UL36 Encoded by Marek’s Disease Virus Exhibits Linkage-Specific Deubiquitinase Activity
Abstract
:1. Introduction
2. Results
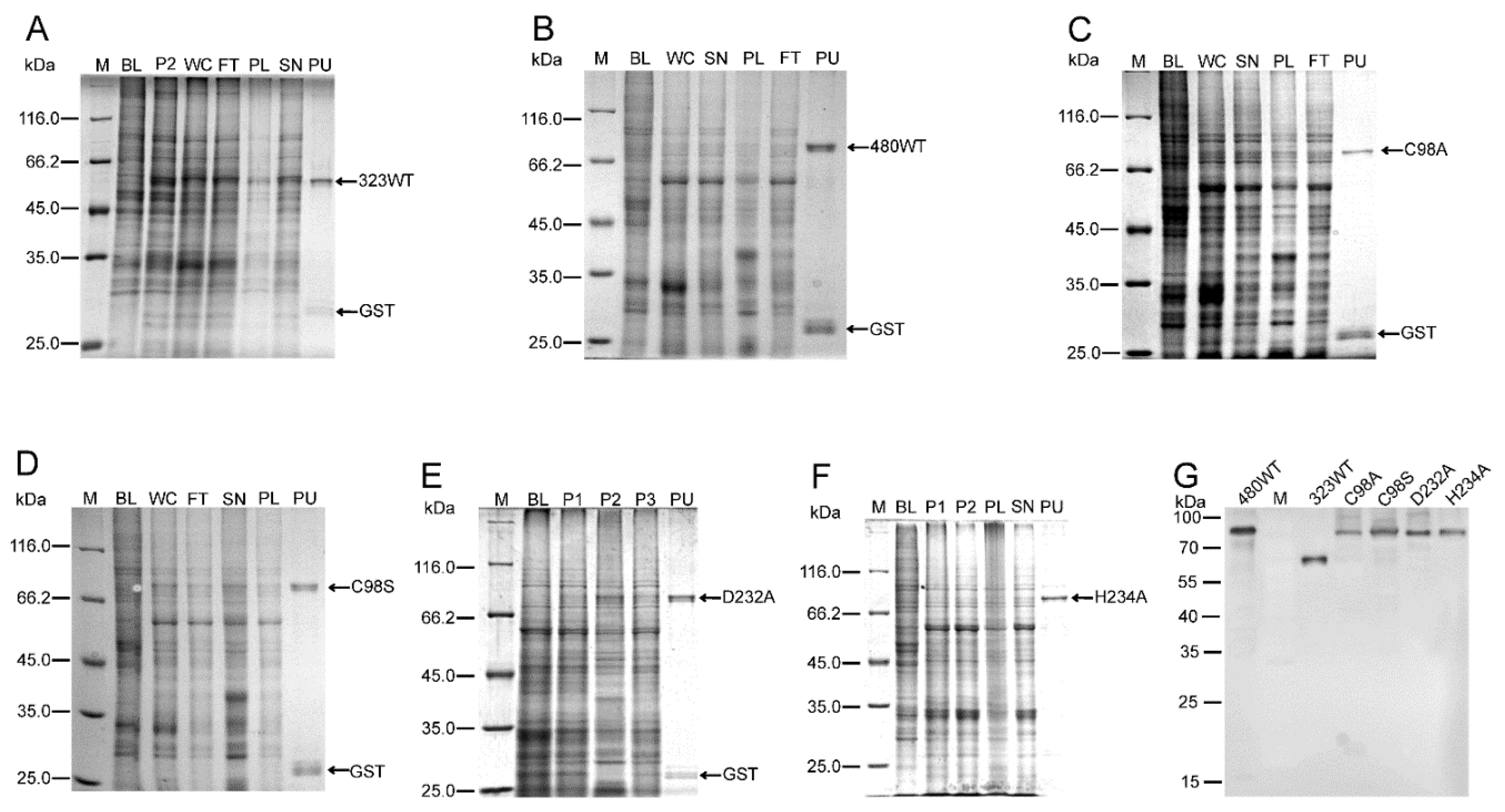
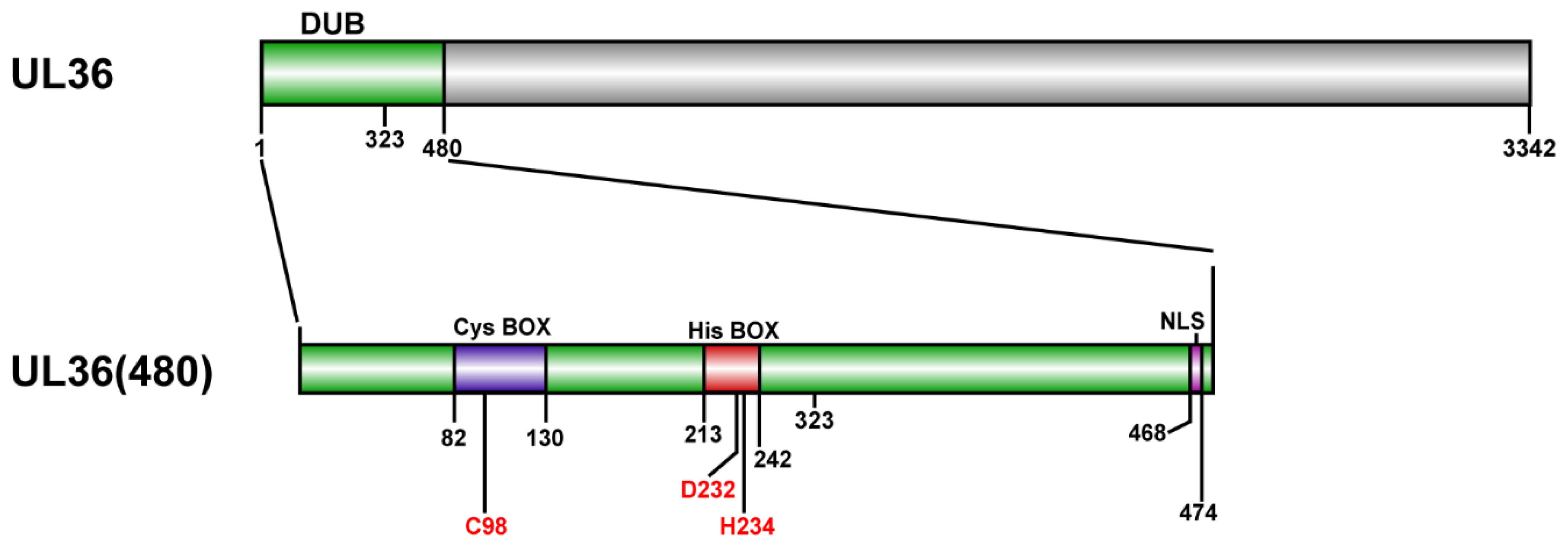
2.1. Purification of MDV-Encoded UL36-DUBs
2.2. Wild Type UL36-DUBs Effectively Hydrolyzed Ubiquitin Substrate
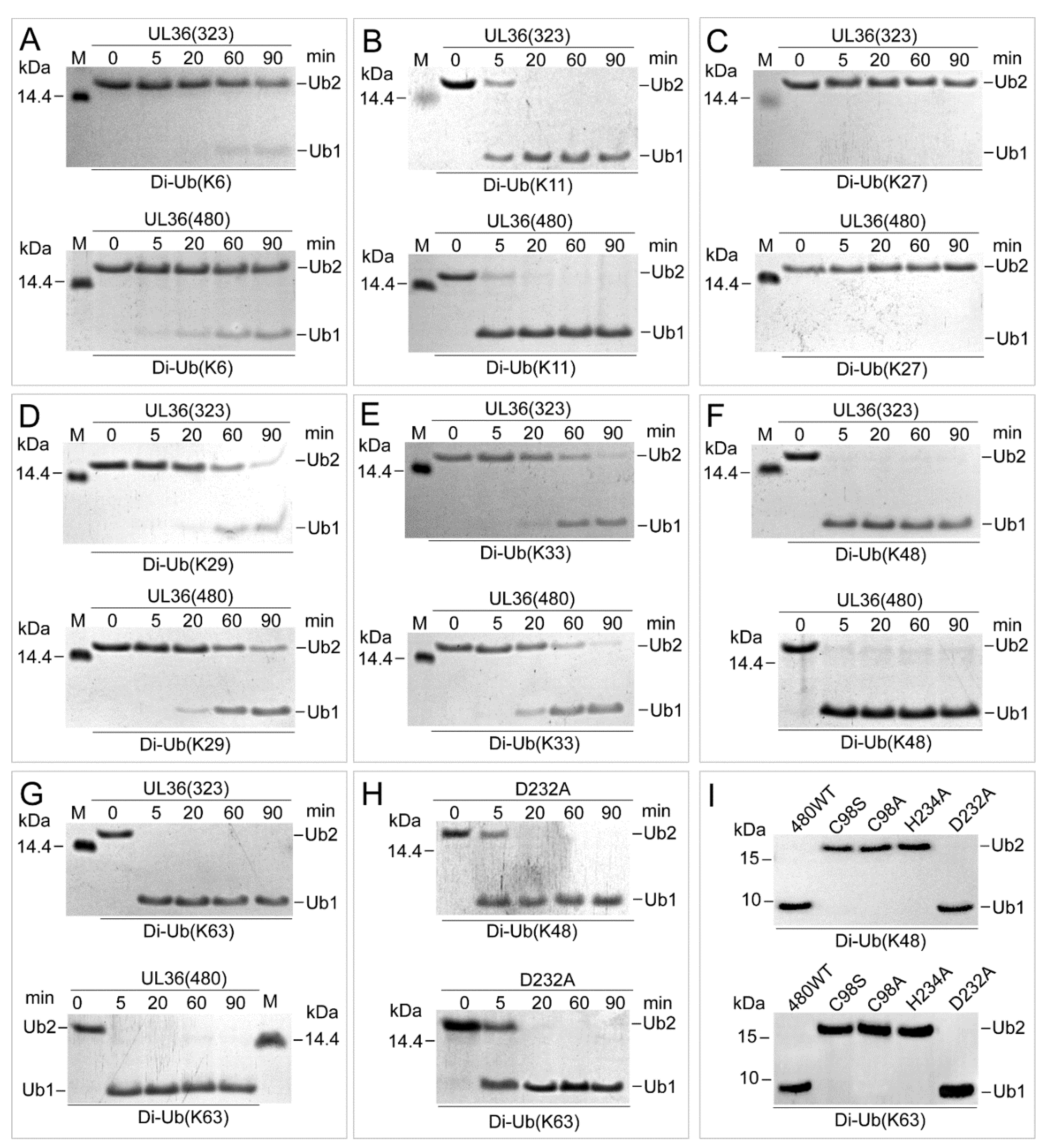
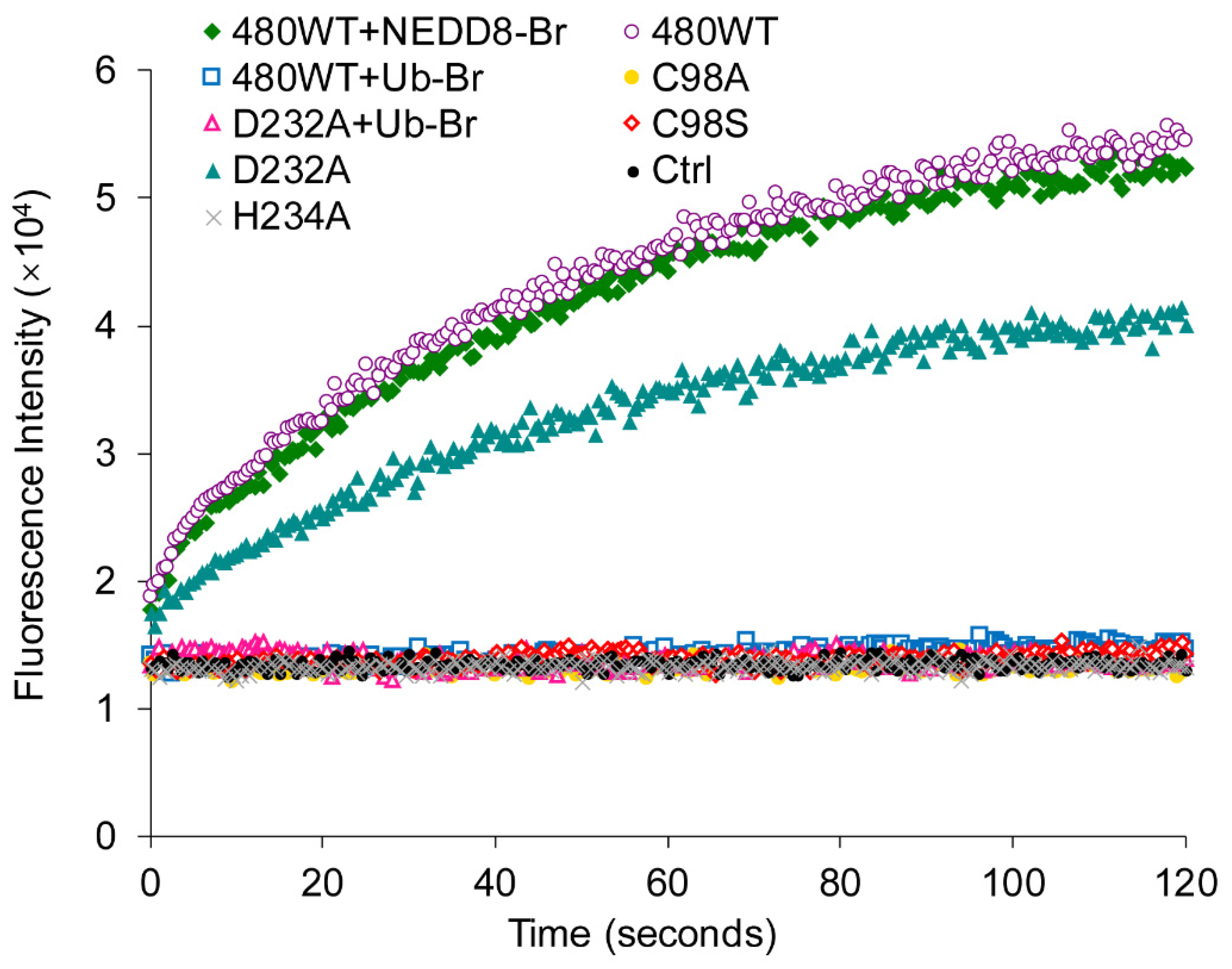
2.3. UL36 Hydrolyzed Ubiquitin Chains in Linkage Preference
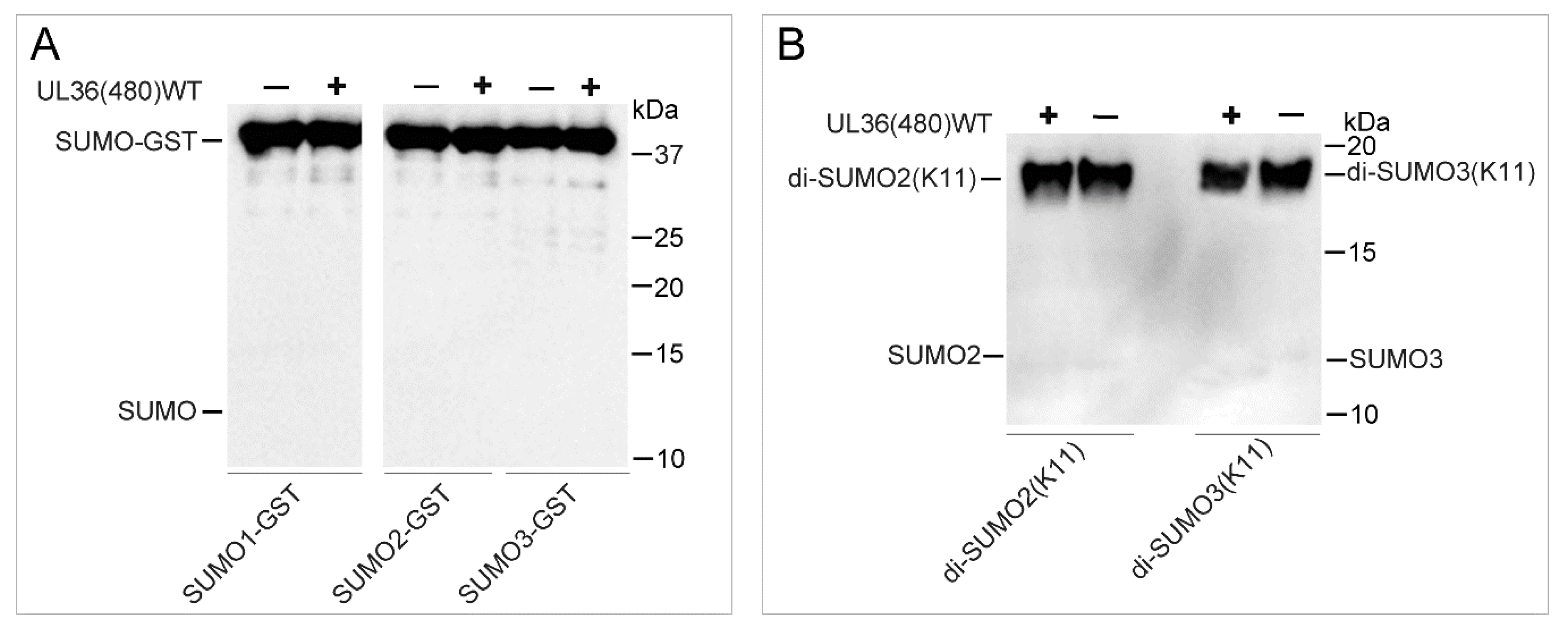
2.4. UL36-DUB Failed to Deconjugate UbL Substrates
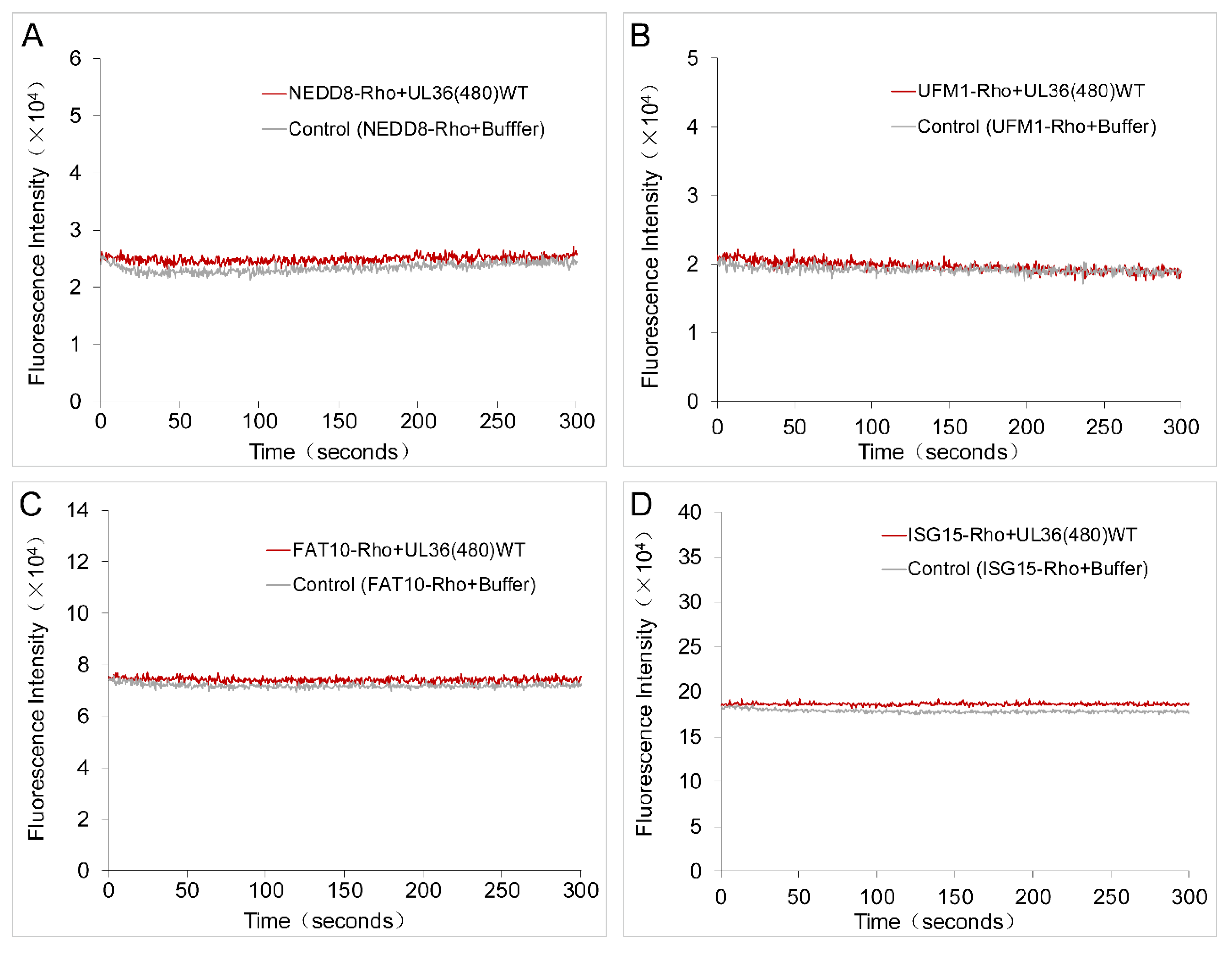
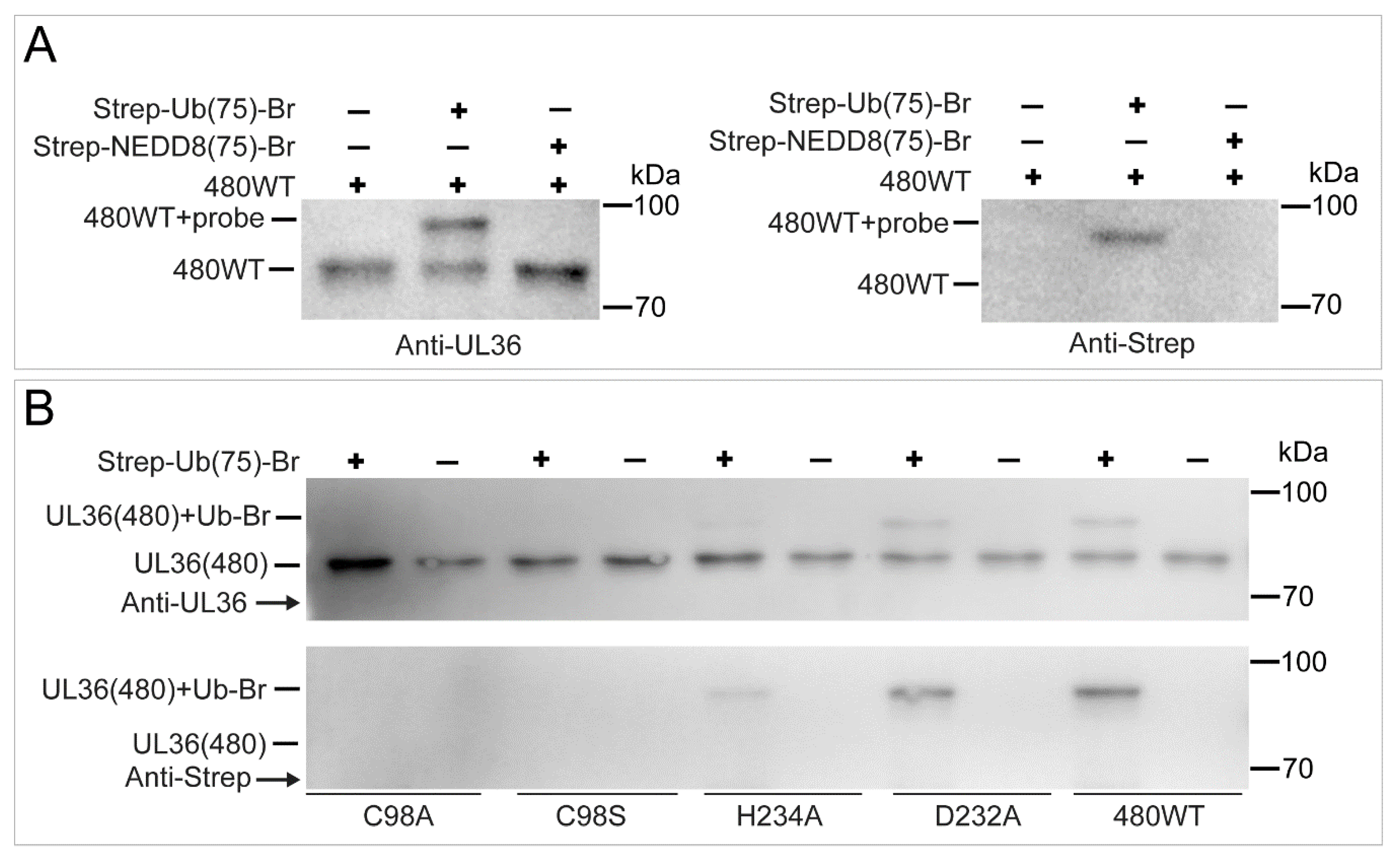
2.5. UL36 Specifically Targets Ub but Not NEDD8
2.6. UL36-DUBs Was Resistant to Some Protease Inhibitors
3. Discussion
4. Materials and Methods
4.1. Construction of Plasmids
4.2. Expression and Purification of UL36-DUBs Proteins
4.3. Investigation of the Kinetics of Deubiquitinating Activity
4.4. Characterization of Ub Substrate Preference of UL36-DUBs
4.5. Characterization of the UbL Substrate Specificity of UL36
4.6. Preparation of Inhibitor Probe
4.7. Determination of the Cross-Reactivity of UL36-DUB via Site-Specific Incorporation
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CCHFV | Crimean–Congo hemorrhagic fever virus |
| CPIC | Complete protease inhibitor cocktail |
| DUB | Deubiquitinase |
| DUGV | Dugbe virus |
| EAV | Equine arteritis virus |
| EBV | Epstein–Barr virus |
| ECL | Enhanced chemiluminescence |
| GaHV-2 | Gallid alphaherpesvirus 2 |
| HCMV | Human cytomegalovirus |
| HSV | Herpes simplex virus |
| KSHV | Kaposi’s sarcoma-associated herpesvirus |
| MD | Marek’s disease |
| MDV | Marek’s disease virus |
| MERS-CoV | Middle East respiratory syndrome-related coronavirus |
| MESNa | Mercaptosulfonate sodium salt |
| MJD | Machado–Josephin domain protease |
| OTU | Ovarian tumor proteases |
| PMSF | Phenylmethanesulfonyl fluoride |
| PRRSV | Porcine reproductive and respiratory syndrome virus |
| PRV | Pseudorabies virus |
| SARS-CoV | Severe acute respiratory syndrome-coronavirus |
| Ub | Ubiquitin |
| Ub-AMC | Ubiquitin 7-amido-4-methylcoumarin |
| UbL | Ubiquitin-like modifier |
| UCH | Ubiquitin C-terminal hydrolase |
| USP | Ubiquitin-specific protease |
References
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 2017, 42, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Al-Hakim, A.K.; Zagorska, A.; Chapman, L.; Deak, M.; Peggie, M.; Alessi, D.R. Control of AMPK-related kinases by USP9X and atypical Lys29/Lys33-linked polyubiquitin chains. Biochem. J. 2008, 411, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobson, A.D.; Zhang, N.-Y.; Xu, P.; Han, K.-J.; Noone, S.; Peng, J.; Liu, C.-W. The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 s proteasome. J. Biol. Chem. 2009, 284, 35485–35494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickliffe, K.E.; Williamson, A.; Meyer, H.-J.; Kelly, A.; Rape, M. K11-linked ubiquitin chains as novel regulators of cell division. Trends Cell Biol. 2011, 21, 656–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Ye, Y. Polyubiquitin chains: Functions, structures, and mechanisms. Cell. Mol. Life Sci. 2008, 65, 2397–2406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Veen, A.G.; Ploegh, H.L. Ubiquitin-like proteins. Annu. Rev. Biochem. 2012, 81, 323–357. [Google Scholar] [CrossRef]
- Clague, M.J.; Urbe, S.; Komander, D. Breaking the chains: Deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 2019, 20, 338–352. [Google Scholar] [CrossRef]
- Turcu, F.E.R.; Ventii, K.H.; Wilkinson, K.D. Regulation and Cellular Roles of Ubiquitin-specific Deubiquitinating Enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef] [Green Version]
- Komander, D. Mechanism, specificity and structure of the deubiquitinases. Subcell. Biochem. 2010, 54, 69–87. [Google Scholar]
- Lee, J.I.; Sollars, P.J.; Baver, S.B.; Pickard, G.E.; Leelawong, M.; Smith, G.A. A Herpesvirus Encoded Deubiquitinase Is a Novel Neuroinvasive Determinant. PLoS Pathog. 2009, 5, e1000387. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, C.M.; Wang, L.; Damania, B. Kaposi’s sarcoma-associated herpesvirus encodes a viral deubiquitinase. J. Virol. 2009, 83, 10224–10233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitehurst, C.B.; Vaziri, C.; Shackelford, J.; Pagano, J.S. Epstein-Barr virus BPLF1 deubiquitinates PCNA and attenuates polymerase eta recruitment to DNA damage sites. J. Virol. 2012, 86, 8097–8106. [Google Scholar] [CrossRef] [Green Version]
- Capodagli, G.C.; McKercher, M.A.; Baker, E.A.; Masters, E.M.; Brunzelle, J.S.; Pegan, S.D. Structural analysis of a viral ovarian tumor domain protease from the Crimean-Congo hemorrhagic fever virus in complex with covalently bonded ubiquitin. J. Virol. 2011, 85, 3621–3630. [Google Scholar] [CrossRef] [Green Version]
- Capodagli, G.C.; Deaton, M.K.; Baker, E.A.; Lumpkin, R.J.; Pegan, S.D. Diversity of ubiquitin and ISG15 specificity among nairoviruses’ viral ovarian tumor domain proteases. J. Virol. 2013, 87, 3815–3827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isaacson, M.K.; Ploegh, H.L. Ubiquitination, Ubiquitin-like Modifiers, and Deubiquitination in Viral Infection. Cell Host Microbe 2009, 5, 559–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calistri, A.; Munegato, D.; Carli, I.; Parolin, C.; Palù, G. The ubiquitin-conjugating system: Multiple roles in viral replication and infection. Cells 2014, 3, 386–417. [Google Scholar] [CrossRef] [Green Version]
- Boodhoo, N.; Gurung, A.; Sharif, S.; Behboudi, S. Marek’s disease in chickens: A review with focus on immunology. Vet. Res. 2016, 47, 119. [Google Scholar] [CrossRef] [Green Version]
- Reddy, S.M.; Izumiya, Y.; Lupiani, B. Marek’s disease vaccines: Current status, and strategies for improvement and development of vector vaccines. Vet. Microbiol. 2017, 206, 113–120. [Google Scholar] [CrossRef]
- Baigent, S.J.; Smith, L.P.; Nair, V.K.; Currie, R.J.W. Vaccinal control of Marek’s disease: Current challenges, and future strategies to maximize protection. Vet. Immunol. Immunopathol. 2006, 112, 78–86. [Google Scholar] [CrossRef]
- Davison, F.; Nair, V. Use of Marek’s disease vaccines: Could they be driving the virus to increasing virulence? Expert Rev. Vaccine 2005, 4, 77–88. [Google Scholar] [CrossRef]
- Padhi, A.; Parcells, M.S. Positive Selection Drives Rapid Evolution of the meq Oncogene of Marek’s Disease Virus. PLoS ONE 2016, 11, e0162180. [Google Scholar] [CrossRef] [PubMed]
- Chbab, N.; Egerer, A.; Veiga, I.; Jarosinski, K.W.; Osterrieder, N. Viral control of vTR expression is critical for efficient formation and dissemination of lymphoma induced by Marek’s disease virus (MDV). Vet. Res. 2010, 41, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, A.T.; Selvaraj, R.K.; Kamil, J.P.; Osterrieder, N.; Kaufer, B.B. Marek’s disease viral interleukin-8 promotes lymphoma formation through targeted recruitment of B cells and CD4+ CD25+ T cells. J. Virol. 2012, 86, 8536–8545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veiga, I.B.; Jarosinski, K.W.; Kaufer, B.B.; Osterrieder, N. Marek’s disease virus (MDV) ubiquitin-specific protease (USP) performs critical functions beyond its enzymatic activity during virus replication. Virology 2013, 437, 110–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McPherson, M.C.; Delany, M.E. Virus and host genomic, molecular, and cellular interactions during Marek’s disease pathogenesis and oncogenesis. Poult. Sci. 2016, 95, 412–429. [Google Scholar] [CrossRef]
- Wozniakowski, G.; Samorek-Salamonowicz, A.E. Molecular evolution of Marek’s disease virus (MDV) field strains in a 40-year time period. Avian Dis. 2014, 58, 550–557. [Google Scholar] [CrossRef]
- Calnek, B.W. Pathogenesis of Marek’s Disease Virus Infection. In Marek’s Disease; Hirai, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2001; pp. 25–55. [Google Scholar]
- Zhou, X.; Wu, S.; Zhou, H.; Wang, M.; Wang, M.; Lü, Y.; Cheng, Z.; Xu, J.; Ai, Y. Marek’s Disease Virus Regulates the Ubiquitylome of Chicken CD4+ T Cells to Promote Tumorigenesis. Int. J. Mol. Sci. 2019, 20, 2089. [Google Scholar] [CrossRef] [Green Version]
- Jarosinski, K.; Kattenhorn, L.; Kaufer, B.; Ploegh, H.; Osterrieder, N. A herpesvirus ubiquitin-specific protease is critical for efficient T cell lymphoma formation. Proc. Natl. Acad. Sci. USA 2007, 104, 20025–20030. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Wang, M.; Zhao, C.; Wu, S.; Yu, P.; Lu, Y.; Wang, T.; Ai, Y. Characterization of the deubiquitination activity and substrate specificity of the chicken ubiquitin-specific protease 1/USP associated factor 1 complex. PLoS ONE 2017, 12, e0186535. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Akutsu, M.; Reyes-Turcu, F.; Enchev, R.I.; Wilkinson, K.D.; Komander, D. Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep. 2011, 12, 350–357. [Google Scholar] [CrossRef] [Green Version]
- Puvar, K.; Iyer, S.; Sheedlo, M.J.; Das, C. Chapter Fifteen—Purification and functional characterization of the DUB domain of SdeA. In Methods in Enzymology; Hochstrasser, M., Ed.; Academic Press: New Haven, CT, USA, 2019; Volume 618, pp. 343–355. [Google Scholar]
- Artavanis-Tsakonas, K.; Misaghi, S.; Comeaux, C.A.; Catic, A.; Spooner, E.; Duraisingh, M.T.; Ploegh, H.L. Identification by functional proteomics of a deubiquitinating/deNeddylating enzyme in Plasmodium falciparum. Mol. Microbiol. 2006, 61, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Gastaldello, S.; Hildebrand, S.; Faridani, O.; Callegari, S.; Palmkvist, M.; Di Guglielmo, C.; Masucci, M.G. A deneddylase encoded by Epstein-Barr virus promotes viral DNA replication by regulating the activity of cullin-RING ligases. Nat. Cell Biol. 2010, 12, 351–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kattenhorn, L.M.; Korbel, G.A.; Kessler, B.M.; Spooner, E.; Ploegh, H.L. A Deubiquitinating Enzyme Encoded by HSV-1 Belongs to a Family of Cysteine Proteases that Is Conserved across the Family Herpesviridae. Mol. Cell 2005, 19, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Ge, X.; Kong, C.; Liu, T.; Liu, A.; Gao, P.; Song, J.; Zhou, L.; Guo, X.; Han, J.; et al. Characterizing the PRRSV nsp2 Deubiquitinase Reveals Dispensability of Cis-Activity for Replication and a Link of nsp2 to Inflammation Induction. Viruses 2019, 11, 896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.T.; Oh, S.E.; Lee, Y.-O.; Gibson, W.; Ahn, J.-H. Cleavage Specificity of the UL48 Deubiquitinating Protease Activity of Human Cytomegalovirus and the Growth of an Active-Site Mutant Virus in Cultured Cells. J. Virol. 2009, 83, 12046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, H.-J.; Rape, M. Enhanced protein degradation by branched ubiquitin chains. Cell 2014, 157, 910–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimeno, I.M. Marek’s disease vaccines: A solution for today but a worry for tomorrow? Vaccine 2008, 26, C31–C41. [Google Scholar] [CrossRef]
- Haq, K.; Schat, K.A.; Sharif, S. Immunity to Marek’s disease: Where are we now? Dev. Comp. Immunol. 2013, 41, 439–446. [Google Scholar] [CrossRef]
- Lee, H.; Lei, H.; Santarsiero, B.D.; Gatuz, J.L.; Cao, S.; Rice, A.J.; Patel, K.; Szypulinski, M.Z.; Ojeda, I.; Ghosh, A.K.; et al. Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. ACS Chem. Biol. 2015, 10, 1456–1465. [Google Scholar] [CrossRef]
- Rhee, S.-Y.; Sankaran, K.; Varghese, V.; Winters, M.A.; Hurt, C.B.; Eron, J.J.; Parkin, N.; Holmes, S.P.; Holodniy, M.; Shafer, R.W. HIV-1 Protease, Reverse Transcriptase, and Integrase Variation. J. Virol. 2016, 90, 6058–6070. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, K.; Kato, R.; Kavlick, M.F.; Nguyen, A.; Maroun, V.; Maeda, K.; Hussain, K.A.; Ghosh, A.K.; Gulnik, S.V.; Erickson, J.W.; et al. A potent human immunodeficiency virus type 1 protease inhibitor, UIC-94003 (TMC-126), and selection of a novel (A28S) mutation in the protease active site. J. Virol. 2002, 76, 1349–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, A. The human immunodeficiency virus protease inhibitor ritonavir is potentially active against urological malignancies. OncoTargets Ther. 2015, 8, 761–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, H.; Gao, H.; Cui, X.; Zhao, Y.; Shi, X.; Li, Q.; Yan, S.; Gao, M.; Wang, M.; Liu, C.; et al. Avirulent Marek’s disease virus type 1 strain 814 vectored vaccine expressing avian influenza (AI) virus H5 haemagglutinin induced better protection than turkey herpesvirus vectored AI vaccine. PLoS ONE 2013, 8, e53340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. DOG 1.0: Illustrator of protein domain structures. Cell Res. 2009, 19, 271–273. [Google Scholar] [CrossRef]
- Yu, P. Expression, Purification and Activity Identification of Chicken SUMO Tagged Protein Substrates. Master’s Thesis, Jilin University, Jilin, China, 2015. [Google Scholar]
- Yu, P.; Zheng, H.; Deng, C.; Zhang, X.; Wu, S.; Wang, M.; Lv, Y.; Ai, Y. Construction and Characterization of Expression Vector Harboring Chicken SUMO1 Fused by His/GST ditags. Chin. J. Vet. Sci. 2015, 35, 1799–1803. [Google Scholar]
- Borodovsky, A.; Ovaa, H.; Kolli, N.; Gan-Erdene, T.; Wilkinson, K.D.; Ploegh, H.L.; Kessler, B.M. Chemistry-Based Functional Proteomics Reveals Novel Members of the Deubiquitinating Enzyme Family. Chem. Biol. 2002, 9, 1149–1159. [Google Scholar] [CrossRef] [Green Version]
- Turnbull, A.P.; Ioannidis, S.; Krajewski, W.W.; Pinto-Fernandez, A.; Heride, C.; Martin, A.C.L.; Tonkin, L.M.; Townsend, E.C.; Buker, S.M.; Lancia, D.R.; et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 2017, 550, 481–486. [Google Scholar] [CrossRef]
- Zheng, H.; Deng, C.; Wang, M.; Jin, W.; Wu, S.; Lv, Y.; Yu, Z.; Ai, Y. Expression and polyclonal antibody preparation of UL36USP protein encoded by MDV. Chin. J. Vet. Sci. 2014, 34, 712–716. [Google Scholar]



| UL36-DUB Proteins | KM (μM) | Vmax (nM/s) | kcat (s−1) | kcat/KM (mM−1s−1) | R2 |
|---|---|---|---|---|---|
| UL36(323)WT | 1.180 ± 0.1490 | 1.693 ± 0.1323 | 0.847 ± 0.1358 | 717.797 ± 60.859 | 0.99 |
| UL36(480)WT | 1.0554 ± 0.1262 | 1.724 ± 0.1233 | 0.862 ± 0.0682 | 816.752 ± 52.949 | 0.99 |
| UL36(480)D232A | 2.444 ± 0.611 | 2.747 ± 0.5190 | 1.374 ± 0.263 | 561.99 ± 28.448 | 0.99 |
| Primer Name | Primer Sequence |
|---|---|
| U-F-Sal I | 5’-ATACGCGTCGACATGACTGACAGCACTGAC-3’ |
| U480-R-Hind III | 5’-ATTCCCAAGCTTGCTCTGGGGGGTCCAGAG-3’ |
| U323-R-Hind III | 5’- ATTCCCAAGCTTGGGGTCGAAGTCAGCGGACA -3’ |
| GST-F-Hind III | 5’-ATTCCCAAGCTTCTGGTTCCGCGTGGATCCATG-3’ |
| GST-R-Pst I | 5’-AGTGCACTGCAGCTGGTTCCGCGTGGATCCATG-3’ |
| C98A-F | 5’-TGCTTCCTCCGTGTCCgcCCTGCGTTCCTCCCTCGCTTTC-3’ |
| C98A-R | 5’- GAAAGCGAGGGAGGAACGCAGGgcGGACACGGAGGAAGCA-3’ |
| C98S-F | 5’- TGCTTCCTCCGTGTCCTcCCTGCGTTCCTCCCTCGCTTTC -3’ |
| C98S-R | 5’- GAAAGCGAGGGAGGAACGCAGGgAGGACACGGAGGAAGCA -3’ |
| D232A-F | 5’-CGACGGTATCTACATCTTCGcCCCTCACGGCCACGGCCAC-3’ |
| D232A-R | 5’-GTGGCCGTGGCCGTGAGGGgCGAAGATGTAGATACCGTCG-3’ |
| H234A-F | 5’- GTATCTACATCTTCGACCCTgcCGGCCACGGCCACATCGGC -3’ |
| H234A-R | 5’- GCCGATGTGGCCGTGGCCGgcAGGGTCGAAGATGTAGATAC -3’ |
| Ub-probe-F | 5’-ACGGAATTCCATATGTGGAGCCACCCGCAGTTCGAAAAAGGCAGC ATGCAGATCTTCGTGAAGACCC -3’ |
| Ub-probe-R | 5’- GCGATTTGCTCTTCCGCATCCGCGCAGGCGCAGTACCAGGTGCAGTGTACT -3’ |
| NEDD8-probe-F | 5’- ACGGAATTCCATATGTGGAGCCACCCGCAGTTCGA -3’ |
| NEDD8-probe-R | 5’- GCGATTTGCTCTTCCGCAACCGCGCAGGGCCAGAACCA -3’ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, J.; Ai, Y.; Zhou, H.; Lv, Y.; Wang, M.; Xu, J.; Yu, C.; Zhang, H.; Wang, M. UL36 Encoded by Marek’s Disease Virus Exhibits Linkage-Specific Deubiquitinase Activity. Int. J. Mol. Sci. 2020, 21, 1783. https://doi.org/10.3390/ijms21051783
Lin J, Ai Y, Zhou H, Lv Y, Wang M, Xu J, Yu C, Zhang H, Wang M. UL36 Encoded by Marek’s Disease Virus Exhibits Linkage-Specific Deubiquitinase Activity. International Journal of Molecular Sciences. 2020; 21(5):1783. https://doi.org/10.3390/ijms21051783
Chicago/Turabian StyleLin, Junyan, Yongxing Ai, Hongda Zhou, Yan Lv, Menghan Wang, Jiacui Xu, Cong Yu, Huanmin Zhang, and Mengyun Wang. 2020. "UL36 Encoded by Marek’s Disease Virus Exhibits Linkage-Specific Deubiquitinase Activity" International Journal of Molecular Sciences 21, no. 5: 1783. https://doi.org/10.3390/ijms21051783
APA StyleLin, J., Ai, Y., Zhou, H., Lv, Y., Wang, M., Xu, J., Yu, C., Zhang, H., & Wang, M. (2020). UL36 Encoded by Marek’s Disease Virus Exhibits Linkage-Specific Deubiquitinase Activity. International Journal of Molecular Sciences, 21(5), 1783. https://doi.org/10.3390/ijms21051783





