Identification of the Core Pollen-Specific Regulation in the Rice OsSUT3 Promoter
Abstract
1. Introduction
2. Results
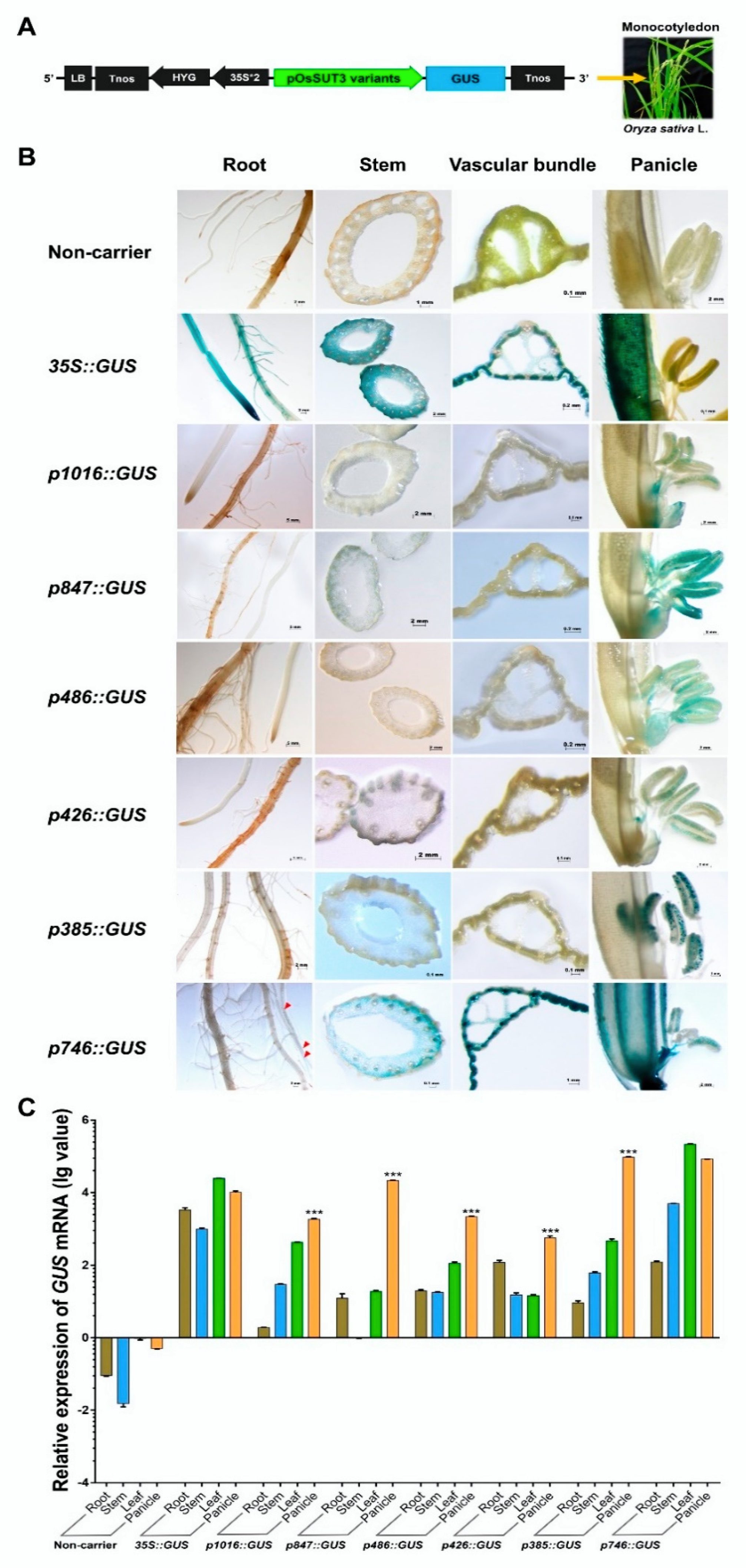
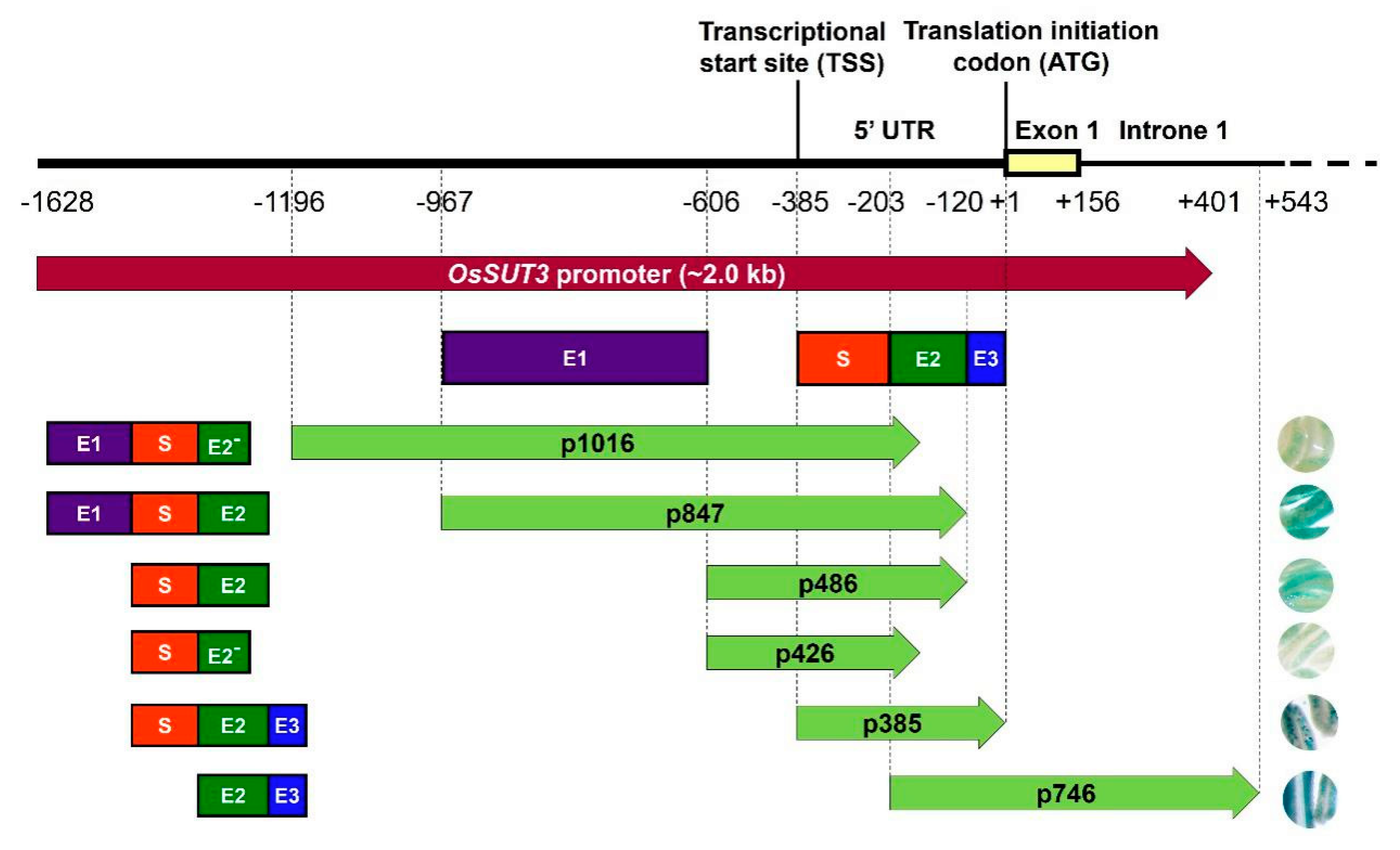
2.1. Pollen-Specific Motifs in the OsSUT3 Promoter and the Deletion
2.2. Identification of the Core Pollen-Specific Region in the OsSUT3 Promoter
2.3. Identification Three Enhancer Regions (E1, E2, and E3) That Control GUS Expression in pOsSUT3
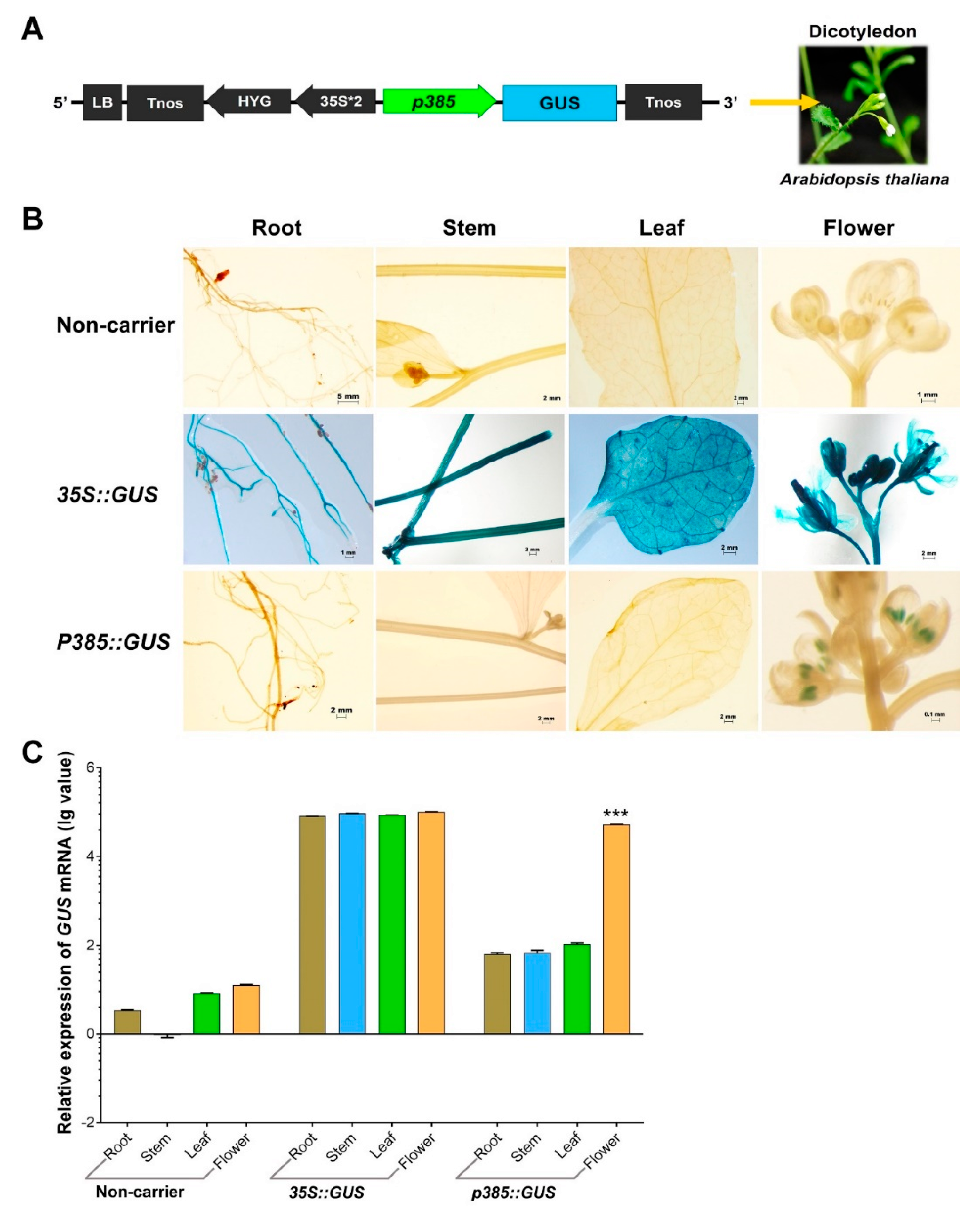
2.4. p385 Can Drive Pollen-Specific Expression in the Dicotyledonous Model Plant Arabidopsis thaliana
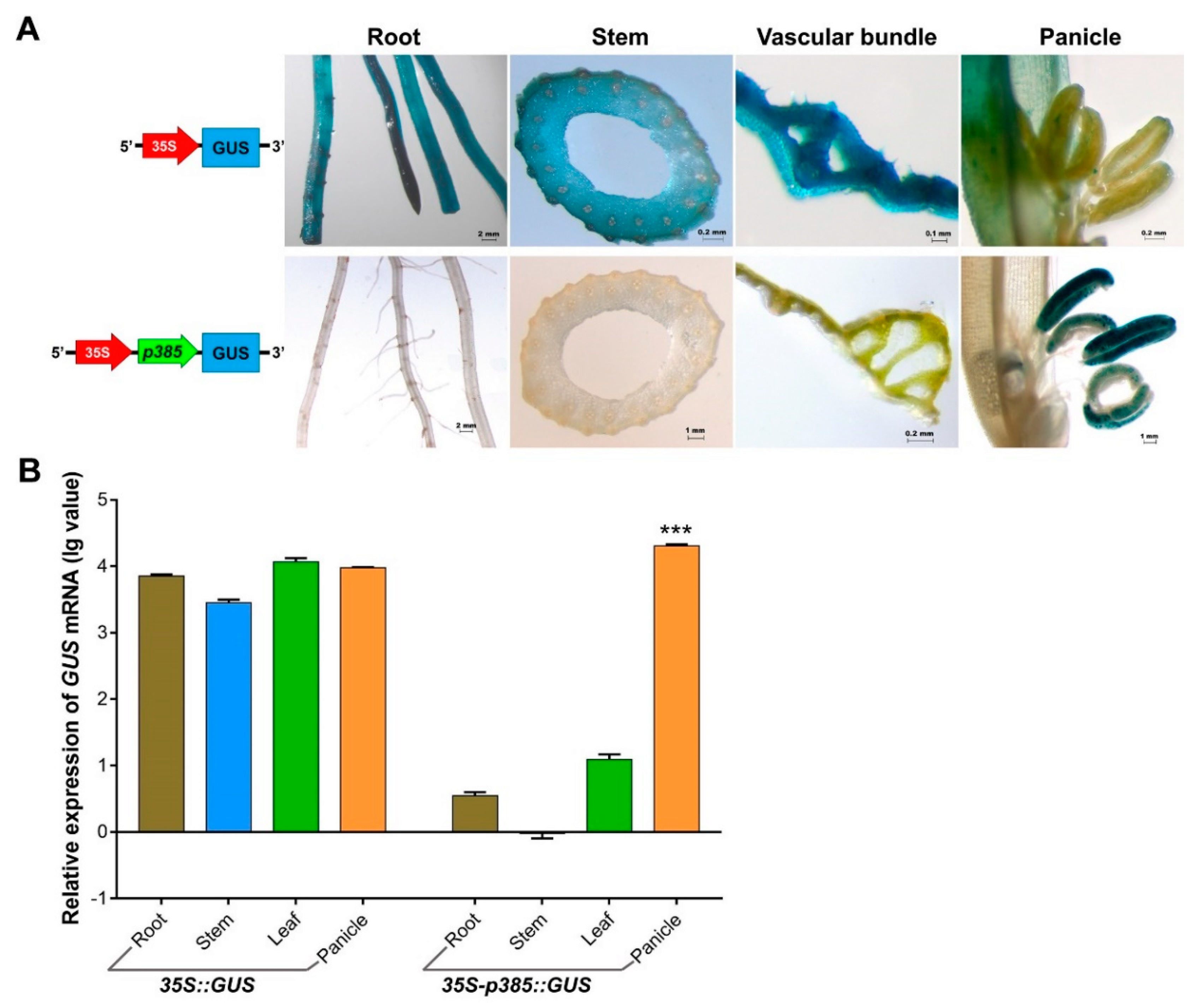
2.5. p385 Alters the Expression Pattern of the CaMV35S Promoter in Rice
3. Discussion
3.1. p385 Is the Core Pollen-Specific Regulatory Region of the OsSUT3 Promoter
3.2. p385 Was Characterized by Two Gene Expression Motifs
3.3. p385 Changes the Expression Pattern of the Constitutive Promoter CaMV35S
4. Materials and Methods
4.1. Sequence Analysis and Promoter Deletion
4.2. Plasmid Construction and Transformation
4.3. Plant Materials and Growth Conditions
4.4. Histochemical Localization of β-Glucuronidase (GUS) Activity
4.5. Genome DNA and Total RNA Extraction
4.6. qRT-PCR
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Odell, J.T.; Nagy, F.; Chua, N.H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 1985, 313, 810–812. [Google Scholar] [CrossRef]
- McElroy, D.; Zhang, W.; Cao, J.; Wu, R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 1990, 2, 163–171. [Google Scholar] [PubMed]
- Christensen, A.H.; Sharrock, R.A.; Quail, P.H. Maize polyubiquitin genes: Structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 1992, 18, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Joshi, J.B.; Geetha, S.; Singh, B.; Kumar, K.K.; Kokiladevi, E.; Arul, L.; Balasubramanian, P.; Sudhakar, D. A maize α-zein promoter drives an endosperm-specific expression of transgene in rice. Physiol. Mol. Biol. Plants 2015, 21, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Anami, S.; Njuguna, E.; Coussens, G.; Aesaert, S.; Van Lijsebettens, M. Higher plant transformation: Principles and molecular tools. Int. J. Dev. Biol. 2013, 57, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Wang, Z.; Liu, Y.; Li, W.; Wu, F.; Lin, X.; Meng, Z. Functional architecture of two exclusively late stage pollen-specific promoters in rice (Oryza sativa L.). Plant Mol. Biol. 2015, 88, 415–428. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhang, X.L.; Nadir, S.; DongChen, W.H.; Guo, X.Q.; Zhang, H.X.; Li, C.Y.; Chen, L.J.; Lee, D.S. A lysM domain-containing gene OsEMSA1 involved in embryo sac development in rice (Oryza sativa L.). Front Plant Sci. 2017, 8, 1596. [Google Scholar] [CrossRef]
- Knoch, E.; Sugawara, S.; Mori, T.; Nakabayashi, R.; Saito, K.; Yonekura-Sakakibara, K. UGT79B31 is responsible for the final modification step of pollen-specific flavonoid biosynthesis in Petunia hybrida. Planta 2018, 247, 779–790. [Google Scholar] [CrossRef]
- Garrido, D.; Busscher, J.; van Tunen, A.J. P Promoter activity of a putative pollen monosaccharide transporter in Petunia hybrida and characterisation of a transposon insertion mutant. Protoplasma 2006, 228, 3–11. [Google Scholar] [CrossRef]
- Choi, H.; Jin, J.Y.; Choi, S.; Hwang, J.U.; Kim, Y.Y.; Suh, M.C.; Lee, Y. An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. Plant J. 2011, 65, 181–193. [Google Scholar] [CrossRef]
- Millwood, R.J.; Moon, H.S.; Poovaiah, C.R.; Muthukumar, B.; Rice, J.H.; Abercrombie, J.M.; Abercrombie, L.L.; Green, W.D.; Stewart, C.N., Jr. Engineered selective plant male sterility through pollen-specific expression of the EcoRI restriction endonuclease. Plant Biotechnol. J. 2016, 14, 1281–1290. [Google Scholar] [CrossRef] [PubMed]
- Khurana, R.; Kapoor, S.; Tyagi, A. Anthology of anther/pollen-specific promoters and transcription factors. Crit. Rev. Plant Sci. 2012, 31, 359–390. [Google Scholar] [CrossRef]
- Turcich, M.P.; Hamilton, D.A.; Mascarenhas, J.P. Isolation and characterization of pollen-specific maize genes with sequence homology to ragweed allergens and pectate lyases. Plant Mol. Biol. 1993, 23, 1061–1065. [Google Scholar] [CrossRef]
- Hanson, D.D.; Hamilton, D.A.; Travis, J.L.; Bashe, D.M.; Mascarenhas, J.P. Characterization of a pollen-specific cDNA clone from Zea mays and its expression. Plant Cell 1989, 1, 173–179. [Google Scholar] [PubMed]
- Guerrero, F.D.; Crossland, L.; Smutzer, G.S.; Hamilton, D.A.; Mascarenhas, J.P. Promoter sequences from a maize pollen-specific gene direct tissue-specific transcription in tobacco. Mol. Gen. Genet. 1990, 224, 161–168. [Google Scholar] [CrossRef]
- Twell, D.; Wing, R.; Yamaguchi, J.; McCormick, S. Isolation and expression of an anther-specific gene from tomato. Mol. Gen. Genet. 1989, 217, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Eyal, Y.; Curie, C.; McCormick, S. Pollen specificity elements reside in 30 bp of the proximal promoters of two pollen-expressed genes. Plant Cell 1995, 7, 373–384. [Google Scholar]
- Bate, N.; Twell, D. Functional architecture of a late pollen promoter: Pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol. Biol. 1998, 37, 859–869. [Google Scholar] [CrossRef]
- Twell, D.; Klein, T.M.; Fromm, M.E.; McCormick, S. Transient expression of chimeric genes delivered into pollen by microprojectile bombardment. Plant Physiol. 1989, 91, 1270–1274. [Google Scholar] [CrossRef]
- Twell, D.; Yamaguchi, J.; Wing, R.A.; Ushiba, J.; McCormick, S. Promoter analysis of genes that are coordinately expressed during pollen development reveals pollen-specific enhancer sequences and shared regulatory elements. Genes Dev. 1991, 5, 496–507. [Google Scholar] [CrossRef]
- Rogers, H.J.; Bate, N.; Combe, J.; Sullivan, J.; Sweetman, J.; Swan, C.; Lonsdale, D.M.; Twell, D. Functional analysis of cis-regulatory elements within the promoter of the tobacco late pollen gene g10. Plant Mol. Biol. 2001, 45, 577–585. [Google Scholar] [CrossRef]
- Yoo, J.; Kohlbrenner, E.; Kim, O.; Hajjar, R.J.; Jeong, D. Enhancing atrial-specific gene expression using a calsequestrin cis-regulatory module 4 with a sarcolipin promoter. J. Gene Med. 2018, 20, e3060. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, M.E.; Shelton, S.; Brockerhoff, S.E.; Hurley, J.B.; Kennedy, B.N. PRE-1, a cis element sufficient to enhance cone- and rod- specific expression in differentiating zebrafish photoreceptors. BMC Dev. Biol. 2011, 11, 3. [Google Scholar] [CrossRef]
- Li, M.; Xie, C.; Song, B.; Ou, Y.; Lin, Y.; Liu, X.; Zhang, H.; Liu, J. Construction of efficient, tuber-specific, and cold-inducible promoters in potato. Plant Sci. 2015, 235, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, S.; Tsuchiya, T.; Toriyama, K.; Hinata, K. Tapetum-specific expression of the Osg6B promoter-β-glucuronidase gene in transgenic rice. Plant Cell Rep. 1997, 16, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Swapna, L.; Khurana, R.; Kumar, S.V.; Tyagi, A.K.; Rao, K.V. Pollen-specific expression of Oryza sativa indica pollen allergen gene (OSIPA) promoter in rice and Arabidopsis transgenic systems. Mol. Biotechnol. 2011, 48, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Khurana, R.; Kathuria, H.; Mukhopadhyay, A.; Kapoor, S.; Tyagi, A.K. A 286 bp upstream regulatory region of a rice anther-specific gene, OSIPP3, confers pollen-specific expression in Arabidopsis. Biotechnol. Lett. 2013, 35, 455–462. [Google Scholar] [CrossRef]
- Wen, K.; Chen, Y.; Zhou, X.; Chang, S.; Feng, H.; Zhang, J.; Chu, Z.; Han, X.; Li, J.; Liu, J.; et al. OsCPK21 is required for pollen late-stage development in rice. J. Plant Physiol. 2019, 240, 153000. [Google Scholar] [CrossRef]
- Jeon, J.S.; Chung, Y.Y.; Lee, S.; Yi, G.H.; Oh, B.G.; An, G. Isolation and characterization of an anther-specific gene, RA8, from rice (Oryza sativa L.). Plant Mol. Biol. 1999, 39, 35–44. [Google Scholar] [CrossRef]
- Oo, M.M.; Bae, H.K.; Nguyen, T.D.; Moon, S.; Oh, S.A.; Kim, J.H.; Soh, M.S.; Song, J.T.; Jung, K.H.; Park, S.K. Evaluation of rice promoters conferring pollen-specific expression in a heterologous system, Arabidopsis. Plant Reprod. 2014, 27, 47–58. [Google Scholar] [CrossRef]
- Aoki, N.; Hirose, T.; Scofield, G.N.; Whitfeld, P.R.; Furbank, R.T. The sucrose transporter gene family in rice. Plant Cell Physiol. 2003, 44, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Zhang, Z.; Miyao, A.; Hirochika, H.; Ohsugi, R.; Terao, T. Disruption of a gene for rice sucrose transporter, OsSUT1, impairs pollen function but pollen maturation is unaffected. J. Exp. Bot. 2010, 61, 3639–3646. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; Li, J.; Luan, Y.F.; Zhang, C.L.; Xu, R.C.; Lv, D.; Tan, Y.L.; Tan, X.L. Cloning and expression analysis of OsSUT3 gene Promoter from Oryza sativa. Mole Plant Breed. 2018, 16, 7225–7233. [Google Scholar]
- Welchen, E.; Viola, I.L.; Kim, H.J.; Prendes, L.P.; Comelli, R.N.; Hong, J.C.; Gonzalez, D.H. A segment containing a G-box and an ACGT motif confers differential expression characteristics and responses to the Arabidopsis Cytc-2 gene, encoding an isoform of cytochrome c. J. Exp. Bot. 2009, 60, 829–845. [Google Scholar] [CrossRef]
- Ravel, C.; Nagy, I.J.; Martre, P.; Sourdille, P.; Dardevet, M.; Balfourier, F.; Pont, C.; Giancola, S.; Praud, S.; Charmet, G. Single nucleotide polymorphism, genetic mapping, and expression of genes coding for the DOF wheat prolamin-box binding factor. Funct. Integr. Genomics. 2006, 6, 310–321. [Google Scholar] [CrossRef]
- Williams, M.E.; Foster, R.; Chua, N.H. Sequences flanking the hexameric G-box core CACGTG affect the specificity of protein binding. Plant Cell 1992, 4, 485–496. [Google Scholar]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999, 17, 287–291. [Google Scholar] [CrossRef]
- Hsieh, T.H.; Lee, J.T.; Charng, Y.Y.; Chan, M.T. Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol. 2002, 130, 618–626. [Google Scholar] [CrossRef]
- Gao, L.; Tian, Y.; Chen, M.C.; Wei, L.; Gao, T.G.; Yin, H.J.; Zhang, J.L.; Kumar, T.; Liu, L.B.; Wang, S.M. Cloning and functional characterization of epidermis-specific promoter MtML1 from Medicago truncatula. J. Biotechnol. 2019, 300, 32–39. [Google Scholar] [CrossRef]
- Smale, S.T.; Kadonaga, J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003, 72, 449–479. [Google Scholar] [CrossRef]
- Grosschedl, R.; Birnstiel, M.L. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc. Natl. Acad. Sci. USA 1980, 77, 7102–7106. [Google Scholar] [CrossRef] [PubMed]
- Benoist, C.; Chambon, P. In vivo sequence requirements of the Sv40 early promotor region. Nature 1981, 290, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Tissier, A. Trichome specific expression: Promoters and their applications. In Transgenic Plants-Advances and Limitations; Ciftci, Y.O., Ed.; InTech: London, UK, 2012; Volume 17, pp. 353–378. [Google Scholar]
- Delaney, S.K.; Orford, S.J.; Martin-Harris, M.; Timmis, J.N. The fiber specificity of the cotton FSltp4 gene promoter is regulated by an AT-rich promoter region and the AT-hook transcription factor GhAT1. Plant Cell Physiol. 2007, 48, 1426–1437. [Google Scholar] [CrossRef]
- Gutiérrez-Alcalá, G.; Calo, L.; Gros, F.; Caissard, J.C.; Gotor, C.; Romero, L.C. A versatile promoter for the expression of proteins in glandular and non-glandular trichomes from a variety of plants. J. Exp. Bot. 2005, 56, 2487–2494. [Google Scholar] [CrossRef][Green Version]
- Yadav, V.K.; Yadav, V.K.; Pant, P.; Singh, S.P.; Maurya, R.; Sable, A.; Sawant, S.V. GhMYB1 regulates SCW stage-specific expression of the GhGDSL promoter in the fibres of Gossypium hirsutum L. Plant Biotechnol. J. 2017, 15, 1163–1174. [Google Scholar] [CrossRef]
- Dong, H.; Liu, L.; Fan, X.; Asghar, S.; Li, Y.; Wang, Y.; Xu, X.; Wu, T.; Zhang, X.; Qiu, C.; et al. The artificial promoter rMdAG2I confers flower-specific activity in Malus. Int. J. Mol. Sci. 2019, 20, 4551. [Google Scholar] [CrossRef]
- Singh, M.; Bhalla, P.L.; Xu, H.; Singh, M.B. Isolation and characterization of a flowering plant male gametic cell-specific promoter. FEBS Lett. 2003, 542, 47–52. [Google Scholar] [CrossRef]
- Engel, M.L.; Holmes-Davis, R.; McCormick, S. Green Sperm. Identification of male gamete promoters in Arabidopsis. Plant Physiol. 2005, 138, 2124–2133. [Google Scholar] [CrossRef]
- Lamb, P.; McKnight, S.L. Diversity and specificity in transcriptional regulation: The benefits of heterotypic dimerization. Trends Biochem. Sci. 1991, 16, 417–422. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Schmidt, R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999, 17, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Rossouw, C.M.; Vergeer, W.P.; du Plooy, S.J.; Bernard, M.P.; Ramirez, F.; de Wet, W.J. DNA sequences in the first intron of the human pro-alpha1 (I) collagen gene enhance transcription. J. Biol. Chem. 1987, 262, 15151–15157. [Google Scholar] [PubMed]
- Schibler, U.; Sierra, F. Alternative promoters in developmental gene expression. Annu. Rev. Genet. 1987, 21, 237–257. [Google Scholar] [CrossRef] [PubMed]
- Ortega, J.L.; Temple, S.J.; Bagga, S.; Ghoshroy, S.; Sengupta-Gopalan, C. Biochemical and molecular characterization of transgenic Lotus japonicus plants constitutively over-expressing a cytosolic glutamine synthetase gene. Planta 2004, 19, 807–818. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seger, M.; Ortega, J.L.; Bagga, S.; Gopalan, C.S. Repercussion of mesophyll-specific overexpression of a soybean cytosolic glutamine synthetase gene in alfalfa (Medicago sativa L.) and tobacco (Nicotiana tabaccum L.). Plant Sci. 2009, 176, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.; Kumar, V.; Bhat, S.R.; Srinivasan, R. Upstream sequences of the LOJ gene leads to identification of a novel enhancer element conferring lateral organ junction-specific expression in Arabidopsis thaliana. Plant Molec. Bio. Rep. 2011, 29, 265–277. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Clarke, J.D. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb. Lab. Protoc. 2009, 4. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar]

| Primer Name | Sequences (from 5′ to 3′) * | Application |
|---|---|---|
| p1016-F | CCCAAGCTTGGGGGTATGTTATAAGGGTCTGGTAGGA | Promoter cloning |
| p1016-R | CATGCCATGGCATGAGAGGAGGGAGCGGTGAGA | Promoter cloning |
| p847-F | CCCAAGCTTGGGCCTTCGTATGTAAGGGACGCT | Promoter cloning |
| p847-R | CATGCCATGGCATGGACCAAGACGACGACGGATA | Promoter cloning |
| p486-F | CCCAAGCTTGGGCCGCAATGAACTCTGCCTAT | Promoter cloning |
| p486-R | CATGCCATGGCATGGACCAAGACGACGACGGATA | Promoter cloning |
| p426-F | CCCAAGCTTGGGCCGCAATGAACTCTGCCTAT | Promoter cloning |
| p426-R | CATGCCATGGCATGGAGGAGGGAGCGGTGAGA | Promoter cloning |
| p746-F | CCCAAGCTTGGGCAATGTCTCACCGCTCCC | Promoter cloning |
| p746-R | CATGCCATGGCATGGCAAACAGGCAGCATAAGAG | Promoter cloning |
| GUS-F | ATCCTCTGGGAACCACTGAACC | Real-time RT-PCR |
| GUS-R | CATCACATTGCTCGCTTCGTTA | Real-time RT-PCR |
| Actin-F | ATCCTTGTATGCTAGCGGTCGA | Real-time RT-PCR |
| Actin-R | ATCCAACCGGAGGATAGCATG | Real-time RT-PCR |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.; Xu, R.; Lv, D.; Zhang, C.; Yang, H.; Zhang, J.; Wen, J.; Li, C.; Tan, X. Identification of the Core Pollen-Specific Regulation in the Rice OsSUT3 Promoter. Int. J. Mol. Sci. 2020, 21, 1909. https://doi.org/10.3390/ijms21061909
Li D, Xu R, Lv D, Zhang C, Yang H, Zhang J, Wen J, Li C, Tan X. Identification of the Core Pollen-Specific Regulation in the Rice OsSUT3 Promoter. International Journal of Molecular Sciences. 2020; 21(6):1909. https://doi.org/10.3390/ijms21061909
Chicago/Turabian StyleLi, Dandan, Rucong Xu, Dong Lv, Chunlong Zhang, Hong Yang, Jianbo Zhang, Jiancheng Wen, Chengyun Li, and Xuelin Tan. 2020. "Identification of the Core Pollen-Specific Regulation in the Rice OsSUT3 Promoter" International Journal of Molecular Sciences 21, no. 6: 1909. https://doi.org/10.3390/ijms21061909
APA StyleLi, D., Xu, R., Lv, D., Zhang, C., Yang, H., Zhang, J., Wen, J., Li, C., & Tan, X. (2020). Identification of the Core Pollen-Specific Regulation in the Rice OsSUT3 Promoter. International Journal of Molecular Sciences, 21(6), 1909. https://doi.org/10.3390/ijms21061909





