Abstract
Premature ovarian failure (POF) is defined as loss of ovarian function in women less than 40 years of age. The causes of POF are diverse and include environmental factors. Di-2-ethylhexyl phthalate (DEHP) is one factor that may cause POF. The ubiquitin-proteasome system maintains intracellular balance by promoting or inhibiting protein degradation. To investigate the differential expressions of deubiquitinating enzyme (DUB) genes in patients with POF, we developed two in vitro POF models by treating A2780 or OVCAR5 with DEHP. Using these models, a multiplex RT-PCR system for DUB genes was applied to identify biomarkers by comparing expression patterns and DUB mRNA levels; multiplex RT-PCR results were validated by qRT-PCR and Western blotting analyses. Observed differential expression levels of several DUB genes including USP12, COPS5, ATXN3L, USP49, and USP34 in A2780 and OVCAR5 cells at the mRNA and protein levels suggest that they should be investigated as potential biomarkers of POF.
1. Introduction
Ubiquitination is an essential process that results in the degradation of dispensable proteins via the ubiquitin-proteasomal pathway (UPP) [1]. Protein ubiquitination is mediated by a series of enzymatic actions by ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3) [1,2]. As its name implies, deubiquitination is the inverse of ubiquitination, and deubiquitinating enzymes (DUBs) play a crucial role in protein stabilization by removing ubiquitin (Ub) from conjugated target proteins by hydrolyzing the isopeptide bonds of Ub-substrates. These ubiquitin-associated systems maintain intracellular protein balance by promoting or inhibiting degradation. The human genome codes for ~100 DUBs, most of which are classified as members of seven subfamilies; that is, ubiquitin C-terminal hydrolases (UCHs), ubiquitin-specific proteases (USPs), JAB1/MPN/Mov34 metalloenzymes (JAMMs), otubain proteases (OTUs), Machado-Joseph disease proteases (MJDs), permutated papain fold peptidases of dsDNA viruses and eukaryote monocyte (PPPDEs), and motif interacting with Ub-containing novel DUB family (MINDY) [3,4,5]. The biological functions of DUBs are not completely understood yet.
Premature ovarian failure (POF) involves loss of ovarian function in women less than 40 years old, which means ovaries do not produce normal amounts of estrogen and ovulation occurs sporadically and results in infertility [6]. POF can be present as primary amenorrhea with no menarche or as secondary amenorrhea with no menstrual period for 6 months or no menstrual period more than three times after the previous menstrual period [6]. The cause of POF is unclear, but it is known that both genetic and environmental factors are involved. Known causes include chromosomal abnormalities, autoimmune diseases, metabolic disorders, enzyme deficiencies, ovarian damage, and genetic diseases [7,8].
Environmental diseases are health disorders caused by physical, chemical, or biological environmental factors, often because of exposure to toxic environmental factors associated with lifestyle and occupational activities. Furthermore, it has been established that POF can be caused by toxic environmental factors [9].
Phthalates are one of the toxic environmental factors that may cause POF [10]. Phthalate esters are alkyl diesters of phthalic acid and are widely used as plasticizers, and are known to adversely affect two essential ovarian processes; that is, folliculogenesis and steroidogenesis [10]. Di-2-ethylhexyl phthalate (DEHP) is a member of the class of phthalates, and the relationship between DEHP and POF has been investigated, showing that the mechanistic link between the two has been elucidated [11,12,13]. One of the previous studies demonstrated that DEHP disrupts estrous cyclicity and PI3K signaling [14].
In order to investigate the differential expression of DUB genes and identify potential biomarkers of POF, we designed a multiplex PCR primer library of DUB genes [15] and treated two in vitro POF models, that is, A2780 and OVCAR5 ovarian cell lines, with DEHP, and confirmed results by qRT-PCR and Western blotting analyses [16]. Our results showed the differential expressions of mRNA and protein for several DUB genes including Ataxin-3-like protein (ATXN3L), Ubiquitin specific peptidase 12 (USP12), Ubiquitin specific peptidase 49 (USP49), COP9 signalosome subunit 5 (COPS5), and Ubiquitin specific peptidase 34 (USP34) in A2780 and OVCAR5 cells.
2. Results
2.1. Differential Expressions of DUBs by DEHP in Ovarian Cells
To investigate DUB genes associated with DEHP, ovarian cell lines were treated with different concentrations of DEHP (Table 1) [16]. Multiplex RT-PCR was used to amplify DNA bands of DUB genes from cDNA synthesized from mRNA of A2780 and OVCAR5 cells treated with DEHP. When the multiplex RT-PCR was performed using all 12 sets of primers in the multiplex RT-PCR library, DNA amplification of DUB genes was detected (Figure 1 and Figure 2). Independent experiments were conducted at least three times. The results showed that the mRNA levels of USP12, COPS5, ATXN3L, and USP49 were increased in A2780 cells (Figure 1 and Figure S1), and that the mRNA level of USP49 was increased and that of USP34 was decreased in OVCAR5 cells (Figure 2 and Figure S2) by DEHP exposure in a dose-dependent manner.

Table 1.
Concentration of di-2-ethylhexyl phthalate (DEHP) treatment.
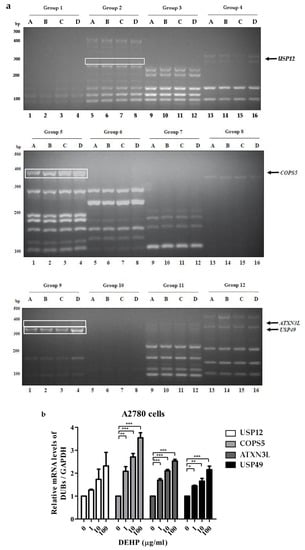
Figure 1.
Deubiquitinating enzyme (DUB) gene screening in A2780 cells treated with DEHP using the multiplex RT-PCR primer library. (a) DUB genes were amplified by multiplex RT-PCR using 12 sets of primer library. (b) mRNA levels of USP12, COPS5, ATXN3L, and USP49 were normalized with respect to GAPDH. The significances of differences were determined by one-way of variance. p-values are presented as * p < 0.05, ** p < 0.01, or *** p < 0.001.
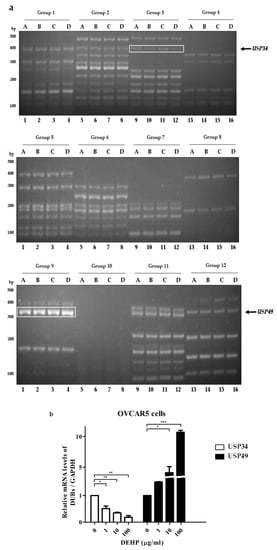
Figure 2.
DUB gene screening in OVCAR5 cells treated with DEHP using the multiplex RT-PCR primer library. (a) DUB genes were amplified by multiplex RT-PCR using 12 sets of primer library. (b) mRNA levels of USP34 and USP49 were normalized with respect to GAPDH. The significances of differences were determined by one-way of variance. p-value are presented as * p < 0.05, ** p < 0.01, or *** p < 0.001.
2.2. DEHP Exposure Influenced the mRNA Levels of Several DUB Genes in Ovarian Cells
To confirm the result of multiplex RT-PCR, RT-PCR and qRT-PCR were performed for selected DUB genes. The mRNA levels of USP12, ATXN3L, COPS5, and USP49 were increased in A2780 cells by DEHP (Figure 3a and Figure S3a), and in OVCAR5 cells, the mRNA level of USP34 was decreased and that of USP49 was increased (Figure 3b and Figure S3b). Furthermore, these results were confirmed by qRT-PCR (Figure 3c and Table S1).
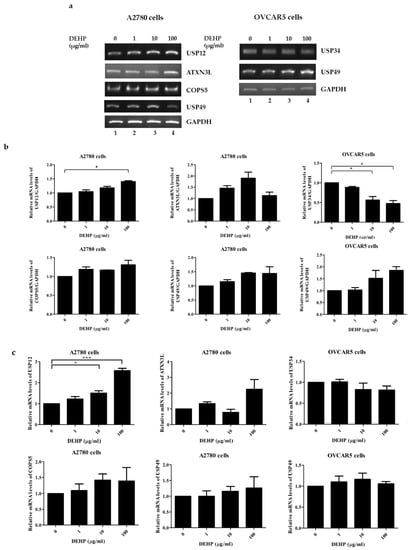
Figure 3.
The mRNA expressions of DUB genes in DEHP-treated cells. (a) mRNA levels were assessed by RT-PCR in A2780 and OVCAR5 cells treated with different concentrations of DEHP. (b) The mRNA levels of DUB genes were normalized with respect to GAPDH. (c) The mRNA levels of DUB genes were examined by qRT-PCR in A2780 and OVCAR5 cells treated with different concentrations of DEHP. The significances of differences were determined by one-way of variance. p-values are presented as * p < 0.05, or *** p < 0.001.
2.3. Up-Regulation of USP49 Protein by DEHP in Ovarian Cells
As the mRNA level of USP49 was found to be upregulated by DEHP in ovarian cells (Figure 3), we examined the effects of DEHP exposure on the protein level of USP49 in both cell lines by Western blotting analysis. As was expected, the protein levels of USP49 were upregulated in both cell lines by DEHP (Figure 4 and Figure S4).
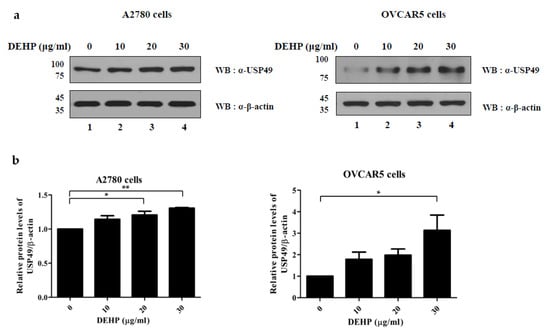
Figure 4.
Expression of USP49 in the DEHP-treated cells. (a) Protein levels of USP49 in A2780 (left) and OVCAR5 (right) cells treated with DEHP were determined by Western blotting. (b) USP49 band intensities were normalized versus β-actin. The significances of differences were determined by one-way of variance. p-values are presented as *p < 0.05, ** p < 0.01.
3. Discussion
Exposure to toxic environmental factors is steadily increasing because they continue to be used in daily life, and as a result, the incidences of the diseases they cause are also increasing. On the other hand, progress is being made to develop products using less toxic materials. POF is a disease associated with toxic environmental factors in addition to genetic factors, and its incidence is also increasing. Therefore, we investigated mechanisms involving the ubiquitin-proteasome system initiated by exposure to DEHP in ovarian cell lines by using multiplex RT-PCR to screen for DUB genes differentially expressed after DEHP exposure.
ATXN3L is a Machado–Joseph disease protease (MJD), which contains the Josephin domain (JD), and JD containing proteins are known to have deubiquitination activity [17]. Furthermore, it has been shown that ATXN3L has greater deubiquitination activity than JD containing proteins [18]. Furthermore, ATXN3L is a DUB of Krüppel-like factor 5 (KLF5) and promotes breast cell proliferation, survival, and tumorigenesis [19]. In the present study, multiplex RT-PCR results showed that ATXN3L expression was increased in A2780 cells after DEHP exposure, suggesting that this would promote POF by promoting oocyte maturation, due to its cell proliferation capability [18].
USP12 is a ubiquitin-specific protease (USP) and a component of a complex containing USP1-associated factor 1 (UAF1) and WDR20. It also regulates androgen receptor (AR) in prostate cancer [20,21,22]. USP12 has been shown to regulate the ubiquitination level of histone H2A and H2B [23] and to deubiquitinate AR and MDM2, which regulate the p53-MDM2-AR-AKT signaling network. In addition, USP12 promotes cell survival, proliferation, tumorigenesis, and cell cycle progression by upregulating BMI-1, c-Myc, and cyclin D2 [24,25,26,27]. Our multiplex RT-PCR results showed that USP12 expression was increased by DEHP in A2780 cells, and thus, the above-mentioned findings of its influence on cell cycle progression and proliferation suggest that it may induce POF by influencing oocyte maturation.
On the other hand, USP49 is also a ubiquitin-specific protease and regulates the initial pre-mRNA splicing by deubiquitinating histone H2B [28]. USP49 is also known to regulate tumorigenesis by regulating the involvement of p53 in DNA damage response and by controlling FK506-binding protein 51 (FKBP51) through the Akt signaling pathway [29,30]. Multiplex RT-PCR and Western blot analysis revealed that USP49 was upregulated by DEHP in A2780 and OVCAR5 cells. These findings suggest that USP49 upregulation by DEHP induces cell death via p53 regulation, which subsequently leads to POF due to follicle depletion.
COPS5 is a JAB1/MPN/Mov34 metalloenzyme. Its expression increases during oocyte maturation and it is required for oocyte meiosis. COPS5 also regulates the maturation-promoting factor (MPF) activity [31]. Furthermore, progesterone receptor (PGR), which regulates ovulation, lies downstream of COPS5 [32,33]. COPS5 has been reported to control proliferation and inhibits the expression of p27 in serous ovarian cancer [34]. Our multiplex RT-PCR results showed that COPS5 expression was increased in A2780 cells exposed to DEHP. Based on the above, COPS5 upregulation would be expected to promote POF by promoting oocyte maturation.
USP34 is a ubiquitin-specific protease that controls the stability of Axin and positively regulates Wnt/β-catenin signaling [35]. USP34 is required for DNA damage repair and regulates E3 ubiquitin-protein ligase RNF168, and recruits the DNA damage repair factors by ubiquitinating DNA double-strand breaks (DSBs) [36]. It has also been shown to negatively regulate T cell receptor (TCR) by inhibiting nuclear factor-κB (NK-κB) activation [37], and to be associated with polycystic ovary syndrome (PCOS), one of the most common endocrine disorders among women [38]. In the present study, multiplex RT-PCR results showed that USP34 was downregulated by DEHP in OVCAR5 cells, which suggests a diminished ability to repair DNA damage and increase the risk of POF due to reductions in follicle numbers.
In summary, this study shows that DEHP changes the mRNA and protein levels of several DUB genes; that are, USP12, ATXN3L, COPS5, USP49, and USP34 in the A2780 and OVCAR5 cells, and that it could potentially participate in the pathogenesis of POF. We suggest an in vivo study to be undertaken in a rodent model to further investigate the link between the DEHP-induced differential expressions of DUB genes and the pathogenesis of POF and to identify potential therapeutic targets.
4. Materials and Methods
4.1. Cell Culture
Human ovarian cancer cell lines, A2780 cells, were grown in the Roswell Park Memorial Institute-1640 medium (11875-093, RPMI-1640, Gibco BRL, Rockville, MD, USA) and OVCAR5 cells were grown in Dulbecco’s modified Eagle’s medium (11965084, DMEM, Gibco BRL, Rockville, MD, USA), containing 10% fetal bovine serum (26140079, FBS, Gibco, Grand Island, NY, USA), and 1% antibiotic-antimycotic reagent (15240062, Gibco, Tewksbury, MA, USA) at 37 °C in 5% CO2 atmosphere.
4.2. RNA Extraction and cDNA Synthesis
For RNA extraction, cells at 80–90% confluence in 60 mm dishes were lysed with TRIzol reagent (15596018, Ambion, Carlsbad, CA, USA). cDNA was prepared by reverse transcription with 1 μg of total RNA using the LaboPass cDNA Synthesis Kit (CMRTK001, Cosmogenetech Inc, Seoul, Korea) according to the manufacturer’s protocols.
4.3. Multiplex RT-PCR and qRT-PCR
For multiplex RT-PCR, we used 2× Multiplex PCR Smart Mix (SMP01-M25h, Solgent, Daejeon, Korea), 200 ng cDNA samples, and GAPDH as a control. PCR products were separated by 2% agarose gel electrophoresis and gels were stained with RedSafe DNA Stain (21141, Chembio, Medford, NY, USA) to visualize amplicons and confirmed the amplification of cDNA bands of expected sizes. Primers for the multiplex RT-PCR library (Table 2) were used to amplify the DUB-specific sequences (Ubiprotein Corp, Seongnam, Korea). mRNA expression levels were normalized versus GAPDH and analyzed using Image J v1.4.3.67 (National Institutes of Health, Bethesda, MD, USA). Expression levels of DUB genes in naïve A2780 and OVCAR5 cells were considered normal. Primers for qRT-PCR (Table 3) were used to compare the quantitative expression of DUB genes identified through multiplex RT-PCR (Ubiprotein Corp, Seongnam, Korea). qRT-PCR was performed using the StepOne Real-Time PCR System (Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer’s instructions, and cDNA was amplified by reverse transcription using SYBR-Green PCR Master Mix (4309155, Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Cycle threshold values of DUB genes were normalized to the endogenous reference gene GAPDH. The expression DUB genes levels were calculated using the 2-∆∆CT method.

Table 2.
Multiplex PCR primer set.

Table 3.
RT-PCR and qRT-PCR primer set.
4.4. Western Blot Analysis
A2780 and OVCAR5 cells treated with DEHP were lysed using a lysis buffer (50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 1 mM EDTA, 10% Glycerol, 1% Triton X-100). After resuspension of cells, samples were incubated for 20 min on ice and centrifuged at 13,000 rpm for 15 min. Then, cell lysates were mixed with 2X SDS loading buffer, boiled, and loaded into the SDS-PAGE gels. Separated proteins were transferred to polyvinylidene fluoride (PVDF) membranes (IPVH00010, Millipore, Billerica, MA, USA), which were incubated overnight with primary antibodies at 4 °C and then with secondary antibodies for 1 h at room temperature. Blots were visualized using the ECL reagent solution (LF-QC0101, Young In Frontier, Seoul, Korea).
4.5. Antibodies
Rabbit anti-USP49 polyclonal antibody (18066-1-AP) was purchased from Proteintech (Proteintech Group, Chicago, IL, USA) and mouse anti-β-actin monoclonal antibody (sc-47778) was purchased from Santa Cruz (Santa Cruz Biotechnology, CA, USA).
4.6. Statistical Analysis
Each experiment was performed at least three times. Densitometric analysis was performed for bands from all RT-PCR and Western blot results using a computer program Image J software (v1.4.3.67, National Institutes of Health, Bethesda, MD, USA) and the t-test using GraphPad Prism version 5 (GraphPad Software, La Jolla, CA, USA). ANOVA was performed by two-way analysis of variance. p-values are presented as * p < 0.05, ** p < 0.01, or *** p < 0.001 as indicated.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/5/1755/s1. Figure S1. Raw data of repeated multiplex RT-PCR used in this study with A2780 cells, Figure S2. Raw data of repeated multiplex RT-PCR used in this study with OVCAR5 cells, Figure S3. Raw data of repeated RT-PCR used in this study with ovarian cells, Figure S4. Raw data of repeated Western blot used in this study with ovarian cells, Table S1. qRT-PCR data used in this study.
Author Contributions
D.-H.L. designed research, performed the experiments, and wrote the manuscript. J.-H.P. and J.C. helped with performed the experiments and analyzed the data. K.-J.L. and B.-S.Y. helped interpretation of the study. K.-H.B. drafted and critically edited the manuscript. All authors read and provided critical feedback and approved the final version of this manuscript.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. NRF-2019R1A6A1A03032888).
Acknowledgments
We would like to thank the members of Baek’s laboratory for their critical comments on the manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Kwon, S.K.; Saindane, M.; Baek, K.H. p53 stability is regulated by diverse deubiquitinating enzymes. Biochim. Biophys Acta Rev. Cancer 2017, 1868, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef] [PubMed]
- Woelk, T.; Sigismund, S.; Penengo, L.; Polo, S. The ubiquitination code: A signalling problem. Cell Div. 2007, 2, 11. [Google Scholar] [CrossRef]
- Todi, S.V.; Paulson, H.L. Balancing act: Deubiquitinating enzymes in the nervous system. Trends Neurosci. 2011, 34, 370–382. [Google Scholar] [CrossRef] [PubMed]
- Abdul Rehman, S.A.; Kristariyanto, Y.A.; Choi, S.Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol. Cell 2016, 63, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Beck-Peccoz, P.; Persani, L. Premature ovarian failure. Orphanet J. Rare. Dis. 2006, 1, 9. [Google Scholar] [CrossRef]
- Jankowska, K. Premature ovarian failure. Prz Menopauzalny 2017, 16, 51–56. [Google Scholar] [CrossRef]
- Ayesha, V.J.; Goswami, D. Premature Ovarian Failure: An Association with Autoimmune Diseases. J. Clin. Diagn. Res. 2016, 10, QC10–QC12. [Google Scholar] [CrossRef]
- Vabre, P.; Gatimel, N.; Moreau, J.; Gayrard, V.; Picard-Hagen, N.; Parinaud, J.; Leandri, R.D. Environmental pollutants, a possible etiology for premature ovarian insufficiency: A narrative review of animal and human data. Environ. Health 2017, 16, 37. [Google Scholar] [CrossRef]
- Tran, D.N.; Jung, E.M.; Yoo, Y.M.; Ahn, C.; Kang, H.Y.; Choi, K.C.; Hyun, S.H.; Dang, V.H.; Pham, T.N.; Jeung, E.B. Depletion of follicles accelerated by combined exposure to phthalates and 4-vinylcyclohexene diepoxide, leading to premature ovarian failure in rats. Reprod. Toxicol. 2018, 80, 60–67. [Google Scholar] [CrossRef]
- Yin, J.; Liu, R.; Jian, Z.; Yang, D.; Pu, Y.; Yin, L.; Wang, D. Di (2-ethylhexyl) phthalate-induced reproductive toxicity involved in dna damage-dependent oocyte apoptosis and oxidative stress in Caenorhabditis elegans. Ecotoxicol. Environ. Saf. 2018, 163, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.C.; Lai, F.N.; Li, L.; Sun, X.F.; Cheng, S.F.; Ge, W.; Wang, Y.F.; Li, L.; Zhang, X.F.; De Felici, M.; et al. Di (2-ethylhexyl) phthalate exposure impairs meiotic progression and DNA damage repair in fetal mouse oocytes in vitro. Cell Death Dis. 2017, 8, e2966. [Google Scholar] [CrossRef]
- Absalan, F.; Saremy, S.; Mansori, E.; Taheri Moghadam, M.; Eftekhari Moghadam, A.R.; Ghanavati, R. Effects of Mono-(2-Ethylhexyl) Phthalate and Di-(2-Ethylhexyl) Phthalate Administrations on Oocyte Meiotic Maturation, Apoptosis and Gene Quantification in Mouse Model. Cell J. 2017, 18, 503–513. [Google Scholar] [PubMed]
- Hannon, P.R.; Peretz, J.; Flaws, J.A. Daily exposure to Di(2-ethylhexyl) phthalate alters estrous cyclicity and accelerates primordial follicle recruitment potentially via dysregulation of the phosphatidylinositol 3-kinase signaling pathway in adult mice. Biol. Reprod. 2014, 90, 136. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kwon, S.K.; Lee, S.Y.; Baek, K.H. Ubiquitin-specific peptidase 5 and ovarian tumor deubiquitinase 6A are differentially expressed in p53+/+ and p53-/- HCT116 cells. Int. J. Oncol. 2018. [Google Scholar] [CrossRef]
- Hannon, P.R.; Brannick, K.E.; Wang, W.; Gupta, R.K.; Flaws, J.A. Di(2-ethylhexyl) phthalate inhibits antral follicle growth, induces atresia, and inhibits steroid hormone production in cultured mouse antral follicles. Toxicol. Appl. Pharmacol. 2015, 284, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Tzvetkov, N.; Breuer, P. Josephin domain-containing proteins from a variety of species are active de-ubiquitination enzymes. Biol. Chem. 2007, 388, 973–978. [Google Scholar] [CrossRef]
- Weeks, S.D.; Grasty, K.C.; Hernandez-Cuebas, L.; Loll, P.J. Crystal structure of a Josephin-ubiquitin complex: Evolutionary restraints on ataxin-3 deubiquitinating activity. J. Biol. Chem. 2011, 286, 4555–4565. [Google Scholar] [CrossRef]
- Ge, F.; Chen, W.; Qin, J.; Zhou, Z.; Liu, R.; Liu, L.; Tan, J.; Zou, T.; Li, H.; Ren, G.; et al. Ataxin-3 like (ATXN3L), a member of the Josephin family of deubiquitinating enzymes, promotes breast cancer proliferation by deubiquitinating Kruppel-like factor 5 (KLF5). Oncotarget 2015, 6, 21369–21378. [Google Scholar] [CrossRef]
- Cohn, M.A.; Kee, Y.; Haas, W.; Gygi, S.P.; D’Andrea, A.D. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J. Biol. Chem. 2009, 284, 5343–5351. [Google Scholar] [CrossRef]
- Kee, Y.; Yang, K.; Cohn, M.A.; Haas, W.; Gygi, S.P.; D’Andrea, A.D. WDR20 regulates activity of the USP12 x UAF1 deubiquitinating enzyme complex. J. Biol. Chem. 2010, 285, 11252–11257. [Google Scholar] [CrossRef] [PubMed]
- McClurg, U.L.; Harle, V.J.; Nabbi, A.; Batalha-Pereira, A.; Walker, S.; Coffey, K.; Gaughan, L.; McCracken, S.R.; Robson, C.N. Ubiquitin-specific protease 12 interacting partners Uaf-1 and WDR20 are potential therapeutic targets in prostate cancer. Oncotarget 2015, 6, 37724–37736. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.Y.; Jones, A.; Yang, C.; Zhai, L.; Smith, A.D.t.; Zhang, Z.; Chandrasekharan, M.B.; Sun, Z.W.; Renfrow, M.B.; Wang, Y.; et al. Regulation of histone H2A and H2B deubiquitination and Xenopus development by USP12 and USP46. J. Biol. Chem. 2011, 286, 7190–7201. [Google Scholar] [CrossRef] [PubMed]
- Burska, U.L.; Harle, V.J.; Coffey, K.; Darby, S.; Ramsey, H.; O’Neill, D.; Logan, I.R.; Gaughan, L.; Robson, C.N. Deubiquitinating enzyme Usp12 is a novel co-activator of the androgen receptor. J. Biol. Chem. 2013, 288, 32641–32650. [Google Scholar] [CrossRef]
- McClurg, U.L.; Chit, N.; Azizyan, M.; Edwards, J.; Nabbi, A.; Riabowol, K.T.; Nakjang, S.; McCracken, S.R.; Robson, C.N. Molecular mechanism of the TP53-MDM2-AR-AKT signalling network regulation by USP12. Oncogene 2018, 37, 4679–4691. [Google Scholar] [CrossRef]
- McClurg, U.L.; Azizyan, M.; Dransfield, D.T.; Namdev, N.; Chit, N.; Nakjang, S.; Robson, C.N. The novel anti-androgen candidate galeterone targets deubiquitinating enzymes, USP12 and USP46, to control prostate cancer growth and survival. Oncotarget 2018, 9, 24992–25007. [Google Scholar] [CrossRef]
- Tang, L.J.; Li, Y.; Liu, Y.L.; Wang, J.M.; Liu, D.W.; Tian, Q.B. USP12 regulates cell cycle progression by involving c-Myc, cyclin D2 and BMI-1. Gene 2016, 578, 92–99. [Google Scholar] [CrossRef]
- Zhang, Z.; Jones, A.; Joo, H.Y.; Zhou, D.; Cao, Y.; Chen, S.; Erdjument-Bromage, H.; Renfrow, M.; He, H.; Tempst, P.; et al. USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes Dev. 2013, 27, 1581–1595. [Google Scholar] [CrossRef]
- Luo, K.; Li, Y.; Yin, Y.; Li, L.; Wu, C.; Chen, Y.; Nowsheen, S.; Hu, Q.; Zhang, L.; Lou, Z.; et al. USP49 negatively regulates tumorigenesis and chemoresistance through FKBP51-AKT signaling. EMBO J. 2017, 36, 1434–1446. [Google Scholar] [CrossRef]
- Tu, R.; Kang, W.; Yang, X.; Zhang, Q.; Xie, X.; Liu, W.; Zhang, J.; Zhang, X.D.; Wang, H.; Du, R.L. USP49 participates in the DNA damage response by forming a positive feedback loop with p53. Cell Death Dis. 2018, 9, 553. [Google Scholar] [CrossRef]
- Kim, E.; Yoon, S.J.; Kim, E.Y.; Kim, Y.; Lee, H.S.; Kim, K.H.; Lee, K.A. Function of COP9 signalosome in regulation of mouse oocytes meiosis by regulating MPF activity and securing degradation. PLoS ONE 2011, 6, e25870. [Google Scholar] [CrossRef] [PubMed]
- Robker, R.L.; Akison, L.K.; Russell, D.L. Control of oocyte release by progesterone receptor-regulated gene expression. Nucl. Recept. Signal. 2009, 7, e012. [Google Scholar] [CrossRef] [PubMed]
- Khan, D.R.; Landry, D.A.; Fournier, E.; Vigneault, C.; Blondin, P.; Sirard, M.A. Transcriptome meta-analysis of three follicular compartments and its correlation with ovarian follicle maturity and oocyte developmental competence in cows. Physiol. Genomics 2016, 48, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhong, A.; Sun, J.; Chen, M.; Xie, S.; Zheng, H.; Wang, Y.; Yu, Y.; Guo, L.; Lu, R. COPS5 inhibition arrests the proliferation and growth of serous ovarian cancer cells via the elevation of p27 level. Biochem. Biophys Res. Commun. 2017, 493, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Lui, T.T.; Lacroix, C.; Ahmed, S.M.; Goldenberg, S.J.; Leach, C.A.; Daulat, A.M.; Angers, S. The ubiquitin-specific protease USP34 regulates axin stability and Wnt/beta-catenin signaling. Mol. Cell Biol. 2011, 31, 2053–2065. [Google Scholar] [CrossRef] [PubMed]
- Sy, S.M.; Jiang, J.; O, W.S.; Deng, Y.; Huen, M.S. The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Res. 2013, 41, 8572–8580. [Google Scholar] [CrossRef] [PubMed]
- Poalas, K.; Hatchi, E.M.; Cordeiro, N.; Dubois, S.M.; Leclair, H.M.; Leveau, C.; Alexia, C.; Gavard, J.; Vazquez, A.; Bidere, N. Negative regulation of NF-kappaB signaling in T lymphocytes by the ubiquitin-specific protease USP34. Cell Commun. Signal. 2013, 11, 25. [Google Scholar] [CrossRef]
- Zhao, S.; Tian, Y.; Zhang, W.; Xing, X.; Li, T.; Liu, H.; Huang, T.; Ning, Y.; Zhao, H.; Chen, Z.J. An association study between USP34 and polycystic ovary syndrome. J. Ovarian. Res. 2015, 8, 30. [Google Scholar] [CrossRef][Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).