Abstract
Altered gene expression is the primary molecular mechanism responsible for the pathological processes of human diseases, including cancer. MicroRNAs (miRNAs) are virtually involved at the post-transcriptional level and bind to 3′ UTR of their target messenger RNA (mRNA) to suppress expression. Dysfunction of miRNAs disturbs expression of oncogenic or tumor-suppressive target genes, which is implicated in cancer pathogenesis. As such, a large number of miRNAs have been found to be downregulated or upregulated in human cancers and to function as oncomiRs or oncosuppressor miRs. Notably, the molecular mechanism underlying the dysregulation of miRNA expression in cancer has been recently uncovered. The genetic deletion or amplification and epigenetic methylation of miRNA genomic loci and the transcription factor-mediated regulation of primary miRNA often alter the landscape of miRNA expression in cancer. Dysregulation of the multiple processing steps in mature miRNA biogenesis can also cause alterations in miRNA expression in cancer. Detailed knowledge of the regulatory mechanism of miRNAs in cancer is essential for understanding its physiological role and the implications of cancer-associated dysfunction and dysregulation. In this review, we elucidate how miRNA expression is deregulated in cancer, paying particular attention to the cancer-associated transcriptional and post-transcriptional factors that execute miRNA programs.
1. Introduction
Normal cells can be progressively developed to the neoplastic stage by acquiring multistep processes of tumorigenesis, and they become malignant, which, in turn, leads to initiate cancer. The study of molecular mechanisms of the initiation and progression of cancer has become a core of cancer research, which can provide a scientific basis for developing the prevention and treatment strategies of cancer patients. Alterations in gene expressions associated with cancer are caused by the dysfunctions of various types of regulators, among which, microRNAs have received great attention in the past decades. MicroRNAs (miRNAs) are ~22 nt small noncoding RNAs that are known to play an important role in the post-transcriptional regulation of messenger RNA (mRNA). miRNAs are typically generated from the nascent primary miRNA (pri-miRNA) transcripts through two sequential cleavage events. The pri-miRNA is initially processed by DROSHA in the nucleus, which releases a hairpin-shaped precursor (pre-miRNA). Pre-miRNA is exported from the nucleus to cytoplasm by exportin 5 (XPO5) and cleaved by DICER. The resulting small RNA duplex is loaded onto the Argonaute (AGO) protein, which preferentially retains only one strand of mature miRNA by removing the other strand [1]. The miRNA-loaded AGO associates with other cofactors, including GW182 (also known as TNRC6A), and constitutes the effector complex called the RNA-induced silencing complex (RISC) [2]. The miRISC (miRNA-induced silencing complex) induces the decay of mRNA and translational suppression through the interaction with the complementary sequences in the 3′-untranslated region (3′-UTR) of target gene mRNA [3,4,5]. The miRNAs target a majority of mRNAs, enabling them to have important regulatory roles in diverse physiological and developmental processes [6]. In particular, miRNA-mediated gene expression control is critical for the cellular response to the environmental stresses, such as starvation, hypoxia, oxidative stress, and DNA damage, thereby being implicated in human diseases such as cancer. Indeed, numerous miRNAs can function as oncogenes (referred to as “oncomiRs”) or tumor suppressors (“oncosuppressor miRs”), and dysregulation of miRNA expression is closely associated with cancer initiation, progression, and metastasis [7,8]. In this review, we summarize how miRNA expression is deregulated in cancer, paying particular attention to the cancer-associated transcriptional and post-transcriptional programs, including transcriptional control, epigenetic methylation of miRNA loci, and dysregulation of the mature miRNA biogenesis pathway. We will further discuss the major genetic and epigenetic mechanisms involved in upregulating or downregulating miRNA expression, in an attempt to elucidate which elements are key to this process in cancer pathogenesis.
2. miRNA Deregulation in Cancer
In the past decades, miRNAs have been demonstrated to be extensively deregulated in human cancers, highlighting their important role in tumor onset, growth, and metastasis. Lu et al. demonstrated the profiling of 217 mammalian miRNAs from normal and human cancer samples and found that miRNA expression is globally suppressed in tumor cells compared to normal cells [9]. In addition to the global downregulation of miRNA expression, Volinia et al. presented the differentially expressed miRNAs in 540 solid tumor samples, indicating that specific alterations of individual miRNA expression were also apparent in tumors [10], since miRNA expression has been deregulated during cancer progression, creating an explicit expression pattern; for instance, the level of miR-21 expression is higher in early stage of diffuse large B-cell lymphoma (DLBCL) than in later stages [11]. Interestingly, some miRNAs are packed into a vesicle-like structure called exosomes for secretion, which can circulate throughout the body and can act differentially in a tissue-dependent manner. These include miR-21, the miR-200 family, and the miR-17∼92 cluster, and these exosomal miRNAs have been proved to be functionally implicated and clinically relevant in cancer [12]. The tumor microenvironment (TME) modulation accounts for the patient heterogeneity of treatment responses [13]. Therefore, miRNA-based exosomes represent one of the dynamic facets of the tumor microenvironment, and exosomal miRNAs in the TME may profoundly impact on tumor progression and therapeutic efficacy.
The role of specific miRNAs in cancer was firstly appreciated by Calin et al. [14]—the deletion of miR-15 and miR-16 genomic loci in the majority of samples from chronic lymphocytic leukemia patients. Cimmino et al. further confirmed that miR-15 and miR-16 induce apoptosis by targeting B cell lymphoma 2 (BCL2) in leukemia [15]. Since then, a large number of studies have reported altered expressions of miRNA in diverse types of cancer, and the implication of those miRNAs in cancer has been investigated by loss-of-function and gain-of-function experiments in animal models and human cancer cell lines. For example, let-7 is downregulated in breast, colon, and lung cancers [16] and proven as an oncosuppressor miR to prevent tumor development by repressing RAS or MYC [17,18]. miR-34a that belongs to a p53-responsive miR-34 family was also observed to be reduced in several types of cancers. The expression level of miRNA-331-3p has been reduced in patients of nasopharyngeal carcinoma, and its overexpression induces apoptosis, resulting in the suppression of cell proliferation [19]. In contrast to those tumor-suppressive miRNAs, other miRNAs are known to be upregulated and have oncogenic roles. These include miR-21 in diverse solid tumors and hematological malignancies [20,21,22], miR-155 [23,24,25] and the miR-17~19b cluster [26,27] in B-cell lymphoma and breast cancer, and miR-106b-5p in lung cancer [28] and metastatic breast carcinoma [29].
Since miRNA expression and function are regulated upon the cellular stress [30], the limited oxygen supply, hypoxia, in the TME can affect the production and function of mature miRNAs. Epidermal growth factor receptor (EGFR) signaling is activated by the hypoxic condition to promote growth and oncogenesis [31]. Interestingly, protein argonaute 2 (AGO2) has been identified to interact with EGFR in serum-starved conditions, and deregulated AGO2 correlates with poor survival in breast cancer patients [32].
Since miRNAs are required to maintain the proper regulation of cellular processes, such as cell proliferation, cell metabolism, and protein synthesis, in normal physiological conditions, their deregulation leads to the abnormal growth and biosynthesis of cells that contribute to tumor development, progression, and metastasis (Table 1). Emerging evidence showing not only the genetic and epigenetic dysregulations of miRNA biogenesis machineries but also the regulatory mechanisms of miRNAs has demonstrated the importance of regulatory mechanisms of miRNA expression in cancer.

Table 1.
Dysregulation of miRNA transcription in cancer.
3. Dysregulation of miRNA Transcription in Cancer
Alterations of miRNA expression in cancer can arise from genomic variations of miRNA genomic loci. For example, the genomic locus of the miR-15/miR-16 cluster is deleted at high frequency in B-cell chronic lymphocytic leukemia (CLL) [72,73]. miR-146a is also repressed as a consequence of the deletion of chromosome 5q in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) [74]. Besides the genomic variation, miRNA expression is also controlled at transcriptional level, which is mediated by transcription factors and the epigenetic control of DNA methylation.
3.1. Modulation of miRNA Expression by Transcription Factors in Cancer
Several studies have provided compelling evidence that alterations in transcriptional activators or repressors cause abnormal pri-miRNA transcription in cancer. For instance, expression of the miR-34 family genes (miR-34a, miR-34b, and miR-34c) are controlled by the transcription factor p53 [26], reflecting the importance of the p53 functional status in predicting miR-34 expression in human cancers. Upon DNA damage and oncogenic stress, p53 is activated and regulates miR-34 transcription, which impacts cell cycle arrest, apoptosis, and senescence [75]. miR-145 is also transcriptionally activated by upregulated p53 to induce apoptosis [33,34,76]. In contrast, the miR-143/145 cluster is repressed by oncogenic RAS signaling that induces tumorigenesis. RAS-responsive element-binding protein 1 (RREB1) leads to the transcriptional repression of the miR-143/145 cluster, and in turn, miR-143/145 suppresses expression of RREB1, forming a tumor-promoting feedback circuit of RAS signaling [35]. In addition to p53 and RREB1, miR-145 is regulated by other transcription factors, including CCAAT/enhancer-binding protein beta (C/EBPβ), beta-catenin/T cell factor 4 (TCF4), and forkhead transcription factors FOXO1 and FOXO3 in human cancers [36,37]. The transcriptional co-factor meningioma 1 (MN1) gene is highly expressed, and its upregulation is inversely correlated with miR-20a and miR-181b transcripts in acute myeloid leukemia (AML) patients [38]. The c-Myc oncogenic transcription factor (MYC) transactivates expression of the miR-17~92 cluster (also known as oncomiR-1), and MYC-activated miR-17~92 promotes cancer progression by controlling expressions of E2F1, connective tissue growth factor (CTGF), thrombospondin 1 (THBS1), and phosphatase and tensin homolog (PTEN) in multiple cancers [39,40,41,42,43,77]. On the contrary, MYC suppresses the expression of genes of oncosuppressor miRs, such as miR-26, miR-29, miR-30, and let-7 family members in lymphoma [44,45,46]. The hypoxia-inducible factor-alpha (HIF1α) transcription factor induces the miR-210 and miR-155 transcription in hypoxia [48,49]. In addition, the zinc-finger E-box-binding homeobox (ZEB) transcription factors, ZEB1 and ZEB2, which are known as key activators to promote the epithelial-mesenchymal transition (EMT), repress transcription of the miR-200 family gene [50]. It is also noted that miR-200c has been identified as a transcriptional target of MYC in nasopharyngeal carcinoma [47]. Activation protein 1 (AP1), Ets family transcription factor PU.1, C/EBPα, nuclear factor I (NFI), and signal transducer and activator of transcription 3 (STAT3) activate miR-21 transcription by binding to the defined miR-21 promoter [54,55]. Therefore, targeting or activating specific transcription factors responsible for the abundance of oncomiRs or oncosuppressor miRs may be promising and innovative approaches to cancer treatment.
Nuclear receptors (NRs) are ligand-activated transcription factors regulating gene expression by binding to the specific DNA sequences or regulatory regions of target genes. Since it has been reported that the NR superfamily contains 48 human members, including the hormone receptors: estrogen receptor (ER), progesterone receptor (PR), androgen receptor (AR), glucocorticoid receptor (GR), and mineralocorticoid receptor (MR) [78], several studies have shown that NRs, especially ER and AR, not only indirectly change miRNA abundance through diverse signaling pathways but also directly regulate the transcriptional activity of miRNAs in cancer. ER binds to the promoter region of the miR-221/222 gene and recruits the NCoR/SMRT co-repressor complex to suppress miR-221/222 expression in breast cancer [51]. ER also inhibits transcription of the miR-515, leading to increased levels of oncogenic sphingosine kinase 1 (SK1) [52]. Like estrogen/ER, androgen/AR can regulate the transcriptional output from the miRNA loci. Indeed, numerous miRNAs have been identified to be directly regulated by androgen/AR during prostate cancer progression; these include oncomiRs, miR-125b, miR-21, miR-221/222, miR-27a, and miR-32 [53] and oncosuppressor miRs, miR-135a [56], and miR-141 [57]. The recruitment of AR to the promoter regions of these miRNAs has been demonstrated with chromatin immunoprecipitation (ChIP) analysis. In addition to ER and AR, other NRs can regulate miRNA expression in cancer. For example, PR can regulate the expression of several miRNAs, including miR-141 [58], miR-23 [59], miR-320 [60], and let-7 in human cancers [61]. Glucocorticoids have been shown to upregulate miR-15, miR-16, and miR-223 through activating both GR and MR in leukemia cell lines [62]. Therefore, increased understanding of the molecular basis of the modulation of miRNA expression by NRs may enable new therapeutic interventions for cancer patients.
3.2. Aberrant miRNA Expression by DNA Methylation Modification in Cancer
In recent years, evidence has been mounting to suggest the epigenetic interaction between DNA methylation modification and miRNA expression in cancer. The transcription of pri-miRNA is also affected by epigenetic control, particularly the methylation of the promoter-associated CpG island. In human bladder cancer, miR-127 is silenced through its promoter hypermethylation, resulting in increased expression of its cognate target, BCL6 [63]. The hypermethylation of the miR-124-1 promoter region is also appreciated in leukemia, lymphoma, breast, colon, and liver cancers, and the epigenetic repression of the miR-124-1 loci leads to the activation of its target, CDK6 [64,65]. The methylation of the miR-129-2 promoter region is found in endometrial and gastric cancers, along with the upregulation of its targets, SOX4 [66]. The frequent inactivation of miR-200 by its CpG methylayion is also found in bladder [69], breast [70], and non-small lung cancers [71]. These results suggest that DNA demethylation can activate expression of miRNAs which may act as tumor suppressors. Recently, we found that the DNA demethylase TET (ten eleven translocation) family members (TET1, TET2, and TET3) can unmask the epigenetically silenced miR-200, while miR-22 antagonizes miR-200 through directly targeting TETs and thereby promotes the metastatic process and EMT in breast cancer [79]. miR-34a and miR-34b/c loci are separately located on the different chromosomes among miR-34 family, but both miR-34a and miR-34b/c are hypermethylated in solid cancers and hematological tumors [67,68]. In CLL, the silencing of miRNAs by global methylation has been extensively studied using genome-wide methylation array and targeted methylation assay [80,81]. In addition to DNA methylation, histone modification has an effect on controlling miRNA expression by chromatin remodeling as well as cooperating DNA methylation modification [82]. Therefore, it is necessary to better understand how different epigenetic components interact with and influence miRNA expression and its output in the pathogenesis of cancer.
4. Dysregulation of Pri-miRNA Processing in Cancer
A class 2 ribonuclease lll enzyme, DROSHA, and its cofactor DGCR8 form a heterotrimeric complex named the “microprocessor”, which processes a stem-loop secondary structure of the nascent pri-miRNA transcript flanked by single-stranded RNA segments. The microprocessor recognizes the terminal loop region and the basal junction between the stem and the basal ssRNA segment and cleaves dsRNA at ~ 11 bp from the basal junction, releasing the hairpin-shaped pre-miRNA [83,84,85]. The aberrant processing of pri-miRNA can affect the overall production of pre-miRNA, as well as the accumulation of miscleaved pri-miRNA (Figure 1). In addition to the genomic mutation of the miRNA sequence, dysregulation of the microprocessor or microprocessor-associated proteins involved in pri-miRNA processing contributes to the global alterations of miRNA expression in cancer (Table 2).
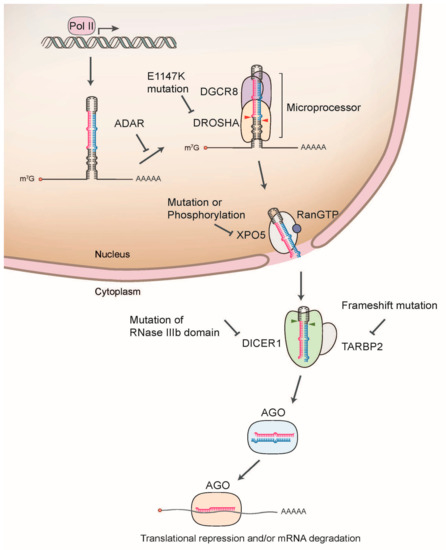
Figure 1.
Schematic illustration of miRNA biogenesis dysregulation in cancer. The nascent primary miRNA (pri-miRNA) is transcribed by RNA Polymerase II (Pol II). The pri-miRNA is initially cleaved by DROSHA/DGCR8 microprocessor complex in the nucleus, which releases a hairpin shaped precursor (pre-miRNA). Pre-miRNA is then exported from the nucleus to the cytoplasm by exportin 5 (XPO5). In the cytoplasm, DICER1 cleaves pre-miRNA to produce the miRNA duplex, which is loaded onto the Argonaute (AGO) protein. AGO preferentially retains one strand mature miRNA and further associates with other cofactors including GW182, forming the effector complex called RNA-induced silencing complex (RISC). The RISC induces the translational suppression and mRNA degradation through the interaction with the complementary sequences in the 3’-untranslated region (3’-UTR) of target mRNA.

Table 2.
Dysregulation of miRNA biogenesis.
4.1. Dysregulation of the Microprocessor in Cancer
The expression and function of the miRNA biogenesis machinery genes are often deregulated in cancer. The gain of DROSHA copy-number is found in more than 50% of advanced cervical squamous cell carcinomas [86], and its expression is upregulated in various types of cancers, which affect the global miRNA profile [87]. By contrast, DROSHA expression has been also shown to be downregulated in many other types of cancers, suggesting its role as a tumor suppressor in different contexts [112]. Although the function of DROSHA is still controversial, either upregulation or downregulation of DROSHA expression alters the global miRNA expression profile, which is correlated with cancer progression and patient survival rate [87]. DROSHA is frequently mutated in Wilms tumors, and mature miRNAs are globally downregulated in those tumors. The recurrent DROSHA E1147K mutation has been verified to hinder the metal binding and affect the processing activity of DROSHA [88,89,90,91]. It has not been yet identified in the functions of other mutations, such as the missense mutation and nonsense mutation of the DROSHA gene found in Wilms tumors. The expression level of DGCR8, another component of the microprocessor, has been found to be increased in various human cancers, including oesophageal, bladder, prostate, and ovarian cancers [92]. Altered expression of DGCR8 is associated with dysregulated miRNA expression and poor patient prognosis [92]. DGCR8 is frequently mutated in Wilms tumors, and the recurrent mutation of E518K in the dsRBD1 domain of DGCR8 results in the decrease of miRNAs [89,91], implicating the importance in controlling the pri-miRNA processing machinery in cancer pathogenesis.
4.2. Dysregulation of the Microprocessor-Associated Proteins in Cancer
The regulators of the microprocessor, such as DROSHA- or DGCR8–associated proteins, pri-miRNA–associated RNA-binding proteins, and cellular signaling components, can also affect pri-miRNA processing. Analysis of a DROSHA-containing large complex has revealed that several microprocessor-associated RNA-binding proteins, including DEAD-box helicases p68 (also known as DDX5) [113] and p72 (also known as DDX17) [114] facilitate pri-miRNA processing. p68/p72 may serve as scaffold proteins to recruit multiple different protein factors to the DROSHA microprocessor. Interestingly, the p53 tumor suppressor protein interacts with the microprocessor complex via p68/p72 and thereby enhances the biogenesis of oncosuppressor miRs, such as miR-16-1, miR-143, and miR-145 [33]. Hippo downstream effector Yes-associated protein (YAP) also regulates pri-miRNA processing. While nuclear YAP sequesters p72, leading to p72 dissociation from the microprocessor complex to suppress miRNAs, YAP retained in the cytoplasm enable p72 to interact with the microprocessor for efficient pri-miRNA processing [115]. Likewise, constitutive activation of YAP1 or inactivation of Hippo-signaling can mediate the global downregulation of miRNAs and promote tumorigenesis. Nuclear factor 90/45 (NF90/NF45) complex impairs the access of the microprocessor to a subset of human pri-miRNAs, including pri-let-7 and pri-miR7-1, leading to the reduction in mature miRNA levels in liver cancer [93,94]. These results expand our knowledge of how pri-miRNA processing is controlled by the microprocessor regulators and cell signaling and of how this causes large perturbations of miRNA expression in cancer.
4.3. Dysregulation of pri-miRNA Editing in Cancer
RNA editing is a main post-transcriptional mechanism that modifies specific nucleotides at the RNA level. Adenosine deaminases acting on RNA (ADARs) are the RNA modification enzymes that convert adenosine (A) to inosine (I) in double-stranded RNAs (dsRNAs). ADAR can edit the dsRNA in the stem region of the pri-miRNA and change the secondary structure, which inhibits its processing by the DROSHA/DGCR8 microprocessor complex and leads to their degradation by endonuclease V [116]. Recent studies have demonstrated that miRNA editing is dysregulated in human cancers, and miRNA-related editing promotes or inhibits tumor development and progression [117]. Likewise, the miRNA editing level varies between different patients and cancer types (either hyperedited or hypoedited pri-miRNAs) [118,119]. The ADARs’ tissue specificity and over/underexpression in different tumor contexts may account for the diverse patterns of pri-miRNA editing in cancer. Nevertheless, the pathophysiological role of pri-miRNA editing events observed in cancer remains largely unexamined.
5. Dysregulation of Pre-miRNA Processing in Cancer
Pre-miRNA generated by the microprocessor in the nucleus is transported into the cytoplasm by a complex of XPO5 and RAN-GTP, a cofactor of XPO5. It is further processed to generate ~ 22 nt small RNA duplexes. DICER1 recognizes 2 nt 3′ overhang of pre-miRNA, 22 nt apart from which the cleavage site is defined [120]. DICER1 associates with the dsRNA-binding protein TARBP2 to increase the stability of the DICER1-RNA complex and enhance the fidelity of miRNA processing. Importantly, genetic mutations and dysregulation of key components in the pre-miRNA processing step cause aberrant miRNA expression in cancer.
5.1. Defect in Pre-miRNA Export in Cancer
Inactivated mutations of XPO5 have been identified in sporadic colon, gastric, and endometrial tumors with microsatellite instability [95]; these mutations cause the defect of pre-miRNA export, leading to the accumulation of pre-miRNA in the nucleus. The genetic alterations of XPO5 are also associated with the risk of breast cancer [121]. Additionally, the MAPK/ERK pathway can suppress pre-miRNA export through phosphorylating XPO5 at Thr345, Ser416, and Ser497 [96]. Phosphorylation of XPO5 correlates with the global downregulation of miRNAs and poor prognosis in patients with hepatocellular carcinoma, providing functional and clinical evidence of the cancer-associated dysregulation of XPO5 for aberrant miRNA processing and tumorigenesis. However, the upstream signaling regulators for pre-miRNA export, via either XPO5 or Ran-GTP, have not yet been identified.
5.2. Dysregulation of DICER1 and TARBP2 in Cancer
Global inhibition of miRNA biogenesis by depletion of DICER1 promotes cell growth and tumorigenesis in human cancer cell lines and mouse models of cancer [122], suggesting the oncogenic role of DICER1 in tumorigenesis. Recurrent somatic and germline DICER1 mutations that change its protein levels and/or impair its function, leading to defective pre-miRNA processing, are frequently found in many types of tumors, including pleuropulmonary blastoma, rhabdomyosarcoma, non-epithelial ovarian cancer, and liver tumor [97,98,99,100,101,102]. In particular, mutations within the RNase IIIb domain of DICER1 markedly reduce the expression of 5p miRNAs (miRNAs derived from the 5′ side of the pre-miRNA) in cancer [92,103]. DICER1-associated regulatory factors are also involved in the dysregulation of pre-miRNA processing. TAp63 suppresses tumorigenesis and metastasis by direct binding to DICER [123], suggesting both genetic mutation and functional inactivation of DICER1 dictate global miRNA expression in tumor malignancy.
The frameshift mutations of TARBP2 are found in sporadic and hereditary carcinomas with microsatellite instability, which correlates with reduced levels of DICER1 and mature miRNAs [106,107]. TARBP2 is also deleted in 15% of adenoid cystic carcinoma [104]. In contrast, TARBP2 is overexpressed in cutaneous melanoma, adrenocortical carcinoma, and metastatic breast and prostate cancers [105], suggesting its specific pivotal role in different cancer types.
5.3. Dysregulation of AGO2 in Cancer
Argonaute 2 (AGO2), the only member of the Argonautes with an intrinsic endonuclease activity, is involved in the accumulation of mature miRNAs [124,125]. As a key regulator of miRNA function and maturation, AGO2 has been found to be overexpressed in various types of human cancers, including breast, gastric, and head and neck cancers [108,109,110,111]. The overexpression of AGO2 may facilitate oncomiRs to repress their targets [126]. Despite identified functions of AGO2 in different types of cancer being contradicted [127], its dysregulation has been implicated in recent years in tumorigenesis.
6. Conclusions
Numerous studies have documented the aberrant expression of miRNAs in cancer and the oncogenic or tumor-suppressor roles of miRNAs. Likewise, the regulatory mechanisms to control the expression of miRNAs are strongly associated with cancer diagnosis, prognosis, and treatment, as well as the pathogenesis of cancer. Different core players and their partners involved in the multiple sequential step process for producing miRNA show deregulated activity and abundance in cancers, some of which are known to be affected by cancer-associated signaling regulators. Nevertheless, current knowledge is still behind a comprehensive understanding of how each miRNA is specifically controlled in specific types of cancer, emphasizing the systemic approach to the multi-layered regulation governing miRNA expression in cancers. We have discussed the series of processes to generate miRNAs and the possible regulatory mechanisms modulating miRNA expression in cancers. Dysregulation of miRNA biogenesis inevitably changes the mRNA profile in a cell, which in turn affects the miRNA expression and function through a feedback loop. Thus, it is necessary to comprehensively investigate the gene expression regulatory networks that cover both miRNA expression and its effect on mRNA targets in the near future. With evolved technologies such as the gene editing system by CRISPR-Cas9 and high-throughput sequencing, the studies to understand the molecular and cellular regulatory mechanisms controlling the expression of miRNAs in cancer will be facilitated and suggest compelling evidence to explore new therapeutic strategies for the treatment of cancer by targeting or restoring the expression profiles of miRNAs.
Author Contributions
This work was conceived and planned by M.L. and S.J.S. The original draft preparation and writing: Z.A.S.; S.S.S.L.; C.M.; M.L.; S.J.S. Review and editing: S.J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science & Information and Communication Technology, Korea (2019M3E5D3073090) to M.H.Lee, and Korea Health Technology R&D Project through the KHIDI funded by the Ministry of Health & Welfare, Korea (Grant number HI15C2679) to S.J.Song.
Acknowledgments
We thank T. Garvey for critical editing of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| mRNA | Messenger RNA |
| miRNA | MicroRNA |
| pri-miRNA | Primary-microRNA |
| pre-miRNA | Precursor-microRNA |
| XPO5 | Exportin 5 |
| AGO2 | Argonaute 2 |
| TNRC6A | Trinucleotide Repeat Containing Adaptor 6A |
| RISC | RNA-induced Silencing Complex |
| miRISC | miRNA-induced silencing complex |
| 3′-UTR | 3′- Untranslated Region |
| DNA | Deoxyribonucleic Acid |
| CLL | Chronic Lymphocytic Leukemia |
| BCL2 | B-cell Lymphoma 2 |
| MDS | Myelodysplastic syndrome |
| AML | Acute Myeloid Leukemia |
| RREB1 | Ras Responsive Element Binding Protein 1 |
| C/EBPβ | Ras Responsive Element Binding Protein 1 |
| C/EBPβ | CCAAT-enhancer-binding Protein Beta |
| TCF4 | Transcription Factor 4 |
| FOXO1 | Forkhead Box Protein O1 |
| FOXO3 | Forkhead Box Protein O3 |
| MN1 | Meniongoma-1 |
| E2F1 | E2F Transcription Factor 1 |
| CTGF | Connective Tissue Growth Factor |
| THBS1 | Thrombospondin 1 |
| PTEN | Phosphatase and Tensin Homolog |
| HIF1-α | Hypoxia-inducible Factor 1-alpha |
| ZEB | Zinc Finger E-box-binding Homeobox 1 |
| EMT | Epithelial-Mesenchymal Transition |
| NRs | Nuclear Receptors |
| ER | Estrogen Receptor |
| GR | Glucocorticoid Receptor |
| MR | Mineralocorticoid Receptor |
| NCoR/SMRT | Nuclear Receptor co-repressor/ Silencing Mediator of Retinoic |
| SK1 | Sphingosine Kinase 1 |
| ChIP | Chromatin Immunoprecipitation |
| BCL6 | B-cell Lymphoma 6 |
| CDK6 | Cyclin-dependent Kinase 6 |
| SOX4 | SRY-Box Transcription Factor 4 |
| CDH1 | Cadherin-1 |
| TET | Ten-eleven Translocation Methylcytosine Dioxygenase |
| PLAG | Pleomorphic adenoma gene |
| CREB | cAMP-response element binding |
| DGCR8 | DiGeorge Critical Region 8 |
| ssRNA | Single-stranded RNA |
| dsRNA | Double-stranded RNA |
| dsRBD1 | dsRNA Binding Domain1 |
| DDX5 | DEAD-box Helicases p68 |
| DDX17 | DEAD-box Helicases p72 |
| YAP | Yes-associated Protein |
| NF90/NF45 | Nuclear Factor 90/45 |
| ADAR | Adenosine Deaminases Acting on RNA |
| RAN-GTP | RAs-related Nuclear protein-GTP |
| TARBP2 | Tar RNA-Binding Protein 2 |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
References
- O′Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. (Lausanne) 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol Sci. 2016, 17. [Google Scholar] [CrossRef]
- Akgul, B.; Erdogan, I. Intracytoplasmic Re-Localization of miRISC Complexes. Front. Genet. 2018, 9, 403. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, S.; Kobayashi, M.; Yoda, M.; Sakaguchi, Y.; Katsuma, S.; Suzuki, T.; Tomari, Y. Hsc70/Hsp90 chaperone machinery mediates ATP-Dependent RISC loading of small RNA duplexes. Mol. Cell 2010, 39, 292–299. [Google Scholar] [CrossRef]
- Robin, C.F.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef]
- Ventura, A.; Jacks, T. MicroRNAs and cancer: Short RNAs go a long way. Cell 2009, 136, 586–591. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Chen, W.; Wang, H.; Chen, H.; Liu, S.; Lu, H.; Kong, D.; Huang, X.; Kong, Q.; Lu, Z. Clinical significance and detection of microRNA-21 in serum of patients with diffuse large B-Cell lymphoma in Chinese population. Eur. J. Haematol. 2014, 92, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef] [PubMed]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [PubMed]
- Thammaiah, C.K.; Jayaram, S. Role of let-7 family microRNA in breast cancer. Noncoding RNA Res. 2016, 1, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Manier, S.; Powers, J.T.; Sacco, A.; Glavey, S.V.; Huynh, D.; Reagan, M.R.; Salem, K.Z.; Moschetta, M.; Shi, J.; Mishima, Y.; et al. The LIN28B/let-7 axis is a novel therapeutic pathway in multiple myeloma. Leukemia 2017, 31, 853–860. [Google Scholar] [CrossRef]
- Xuefang, Z.; Ruinian, Z.; Liji, J.; Chun, Z.; Qiaolan, Z.; Jun, J.; Yuming, C.; Junrong, H. miR-331-3p Inhibits Proliferation and Promotes Apoptosis of Nasopharyngeal Carcinoma Cells by Targeting elf4B-PI3K-AKT Pathway. Technol. Cancer Res. Treat. 2020, 19. [Google Scholar] [CrossRef]
- Medina, P.P.; Nolde, M.; Slack, F.J. OncomiR addiction in an in vivo model of microRNA-21-Induced pre-B-Cell lymphoma. Nature 2010, 467, 86–90. [Google Scholar] [CrossRef]
- Wang, W.; Li, J.; Zhu, W.; Gao, C.; Jiang, R.; Li, W.; Hu, Q.; Zhang, B. MicroRNA-21 and the clinical outcomes of various carcinomas: A systematic review and meta-Analysis. BMC Cancer 2014, 14, 819. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.X.; Huang, X.F.; Shao, Q.; Huang, M.Y.; Deng, L.; Wu, Q.L.; Zeng, Y.X.; Shao, J.Y. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008, 14, 2348–2360. [Google Scholar] [CrossRef] [PubMed]
- Ahmadvand, M.; Eskandari, M.; Pashaiefar, H.; Yaghmaie, M.; Manoochehrabadi, S.; Khakpour, G.; Sheikhsaran, F.; Montazer Zohour, M. Over expression of circulating miR-155 predicts prognosis in diffuse large B-Cell lymphoma. Leuk Res. 2018, 70, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, H.W.; Lu, M.H.; He, X.H.; Li, Y.; Gu, H.; Liu, M.F.; Wang, E.D. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010, 70, 3119–3127. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Yu, Y.; Zhu, M.; Jing, W.; Yu, M.; Chai, H.; Liang, C.; Tu, J. Inhibition of miR-155, a therapeutic target for breast cancer, prevented in cancer stem cell formation. Cancer Biomark 2018, 21, 383–392. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, X.; Lim, L.P.; de Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef]
- Li, J.; Lai, Y.; Ma, J.; Liu, Y.; Bi, J.; Zhang, L.; Chen, L.; Yao, C.; Lv, W.; Chang, G.; et al. miR-17-5p suppresses cell proliferation and invasion by targeting ETV1 in triple-Negative breast cancer. BMC Cancer 2017, 17, 745. [Google Scholar] [CrossRef]
- Schrijver, W.A.; van Diest, P.J.; Dutch Distant Breast Cancer Metastases, C.; Moelans, C.B. Unravelling site-specific breast cancer metastasis: A microRNA expression profiling study. Oncotarget 2017, 8, 3111–3123. [Google Scholar] [CrossRef]
- Lee, J.; Kim, H.E.; Song, Y.S.; Cho, E.Y.; Lee, A. miR-106b-5p and miR-17-5p could predict recurrence and progression in breast ductal carcinoma in situ based on the transforming growth factor-Beta pathway. Breast Cancer Res. Treat. 2019, 176, 119–130. [Google Scholar] [CrossRef]
- Leung, A.K.; Sharp, P.A. MicroRNA functions in stress responses. Mol. Cell 2010, 40, 205–215. [Google Scholar] [CrossRef]
- Franovic, A.; Gunaratnam, L.; Smith, K.; Robert, I.; Patten, D.; Lee, S. Translational up-Regulation of the EGFR by tumor hypoxia provides a nonmutational explanation for its overexpression in human cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 13092–13097. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Xia, W.; Khotskaya, Y.B.; Huo, L.; Nakanishi, K.; Lim, S.O.; Du, Y.; Wang, Y.; Chang, W.C.; Chen, C.H. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 2013, 497, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Yamagata, K.; Sugimoto, K.; Iwamoto, T.; Kato, S.; Miyazono, K. Modulation of microRNA processing by p53. Nature 2009, 460, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sun, Q.; Zhang, Z.; Ge, S.; Han, Z.G.; Chen, W.T. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene 2013, 32, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Kent, O.A.; Fox-Talbot, K.; Halushka, M.K. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene 2013, 32, 2576–2585. [Google Scholar] [CrossRef] [PubMed]
- Gan, B.; Lim, C.; Chu, G.; Hua, S.; Ding, Z.; Collins, M.; Hu, J.; Jiang, S.; Fletcher-Sananikone, E.; Zhuang, L.; et al. FoxOs enforce a progression checkpoint to constrain mTORC1-Activated renal tumorigenesis. Cancer Cell 2010, 18, 472–484. [Google Scholar] [CrossRef]
- Zeinali, T.; Mansoori, B.; Mohammadi, A.; Baradaran, B. Regulatory mechanisms of miR-145 expression and the importance of its function in cancer metastasis. Biomed. Pharm. 2019, 109, 195–207. [Google Scholar] [CrossRef]
- Seipel, K.; Messerli, C.; Wiedemann, G.; Bacher, U.; Pabst, T. MN1, FOXP1 and hsa-miR-181a-5p as prognostic markers in acute myeloid leukemia patients treated with intensive induction chemotherapy and autologous stem cell transplantation. Leuk Res. 2020, 89, 106296. [Google Scholar] [CrossRef]
- Dews, M.; Homayouni, A.; Yu, D.; Murphy, D.; Sevignani, C.; Wentzel, E.; Furth, E.E.; Lee, W.M.; Enders, G.H.; Mendell, J.T.; et al. Augmentation of tumor angiogenesis by a Myc-Activated microRNA cluster. Nat. Genet. 2006, 38, 1060–1065. [Google Scholar] [CrossRef]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef]
- O′Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef]
- Ventura, A.; Young, A.G.; Winslow, M.M.; Lintault, L.; Meissner, A.; Erkeland, S.J.; Newman, J.; Bronson, R.T.; Crowley, D.; Stone, J.R.; et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008, 132, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Choi, P.S.; Casey, S.C.; Dill, D.L.; Felsher, D.W. MYC through miR-17-92 Suppresses Specific Target Genes to Maintain Survival, Autonomous Proliferation, and a Neoplastic State. Cancer Cell 2014, 26, 262–272. [Google Scholar] [CrossRef]
- Chang, T.C.; Yu, D.; Lee, Y.S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008, 40, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, J.J.; Domingo-Fernandez, R.; Ebus, M.E.; Lindner, S.; Koster, J.; Drabek, K.; Mestdagh, P.; van Sluis, P.; Valentijn, L.J.; van Nes, J.; et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 2012, 44, 1199–1206. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, X.; Fiskus, W.; Lin, J.; Lwin, T.; Rao, R.; Zhang, Y.; Chan, J.C.; Fu, K.; Marquez, V.E.; et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer Cell 2012, 22, 506–523. [Google Scholar] [CrossRef]
- Chen, P.; Guo, X.; Zhang, L.; Zhang, W.; Zhou, Q.; Tian, Z.; Zheng, Y.; Liao, Q.; Wang, H.; Li, G. MiR-200c is a cMyc-Activated miRNA that promotes nasopharyngeal carcinoma by downregulating PTEN. Oncotarget 2017, 8, 5206–5218. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Ding, L.; Bennewith, K.L.; Tong, R.T.; Welford, S.M.; Ang, K.K.; Story, M.; Le, Q.T.; Giaccia, A.J. Hypoxia-Inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell 2009, 35, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Nallamshetty, S.; Chan, S.Y.; Loscalzo, J. Hypoxia: A master regulator of microRNA biogenesis and activity. Free Radic Biol. Med. 2013, 64, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Guan, T.; Dominguez, C.X.; Amezquita, R.A.; Laidlaw, B.J.; Cheng, J.; Henao-Mejia, J.; Williams, A.; Flavell, R.A.; Lu, J.; Kaech, S.M. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8(+) T cell fates. J. Exp. Med. 2018, 215, 1153–1168. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, G.; Gasparini, P.; Piovan, C.; Ngankeu, A.; Garofalo, M.; Taccioli, C.; Iorio, M.V.; Li, M.; Volinia, S.; Alder, H.; et al. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J. Natl. Cancer Inst. 2010, 102, 706–721. [Google Scholar] [CrossRef] [PubMed]
- Pinho, F.G.; Frampton, A.E.; Nunes, J.; Krell, J.; Alshaker, H.; Jacob, J.; Pellegrino, L.; Roca-Alonso, L.; de Giorgio, A.; Harding, V.; et al. Downregulation of microRNA-515-5p by the estrogen receptor modulates sphingosine kinase 1 and breast cancer cell proliferation. Cancer Res. 2013, 73, 5936–5948. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.I.; Misawa, A.; Inoue, S. Significance of microRNAs in Androgen Signaling and Prostate Cancer Progression. Cancers (Basel) 2017, 102. [Google Scholar] [CrossRef] [PubMed]
- Loffler, D.; Brocke-Heidrich, K.; Pfeifer, G.; Stocsits, C.; Hackermuller, J.; Kretzschmar, A.K.; Burger, R.; Gramatzki, M.; Blumert, C.; Bauer, K.; et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-Mediated induction of microRNA-21 through a highly conserved enhancer. Blood 2007, 110, 1330–1333. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Wang, Z.X.; Wang, R. MicroRNA-21: A novel therapeutic target in human cancer. Cancer Biol. Ther. 2010, 10, 1224–1232. [Google Scholar] [CrossRef] [PubMed]
- Coarfa, C.; Fiskus, W.; Eedunuri, V.K.; Rajapakshe, K.; Foley, C.; Chew, S.A.; Shah, S.S.; Geng, C.; Shou, J.; Mohamed, J.S.; et al. Comprehensive proteomic profiling identifies the androgen receptor axis and other signaling pathways as targets of microRNAs suppressed in metastatic prostate cancer. Oncogene 2016, 35, 2345–2356. [Google Scholar] [CrossRef]
- Liu, C.; Liu, R.; Zhang, D.; Deng, Q.; Liu, B.; Chao, H.P.; Rycaj, K.; Takata, Y.; Lin, K.; Lu, Y.; et al. MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat. Commun. 2017, 8, 14270. [Google Scholar] [CrossRef]
- Finlay-Schultz, J.; Cittelly, D.M.; Hendricks, P.; Patel, P.; Kabos, P.; Jacobsen, B.M.; Richer, J.K.; Sartorius, C.A. Progesterone downregulation of miR-141 contributes to expansion of stem-Like breast cancer cells through maintenance of progesterone receptor and Stat5a. Oncogene 2015, 34, 3676–3687. [Google Scholar] [CrossRef]
- Cittelly, D.M.; Finlay-Schultz, J.; Howe, E.N.; Spoelstra, N.S.; Axlund, S.D.; Hendricks, P.; Jacobsen, B.M.; Sartorius, C.A.; Richer, J.K. Progestin suppression of miR-29 potentiates dedifferentiation of breast cancer cells via KLF4. Oncogene 2013, 32, 2555–2564. [Google Scholar] [CrossRef]
- Xia, H.F.; Jin, X.H.; Song, P.P.; Cui, Y.; Liu, C.M.; Ma, X. Temporal and spatial regulation of miR-320 in the uterus during embryo implantation in the rat. Int. J. Mol. Sci. 2010, 11, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Wendler, A.; Keller, D.; Albrecht, C.; Peluso, J.J.; Wehling, M. Involvement of let-7/miR-98 microRNAs in the regulation of progesterone receptor membrane component 1 expression in ovarian cancer cells. Oncol. Rep. 2011, 25, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Rainer, J.; Ploner, C.; Jesacher, S.; Ploner, A.; Eduardoff, M.; Mansha, M.; Wasim, M.; Panzer-Grumayer, R.; Trajanoski, Z.; Niederegger, H.; et al. Glucocorticoid-Regulated microRNAs and mirtrons in acute lymphoblastic leukemia. Leukemia 2009, 23, 746–752. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Liang, G.; Egger, G.; Friedman, J.M.; Chuang, J.C.; Coetzee, G.A.; Jones, P.A. Specific activation of microRNA-127 with downregulation of the proto-Oncogene BCL6 by chromatin-Modifying drugs in human cancer cells. Cancer Cell 2006, 9, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.J.; Wang, Q.Y.; Zhou, C.X.; Yin, Q.Q.; He, M.; Yu, X.T.; Cao, D.X.; Chen, G.Q.; He, J.R.; Zhao, Q. MiR-124 targets Slug to regulate epithelial-Mesenchymal transition and metastasis of breast cancer. Carcinogenesis 2013, 34, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Lujambio, A.; Ropero, S.; Ballestar, E.; Fraga, M.F.; Cerrato, C.; Setien, F.; Casado, S.; Suarez-Gauthier, A.; Sanchez-Cespedes, M.; Git, A.; et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007, 67, 1424–1429. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.W.; Wu, C.W.; Hu, L.Y.; Li, S.C.; Liao, Y.L.; Lai, C.H.; Kao, H.W.; Fang, W.L.; Huang, K.H.; Chan, W.C. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int. J. Cancer 2011, 129, 2600–2610. [Google Scholar] [CrossRef]
- Schmid, G.; Notaro, S.; Reimer, D.; Abdel-Azim, S.; Duggan-Peer, M.; Holly, J.; Fiegl, H.; Rossler, J.; Wiedemair, A.; Concin, N.; et al. Expression and promotor hypermethylation of miR-34a in the various histological subtypes of ovarian cancer. BMC Cancer 2016, 16, 102. [Google Scholar] [CrossRef]
- Wong, M.Y.; Yu, Y.; Walsh, W.R.; Yang, J.L. microRNA-34 family and treatment of cancers with mutant or wild-Type p53 (Review). Int J. Oncol. 2011, 38, 1189–1195. [Google Scholar] [CrossRef]
- Shindo, T.; Niinuma, T.; Nishiyama, N.; Shinkai, N.; Kitajima, H.; Kai, M.; Maruyama, R.; Tokino, T.; Masumori, N.; Suzuki, H. Epigenetic silencing of miR-200b is associated with cisplatin resistance in bladder cancer. Oncotarget 2018, 9, 24457–24469. [Google Scholar] [CrossRef]
- Davalos, V.; Moutinho, C.; Villanueva, A.; Boque, R.; Silva, P.; Carneiro, F.; Esteller, M. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 2012, 31, 2062–2074. [Google Scholar] [CrossRef]
- Ceppi, P.; Mudduluru, G.; Kumarswamy, R.; Rapa, I.; Scagliotti, G.V.; Papotti, M.; Allgayer, H. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-Small cell lung cancer. Mol. Cancer Res. 2010, 8, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Allegra, D.; Bilan, V.; Garding, A.; Dohner, H.; Stilgenbauer, S.; Kuchenbauer, F.; Mertens, D.; Zucknick, M. Defective DROSHA processing contributes to downregulation of MiR-15/-16 in chronic lymphocytic leukemia. Leukemia 2014, 28, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.; Croce, C.M. Role of miR-15/16 in CLL. Cell Death Differ. 2015, 22, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.L.; Starczynowski, D.T. Role of microRNA-146a in normal and malignant hematopoietic stem cell function. Front. Genet. 2014, 5, 219. [Google Scholar] [CrossRef]
- Navarro, F.; Lieberman, J. miR-34 and p53: New Insights into a Complex Functional Relationship. PLoS ONE 2015, 10, e0132767. [Google Scholar] [CrossRef]
- Seong, O.S.; Chen, Y.; Zaman, M.S.; Hirata, H.; Yamamura, S.; Shahryari, V.; Liu, J.; Tabatabai, Z.L.; Kakar, S.; Deng, G.; et al. MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis 2011, 32, 772–778. [Google Scholar] [CrossRef]
- Mihailovich, M.; Bremang, M.; Spadotto, V.; Musiani, D.; Vitale, E.; Varano, G.; Zambelli, F.; Mancuso, F.M.; Cairns, D.A.; Pavesi, G.; et al. miR-17-92 fine-tunes MYC expression and function to ensure optimal B cell lymphoma growth. Nat. Commun. 2015, 6, 8725. [Google Scholar] [CrossRef]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schutz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P.; et al. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835–839. [Google Scholar] [CrossRef]
- Song, S.J.; Poliseno, L.; Song, M.S.; Ala, U.; Webster, K.; Ng, C.; Beringer, G.; Brikbak, N.J.; Yuan, X.; Cantley, L.C.; et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-Family-Dependent chromatin remodeling. Cell 2013, 154, 311–324. [Google Scholar] [CrossRef]
- Baer, C.; Claus, R.; Frenzel, L.P.; Zucknick, M.; Park, Y.J.; Gu, L.; Weichenhan, D.; Fischer, M.; Pallasch, C.P.; Herpel, E.; et al. Extensive promoter DNA hypermethylation and hypomethylation is associated with aberrant microRNA expression in chronic lymphocytic leukemia. Cancer Res. 2012, 72, 3775–3785. [Google Scholar] [CrossRef]
- Pallasch, C.P.; Patz, M.; Park, Y.J.; Hagist, S.; Eggle, D.; Claus, R.; Debey-Pascher, S.; Schulz, A.; Frenzel, L.P.; Claasen, J.; et al. miRNA deregulation by epigenetic silencing disrupts suppression of the oncogene PLAG1 in chronic lymphocytic leukemia. Blood 2009, 114, 3255–3264. [Google Scholar] [CrossRef]
- Yeoh, G.; Barton, S.; Kaestner, K. The International Journal of Biochemistry & Cell Biology. Preface. Int. J. Biochem. Cell Biol. 2011, 43, 172. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.H.; Nam, J.W.; Heo, I.; Rhee, J.K.; Sohn, S.Y.; Cho, Y.; Zhang, B.T.; Kim, V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef]
- Muralidhar, B.; Winder, D.; Murray, M.; Palmer, R.; Barbosa-Morais, N.; Saini, H.; Roberts, I.; Pett, M.; Coleman, N. Functional evidence that Drosha overexpression in cervical squamous cell carcinoma affects cell phenotype and microRNA profiles. J. Pathol. 2011, 224, 496–507. [Google Scholar] [CrossRef]
- Gurtner, A.; Falcone, E.; Garibaldi, F.; Piaggio, G. Dysregulation of microRNA biogenesis in cancer: The impact of mutant p53 on Drosha complex activity. J. Exp. Clin. Cancer Res. 2016, 35, 45. [Google Scholar] [CrossRef]
- Rakheja, D.; Chen, K.S.; Liu, Y.; Shukla, A.A.; Schmid, V.; Chang, T.C.; Khokhar, S.; Wickiser, J.E.; Karandikar, N.J.; Malter, J.S.; et al. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat. Commun. 2014, 2, 4802. [Google Scholar] [CrossRef]
- Torrezan, G.T.; Ferreira, E.N.; Nakahata, A.M.; Barros, B.D.; Castro, M.T.; Correa, B.R.; Krepischi, A.C.; Olivieri, E.H.; Cunha, I.W.; Tabori, U.; et al. Recurrent somatic mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat. Commun. 2014, 5, 4039. [Google Scholar] [CrossRef]
- Walz, A.L.; Ooms, A.; Gadd, S.; Gerhard, D.S.; Smith, M.A.; Guidry Auvil, J.M.; Meerzaman, D.; Chen, Q.R.; Hsu, C.H.; Yan, C.; et al. Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell 2015, 27, 286–297. [Google Scholar] [CrossRef]
- Wegert, J.; Ishaque, N.; Vardapour, R.; Georg, C.; Gu, Z.; Bieg, M.; Ziegler, B.; Bausenwein, S.; Nourkami, N.; Ludwig, N.; et al. Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-Risk blastemal type Wilms tumors. Cancer Cell 2015, 27, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Kashima, R. Dysregulation of microRNA biogenesis machinery in cancer. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, T.; Todaka, H.; Sugiyama, Y.; Ono, M.; Tamaki, N.; Hatano, E.; Takezaki, Y.; Hanazaki, K.; Miwa, T.; Lai, S.; et al. Suppression of MicroRNA-7 (miR-7) Biogenesis by Nuclear Factor 90-Nuclear Factor 45 Complex (NF90-NF45) Controls Cell Proliferation in Hepatocellular Carcinoma. J. Biol. Chem. 2016, 291, 21074–21084. [Google Scholar] [CrossRef] [PubMed]
- Grasso, G.; Higuchi, T.; Barbier, J.; Helsmoortel, M.; Lorenzi, C.; Sanchez, G.; Bello, M.; Ritchie, W.; Sakamoto, S.; Kiernan, R. NF90 Modulates Processing of a Subset of Human Pri-miRNAs. BioRxiv 2020. [Google Scholar] [CrossRef]
- Melo, S.A.; Moutinho, C.; Ropero, S.; Calin, G.A.; Rossi, S.; Spizzo, R.; Fernandez, A.F.; Davalos, V.; Villanueva, A.; Montoya, G.; et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 2010, 18, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.L.; Cui, R.; Zhou, J.; Teng, K.Y.; Hsiao, Y.H.; Nakanishi, K.; Fassan, M.; Luo, Z.; Shi, G.; Tili, E.; et al. ERK Activation Globally Downregulates miRNAs through Phosphorylating Exportin-5. Cancer Cell 2016, 30, 723–736. [Google Scholar] [CrossRef]
- Caruso, S.; Calderaro, J.; Letouze, E.; Nault, J.C.; Couchy, G.; Boulai, A.; Luciani, A.; Zafrani, E.S.; Bioulac-Sage, P.; Seror, O.; et al. Germline and somatic DICER1 mutations in familial and sporadic liver tumors. J. Hepatol. 2017, 66, 734–742. [Google Scholar] [CrossRef]
- Fernandez-Martinez, L.; Villegas, J.A.; Santamaria, I.; Pitiot, A.S.; Alvarado, M.G.; Fernandez, S.; Torres, H.; Paredes, A.; Blay, P.; Balbin, M. Identification of somatic and germ-Line DICER1 mutations in pleuropulmonary blastoma, cystic nephroma and rhabdomyosarcoma tumors within a DICER1 syndrome pedigree. BMC Cancer 2017, 17, 146. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Priest, J.R.; Duchaine, T.F. DICER1: Mutations, microRNAs and mechanisms. Nat. Rev. Cancer 2014, 14, 662–672. [Google Scholar] [CrossRef]
- Heravi-Moussavi, A.; Anglesio, M.S.; Cheng, S.W.; Senz, J.; Yang, W.; Prentice, L.; Fejes, A.P.; Chow, C.; Tone, A.; Kalloger, S.E.; et al. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N. Engl. J. Med. 2012, 366, 234–242. [Google Scholar] [CrossRef]
- Hill, D.A.; Ivanovich, J.; Priest, J.R.; Gurnett, C.A.; Dehner, L.P.; Desruisseau, D.; Jarzembowski, J.A.; Wikenheiser-Brokamp, K.A.; Suarez, B.K.; Whelan, A.J.; et al. DICER1 mutations in familial pleuropulmonary blastoma. Science 2009, 325, 965. [Google Scholar] [CrossRef]
- Stewart, C.J.; Charles, A.; Foulkes, W.D. Gynecologic Manifestations of the DICER1 Syndrome. Surg. Pathol. Clin. 2016, 9, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Anglesio, M.S.; Wang, Y.; Yang, W.; Senz, J.; Wan, A.; Heravi-Moussavi, A.; Salamanca, C.; Maines-Bandiera, S.; Huntsman, D.G.; Morin, G.B. Cancer-Associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse-Strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J. Pathol. 2013, 229, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.S.; Kannan, K.; Roy, D.M.; Morris, L.G.; Ganly, I.; Katabi, N.; Ramaswami, D.; Walsh, L.A.; Eng, S.; Huse, J.T.; et al. The mutational landscape of adenoid cystic carcinoma. Nat. Genet. 2013, 45, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Fish, L.; Navickas, A.; Culbertson, B.; Xu, Y.; Nguyen, H.C.B.; Zhang, S.; Hochman, M.; Okimoto, R.; Dill, B.D.; Molina, H.; et al. Nuclear TARBP2 Drives Oncogenic Dysregulation of RNA Splicing and Decay. Mol. Cell 2019, 75, 967.e9–981.e9. [Google Scholar] [CrossRef] [PubMed]
- Garre, P.; Perez-Segura, P.; Diaz-Rubio, E.; Caldes, T.; de la Hoya, M. Reassessing the TARBP2 mutation rate in hereditary nonpolyposis colorectal cancer. Nat. Genet. 2010, 42, 817–818. [Google Scholar] [CrossRef]
- Melo, S.A.; Ropero, S.; Moutinho, C.; Aaltonen, L.A.; Yamamoto, H.; Calin, G.A.; Rossi, S.; Fernandez, A.F.; Carneiro, F.; Oliveira, C.; et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat. Genet. 2009, 41, 365–370. [Google Scholar] [CrossRef]
- Casey, M.C.; Prakash, A.; Holian, E.; McGuire, A.; Kalinina, O.; Shalaby, A.; Curran, C.; Webber, M.; Callagy, G.; Bourke, E.; et al. Quantifying Argonaute 2 (Ago2) expression to stratify breast cancer. BMC Cancer 2019, 19, 712. [Google Scholar] [CrossRef]
- Yang, F.Q.; Huang, J.H.; Liu, M.; Yang, F.P.; Li, W.; Wang, G.C.; Che, J.P.; Zheng, J.H. Argonaute 2 is up-regulated in tissues of urothelial carcinoma of bladder. Int. J. Clin. Exp. Pathol. 2014, 7, 340–347. [Google Scholar]
- Zhang, J.; Fan, X.S.; Wang, C.X.; Liu, B.; Li, Q.; Zhou, X.J. Up-Regulation of Ago2 expression in gastric carcinoma. Med. Oncol. 2013, 30, 628. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, B.; Chen, X.; Li, W.; Dong, P. AGO2 involves the malignant phenotypes and FAK/PI3K/AKT signaling pathway in hypopharyngeal-Derived FaDu cells. Oncotarget 2017, 8, 54735–54746. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Min, H.; Ha, J.Y.; Kim, B.H.; Choi, M.S.; Kim, S. Dysregulation of the miRNA biogenesis components DICER1, DROSHA, DGCR8 and AGO2 in clear cell renal cell carcinoma in both a Korean cohort and the cancer genome atlas kidney clear cell carcinoma cohort. Oncol. Lett. 2019, 18, 4337–4345. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Huang, J.; Hu, Z. RNA helicase DDX5 regulates microRNA expression and contributes to cytoskeletal reorganization in basal breast cancer cells. Mol. Cell Proteom. 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.P.; Terrone, S.; Giraud, G.; Benoit-Pilven, C.; Cluet, D.; Combaret, V.; Mortreux, F.; Auboeuf, D.; Bourgeois, C.F. The RNA helicase DDX17 controls the transcriptional activity of REST and the expression of proneural microRNAs in neuronal differentiation. Nucleic Acids Res. 2018, 46, 7686–7700. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Triboulet, R.; Mohseni, M.; Schlegelmilch, K.; Shrestha, K.; Camargo, F.D.; Gregory, R.I. Hippo signaling regulates microprocessor and links cell-Density-Dependent miRNA biogenesis to cancer. Cell 2014, 156, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17, 83–96. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, H. When microRNAs meet RNA editing in cancer: A nucleotide change can make a difference. Bioessays 2018, 40. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Wu, Y.; Zhang, X.; Liao, Y.; Sibanda, V.L.; Liu, W.; Guo, A.Y. Comprehensive analysis of human small RNA sequencing data provides insights into expression profiles and miRNA editing. RNA Biol. 2014, 11, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, X.; Yu, S.; Jeong, K.J.; Zhou, Z.; Han, L.; Tsang, Y.H.; Li, J.; Chen, H.; Mangala, L.S.; et al. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res. 2017, 27, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; He, J.; Pu, W.; Peng, Y. The Role of Exportin-5 in MicroRNA Biogenesis and Cancer. Genom. Proteom. Bioinform. 2018, 16, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Moya, J.; Wert-Lamas, L.; Riesco-Eizaguirre, G.; Santisteban, P. Impaired microRNA processing by DICER1 downregulation endows thyroid cancer with increased aggressiveness. Oncogene 2019, 38, 5486–5499. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Chakravarti, D.; Cho, M.S.; Liu, L.; Gi, Y.J.; Lin, Y.L.; Leung, M.L.; El-Naggar, A.; Creighton, C.J.; Suraokar, M.B.; et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 2010, 467, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Hutvagner, G.; Simard, M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008, 9, 22–32. [Google Scholar] [CrossRef]
- Janas, M.M.; Wang, B.; Harris, A.S.; Aguiar, M.; Shaffer, J.M.; Subrahmanyam, Y.V.; Behlke, M.A.; Wucherpfennig, K.W.; Gygi, S.P.; Gagnon, E.; et al. Alternative RISC assembly: Binding and repression of microRNA-mRNA duplexes by human Ago proteins. RNA 2012, 18, 2041–2055. [Google Scholar] [CrossRef]
- Zhang, J.; Jin, H.; Liu, H.; Lv, S.; Wang, B.; Wang, R.; Liu, H.; Ding, M.; Yang, Y.; Li, L.; et al. MiRNA-99a directly regulates AGO2 through translational repression in hepatocellular carcinoma. Oncogenesis 2014, 3, e97. [Google Scholar] [CrossRef]
- Ye, Z.; Jin, H.; Qian, Q. Argonaute 2: A Novel Rising Star in Cancer Research. J. Cancer 2015, 6, 877–882. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).