, N.B. Monomeric 14-3-3ζ has a chaperone-like activity and is stabilized by phosphorylated HspB6. Biochemistry 2012, 51, 6127–6138. [Google Scholar] [CrossRef]
- Sluchanko, N.N.; Roman, S.G.; Chebotareva, N.A.; Gusev, N.B. Chaperone-like activity of monomeric human 14-3-3ζ on different protein substrates. Arch. Biochem. Biophys. 2014, 549, 32–39. [Google Scholar] [CrossRef]
- Shiraki, K.; Kudou, M.; Fujiwara, S.; Imanaka, T.; Takagi, M.J. Biophysical effect of amino acids on the prevention of protein aggregation. J. Biochem. 2002, 132, 591–595. [Google Scholar] [CrossRef]
- Arakawa, T.; Kita, Y.; Tsumoto, K.; Ejima, D.; Fukata, H. Aggregation suppression of proteins by arginine during thermal unfolding. Protein Pept. Lett. 2006, 13, 921–927. [Google Scholar] [CrossRef]
- Borzova, V.A.; Markossian, K.A.; Kara, D.A.; Chebotareva, N.A.; Makeeva, V.F.; Poliansky, N.B.; Muranov, K.O.; Kurganov, B.I. Quantification of anti-aggregation activity of chaperones: A test-system based on dithiothreitol-induced aggregation of bovine serum albumin. PLoS ONE 2013, 8, e74367. [Google Scholar] [CrossRef]
- Haghighi-Poodeh, S.; Kurganov, B.; Navidpour, L.; Yaghmaei, P.; Ebrahim-Habibi, A. Characterization of arginine preventive effect on heat-induced aggregation of insulin. Int. J. Biol. Macromol. 2020, 145, 1039–1048. [Google Scholar] [CrossRef]
- Tsumoto, K.; Ejima, D.; Kita, Y.; Arakawa, T. Review: Why is arginine effective in suppressing aggregation? Protein Pept. Lett. 2005, 12, 613–619. [Google Scholar] [CrossRef]
- Arakawa, T.; Ejima, D.; Tsumoto, K.; Obeyama, N.; Tanaka, Y.; Kita, Y.; Timasheff, S.N. Suppression of protein interactions by arginine: A proposed mechanism of the arginine effects. Biophys. Chem. 2007, 127, 1–8. [Google Scholar] [CrossRef]
- Shiraki, K.; Hirano, A.; Kita, Y.; Koyama, A.H.; Arakawa, T. Potential application of arginine in interaction analysis. Drug Discov. Ther. 2010, 4, 326–333. [Google Scholar]
- Kamerzell, T.J.; Esfandiary, R.; Joshi, S.B.; Middaugh, C.R.; Volkin, D.B. Protein-excipient interactions: Mechanisms and biophysical characterization applied to protein formulation development. Adv. Drug Deliv. Rev. 2011, 63, 1118–1159. [Google Scholar] [CrossRef] [PubMed]
- Eronina, T.B.; Chebotareva, N.A.; Sluchanko, N.N.; Mikhaylova, V.V.; Makeeva, V.F.; Roman, S.G.; Kleymenov, S.Y.; Kurganov, B.I. Dual effect of arginine on aggregation of phosphorylase kinase. Int. J. Biol. Macromol. 2014, 68, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, E.; Safenkova, I.; Stein-Margolina, B.; Shubin, V.; Gurvits, B. L-arginine induces protein aggregation and transformation of supramolecular structures of the aggregates. Amino Acids 2013, 45, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Eronina, T.B.; Mikhaylova, V.V.; Chebotareva, N.A.; Shubin, V.V.; Kurganov, B.I. Effect of ionic strength and arginine on aggregation of UV-irradiated muscle glycogen phosphorylase b. Int. J. Biol. Macromol. 2018, 118, 1193–1202. [Google Scholar] [CrossRef]
- Srinivas, V.; Raman, B.; Rao, K.S.; Ramakrishna, T.; Rao, C.M. Structural perturbation and enhancement of the chaperone-like activity of alpha-crystallin by arginine hydrochloride. Protein Sci. 2003, 12, 1262–1270. [Google Scholar] [CrossRef]
- Srinivas, V.; Raman, B.; Rao, K.S.; Ramakrishna, T.; Rao, C.M. Arginine hydrochloride enhances the dynamics of subunit assembly and the chaperone-like activity of alpha-crystallin. Mol. Vis. 2005, 11, 249–255. [Google Scholar]
- Ecroyd, H.; Carver, J.A. The effect of small molecules in modulating the chaperone activity of alphaB-crystallin against ordered and disordered protein aggregation. FEBS J. 2008, 275, 935–947. [Google Scholar] [CrossRef]
- Mikhaylova, V.V.; Eronina, T.B.; Chebotareva, N.A.; Kleymenov, S.Y.; Shubin, V.V.; Kurganov, B.I. A thermal after-effect of UV irradiation of muscle glycogen phosphorylase b. PLoS ONE 2017, 12, e0189125. [Google Scholar] [CrossRef]
- Eronina, T.B.; Chebotareva, N.A.; Roman, S.G.; Kleymenov, S.Y.; Makeeva, V.F.; Polansky, N.B.; Muranov, K.O.; Kurganov, B.I. Thermal denaturation and aggregation of apoform of glycogen phosphorylase b. Effect of crowding agents and chaperones. Biopolymers 2014, 101, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Burstein, E.A.; Emelyanenko, V.I. Log-normal description of fluorescence spectra of organic fluorophores. Photochem. Photobiol. 1996, 64, 316–320. [Google Scholar] [CrossRef]
- Bustad, H.J.; Underhaug, J.; Halskau, Ø., Jr.; Martinez, A. The binding of 14-3-3 to membranes studied by intrinsic fluorescence spectroscopy. FEBS Lett. 2011, 585, 1163–1168. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Permyakov, E.A. The Method of Intrinsic Protein Luminescence; Chapter 4.12; Nauka: Moscow, Russia, 2003. [Google Scholar]
- Borzova, V.A.; Markossian, K.A.; Kleymenov, S.Y.; Kurganov, B.I. A Change in the aggregation pathway of bovine serum albumin in the presence of arginine and its derivatives. Sci. Rep. 2017, 7, 3984. [Google Scholar] [CrossRef] [PubMed]
- Kara, D.A.; Borzova, V.A.; Markossian, K.A.; Kleymenov, S.Y.; Kurganov, B.I. A change in the pathway of dithiothreitol-induced aggregation of bovine serum albumin in the presence of polyamines and arginine. Int. J. Biol. Macromol. 2017, 104, 889–899. [Google Scholar] [CrossRef]
- Yang, H.; Huang, S.; Dai, H.; Gong, Y.; Zheng, C.; Chang, Z. The mycobacterium tuberculosis small heat shock protein Hsp16.3 exposes hydrophobic surfaces at mild conditions: Conformational flexibility and molecular chaperone activity. Protein Sci. 1999, 8, 174–179. [Google Scholar] [CrossRef]
- Raman, B.; Rao, C.M. Chaperone-like activity and temperature-induced structural changes of alpha-crystallin. J. Biol. Chem. 1997, 272, 23559–23564. [Google Scholar] [CrossRef]
- Mizobata, T.; Kawata, Y. The guanidine-induced conformational changes of the chaperonin GroEL from Escherichia coli. Evidence for the existence of an unfolding intermediate state. Biochim. Biophys. Acta 1994, 1209, 83–88. [Google Scholar] [CrossRef]
- Burstein, E.A.; Vedenkina, N.S.; Ivkova, M.N. Fluorescence and the location of tryptophan residues in protein molecules. Photochem. Photobiol. 1973, 18, 263–279. [Google Scholar] [CrossRef]
- Tsumoto, K.; Umetsu, M.; Kumagai, I.; Ejima, D.; Philo, J.S.; Arakaw, T. Role of arginine in protein refolding, solubilization, and purification. Biotechnol. Prog. 2004, 20, 1301–1308. [Google Scholar] [CrossRef]
- Dreiza, C.M.; Komalavilas, P.; Furnish, E.J.; Flynn, C.R.; Sheller, M.R.; Smoke, C.C.; Lopes, L.B.; Brophy, C.M. The small heat shock protein, HSPB6, in muscle function and disease. Cell Stress Chaperones 2010, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sluchanko, N.N.; Chebotareva, N.A.; Gusev, N.B. Modulation of 14-3-3/phosphotarget interaction by physiological concentrations of phosphate and glycerophosphates. PLoS ONE 2013, 8, e72597. [Google Scholar] [CrossRef] [PubMed]
- Eronina, T.B.; Chebotareva, N.A.; Roman, S.G.; Makeeva, V.F.; Kleymenov, S.Y.; Kurganov, B.I. Effect of proline on thermal inactivation, denaturation and aggregation of glycogen phosphorylase b from rabbit skeletal muscle. Biophys. Chem. 2009, 141, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Eronina, T.; Borzova, V.; Maloletkina, O.; Kleymenov, S.; Asryants, R.; Markossian, K.; Kurganov, B. A protein aggregation based test for screening of the agents affecting thermostability of proteins. PLoS ONE 2011, 6, e22154. [Google Scholar] [CrossRef]
- Matyushenko, A.M.; Kleymenov, S.Y.; Susorov, D.S.; Levitsky, D.I. Thermal unfolding of homodimers and heterodimers of different skeletal-muscle isoforms of tropomyosin. Biophys. Chem. 2018, 243, 1–7. [Google Scholar] [CrossRef]
- Wang, K.; Kurganov, B.I. Kinetics of heat- and acidification-induced aggregation of firefly luciferase. Biophys. Chem. 2003, 106, 97–109. [Google Scholar] [CrossRef]
- Ghahramani, M.; Yousefi, R.; Krivandin, A.; Muranov, K.; Kurganov, B.; Moosavi-Movahedi, A.A. Kinetic data analysis of chaperone-like activity of Wt, R69C and D109H αB-crystallins. Data Brief. 2020, 28, 104922. [Google Scholar] [CrossRef]
- Ghahramani, M.; Yousefi, R.; Niazi, A.; Kurganov, B. The congenital cataract-causing mutations P20R and A171T associate with the important changes in amyloidogenic feature, structure and chaperone-like activity of human αB-crystallin. Biopolymers 2020, e23350. [Google Scholar] [CrossRef]
- Borzova, V.A.; Markossian, K.A.; Kara, D.A.; Kurganov, B. Kinetic regime of dithiothreitol-induced aggregation of bovine serum albumin. Int. J. Biol. Macromol. 2015, 80, 130–138. [Google Scholar] [CrossRef]
- Eronina, T.B.; Mikhaylova, V.V.; Chebotareva, N.A.; Kurganov, B.I. Kinetic regime of thermal aggregation of holo- and apoglycogen phosphorylases b. Int. J. Biol. Macromol. 2016, 92, 1252–1257. [Google Scholar] [CrossRef]
| Additives | Rh* (nm) |
|---|---|
| No additives | 40 ± 2 |
| + Arg 0.1 M | 49 ± 2 |
| + HspB6 | 49 ± 2 |
| + Arg 0.1 M + HspB6 | 46 ± 2 |
| + 14-3-3ζm | 59 ± 6 |
| + Arg 0.1 M + 14-3-3ζm | 44 ± 5 |
| Sample | Tmax, °C | ΔHcal, kJ·mol−1 |
|---|---|---|
| HspB6 | 60.7 ± 0.1 | 86 ± 8 |
| HspB6 + 0.1 M Arg | 58.9 ± 0.1 | 75 ± 7 |
| 14-3-3ζm | 50.7 ± 0.1 | 136 ± 12 |
| 14-3-3ζm + 0.1 M Arg | 49.1 ± 0.1 | 120 ± 11 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikhaylova, V.V.; Eronina, T.B.; Chebotareva, N.A.; Shubin, V.V.; Kalacheva, D.I.; Kurganov, B.I. Effect of Arginine on Chaperone-Like Activity of HspB6 and Monomeric 14-3-3ζ. Int. J. Mol. Sci. 2020, 21, 2039. https://doi.org/10.3390/ijms21062039
Mikhaylova VV, Eronina TB, Chebotareva NA, Shubin VV, Kalacheva DI, Kurganov BI. Effect of Arginine on Chaperone-Like Activity of HspB6 and Monomeric 14-3-3ζ. International Journal of Molecular Sciences. 2020; 21(6):2039. https://doi.org/10.3390/ijms21062039
Chicago/Turabian StyleMikhaylova, Valeriya V., Tatiana B. Eronina, Natalia A. Chebotareva, Vladimir V. Shubin, Daria I. Kalacheva, and Boris I. Kurganov. 2020. "Effect of Arginine on Chaperone-Like Activity of HspB6 and Monomeric 14-3-3ζ" International Journal of Molecular Sciences 21, no. 6: 2039. https://doi.org/10.3390/ijms21062039
APA StyleMikhaylova, V. V., Eronina, T. B., Chebotareva, N. A., Shubin, V. V., Kalacheva, D. I., & Kurganov, B. I. (2020). Effect of Arginine on Chaperone-Like Activity of HspB6 and Monomeric 14-3-3ζ. International Journal of Molecular Sciences, 21(6), 2039. https://doi.org/10.3390/ijms21062039





