Abstract
Few studies have been conducted regarding the biological function and regulation role of gga-miR-221-5p in the liver. We compared the conservation of miR-221-5p among species and investigated the expression pattern of gga-miR-221-5p, validating the direct target genes of gga-miR-221-5p by dual luciferase reporter assay, the biological function of gga-miR-221-5p in the liver was studied by gga-miR-221-5p overexpression and inhibition. Furthermore, we explored the regulation of gga-miR-221-5p and its target genes by treatment with estrogen and estrogen antagonists in vivo and in vitro. The results showed that miR-221-5p was highly conserved among species, expressed in all tested tissues and significantly downregulated in peak-laying hen liver compared to pre-laying hen liver. Gga-miR-221-5p could directly target the expression of elongase of very long chain fatty acids 6 (ELOVL6) and squalene epoxidase (SQLE) genes to affect triglyceride and total cholesterol content in the liver. 17β-estradiol could significantly inhibit the expression of gga-miR-221-5p but promote the expression of ELOVL6 and SQLE genes. In conclusion, the highly conservative gga-miR-221-5p could directly target ELOVL6 and SQLE mRNAs to affect the level of intracellular triglyceride and total cholesterol. Meanwhile, 17β-estradiol could repress the expression of gga-miR-221-5p but increase the expression of ELOVL6 and SQLE, therefore promoting the synthesis of intracellular triglyceride and cholesterol levels in the liver of egg-laying chicken.
1. Introduction
MicroRNAs (miRNAs) are small, endogenous, non-coding single-strand RNAs that post-transcriptionally regulate gene expression by binding to the 3′ untranslated regions (3′UTR) of potentially hundreds of mRNAs. miRNAs are about 22 nucleotides (nt) in length [1,2], and the 2nd–8th nt of the 5′ end of miRNA, named the seed region, are crucial for recognizing and binding to target genes [3,4]. miRNAs regulate their target mRNAs by mRNA molecule degradation or translation inhibition [5,6].
Increasingly many investigations suggest that miRNAs are involved in various biological processes. Recent studies report that miRNAs exert a biological function in metabolic and endocrine pathways [7] and adipogenesis [8,9]. Emerging evidence shows that miRNAs act as key regulators for hepatic lipid metabolism. For example, the liver-specific miR-122 in mammals is abundantly expressed in liver [10] and can control the hepatic fatty acids and cholesterol synthesis by repressing the expression of mRNAs involved in the cholesterol biosynthesis pathway [11]. miR-33 can regulate the biological metabolism process of the liver by affecting cholesterol efflux and high-density lipoprotein metabolism [12]. Gga-miR-22-3p and gga-miR-101-2-5p are involved in the hepatic lipid metabolism by targeting the key factors Elovl6 and Apolipoprotein B (ApoB) of the lipid metabolism pathway [13,14]. The differential expression of miRNAs has been reported in the liver tissues of pre-laying and peak-laying hens [15], and obese and non-obese human individuals [16,17]. Most of the studies about miRNA functions are mainly focused on mammals, however, the action mechanisms of most miRNAs in chicken are unclear.
The liver plays a key role in lipid metabolism. Especially in poultry, the liver is the hub of fatty acid synthesis and lipid circulation through lipoprotein synthesis [18]. With the onset of egg production, estrogen shifts hepatocytic lipoprotein production to assemble into triacylglycerol-rich yolk very low density lipoprotein (VLDL) [19]. Egg laying is the most important economic trait in hens, and this physiological process is highly influenced by chicken liver function. Our recent study displayed that, compared with pre-laying (20 weeks) hens, gga-miR-221-5p was significantly downregulated in the liver tissue of peak-laying (30 weeks) hens [15] and gga-miR-221-5p was significantly downregulated in the liver tissue of estrogen-treated hens (10 weeks) compared with 10-week-old hens’ livers (unpublished data) [20]. Therefore, it is of great significance to investigate the biological function of gga-miR-221-5p, which might be relevant to hepatic lipid metabolism.
To date, most of the studies about miR-221-5p have focused on diseases in mammals and poultry. MiR-221-5p is located in the intergenic region and is mainly reported to be a known player in oncogenesis, affecting the proliferation and behavior of various cell types [21,22,23,24]. It is suggested that miR-221 also participates in glucose/fat metabolism. miR-221 may contribute to the development of the insulin resistance that typically accompanies obesity by regulating the peroxisome proliferator activated receptor (PPAR) signaling pathway and directly down-regulating adiponectin receptor 1 (ADIPOR1) and ETS proto-oncogene 1, transcription factor (ETS1) [25]. Obese mice show high miR-221 levels and low AdipoR1 levels [26]. MiR-221-5p is suggested to act as a Marek’s disease tumor suppressor [27]. The expression of miR-221 is significantly upregulated in ALV-infected chicken [28]. In addition, gga-miR-221 is implied to be a tumor formation-relevant gene in chicken [29]. However, little is known about the regulation role of gga-miR-221 in the chicken lipid metabolism. Therefore, in this study, we have systematically investigated the biological role and the regulation role of gga-miR-221-5p in the lipid metabolism of chicken.
2. Results
2.1. Conservative Analysis of miR-221-5p Among Species
The mature miR-221-5p sequences derived from different species were collected, and the conservatism of this among species was aligned. The alignment result showed that both the mature miR-221-5p sequences and seed regions were highly conserved among Gallus gallus, Capra hircus, Homo sapiens, Columba livia, Rattus norvegicus, Mus musculus, Sus scrofa, Cricetulus griseus, Macaca mulatta, Danio rerio, Dromaius novaehollandiae, Oryctolagus cuniculus, and Crocodylus porosus. Only one or three bases in the 3’ end of mature sequences were not matched between the two species Gadus morhua and Xenopus laevis and the others (Figure 1).

Figure 1.
Alignment of miR-221-5p mature sequence among species. Note: Gga: Gallus gallus; Chi: Capra hircus; Has: Homo sapiens; Cli: Columba livia; Rno: Rattus norvegicus; Mmu: Mus musculus; Ssc: Sus scrofa; Cgr: Cricetulus griseus; Mml: Macaca mulatta; Dre: Danio rerio; Dno: Dromaius novaehollandiae; Ocu: Oryctolagus cuniculus; Cpo: Crocodylus porosus; Gmo: Gadus morhua; Xla: Xenopus laevis.
2.2. Spatiotemporal Expression Characteristics of gga-miR-221-5p
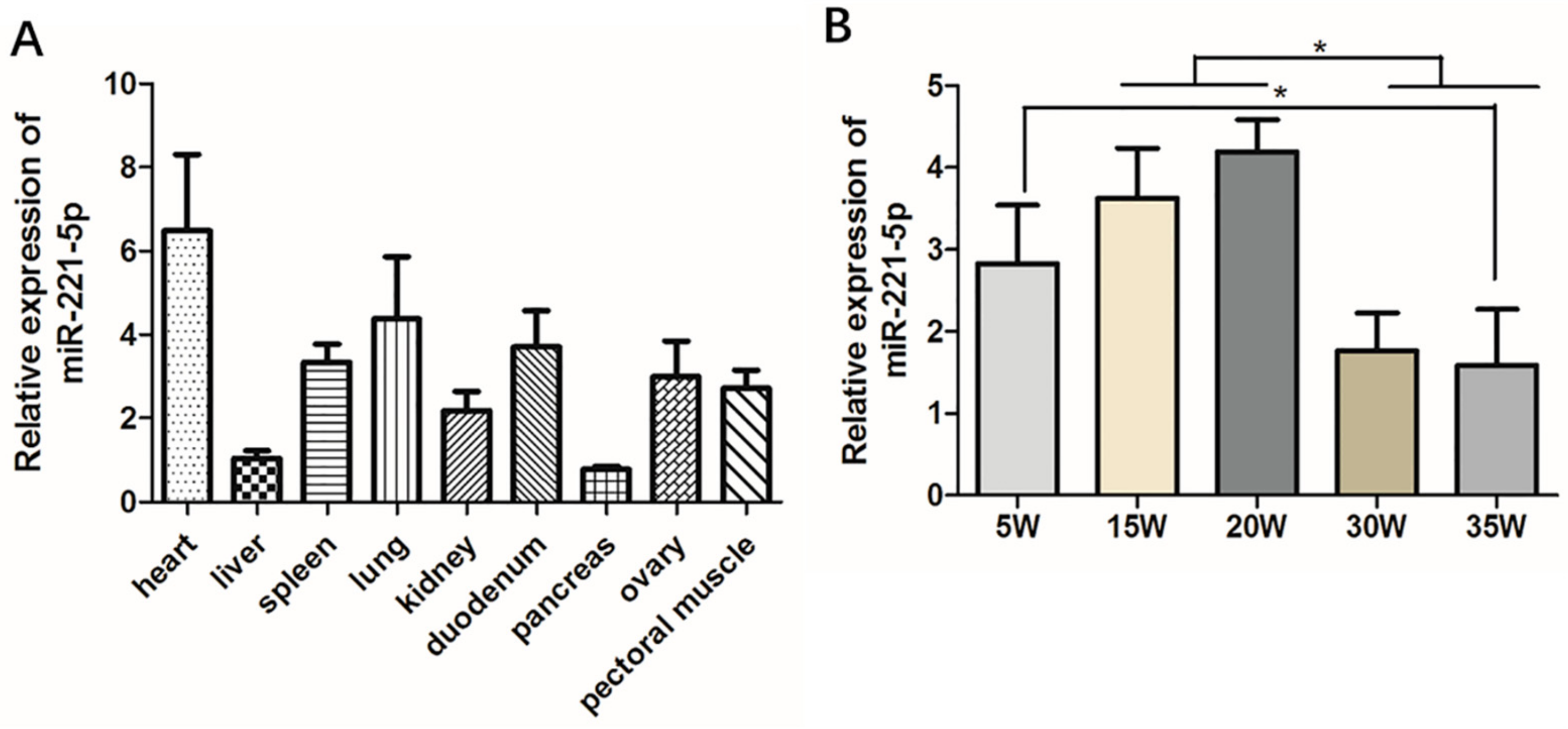
In order to initially explore the basic biological expression characteristics of gga-miR-221-5p, tissue expression distributions of gga-miR-221-5p were detected in various tissues of 30-week-old chickens. It was shown that gga-miR-221-5p was broadly expressed in different tissues (Figure 2A). The relative expression level of gga-miR-221-5p was highest in the heart and lowest in the liver and pancreas. Spatio-temporal expression pattern of gga-miR-221-5p in livers at different stages was detected (Figure 2B). The results showed that the relative expression levels of gga-miR-221-5p in the livers of chicken at the age of 15 and 20 weeks were significantly higher than at 30 and 35 weeks (p < 0.05). The expression difference was not significant among 5, 15 and 20 weeks (p > 0.05). There was also no significant difference existence between 30 and 35 weeks (p > 0.05).
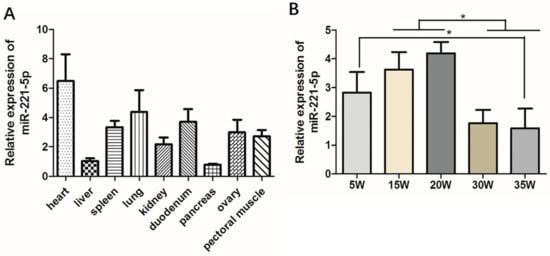
Figure 2.
Spatiotemporal expression profile of gga-miR-221-5p. (A) The relative expression of gga-miR-221-5p in different tissues. (B) The relative expression of gga-miR-221-5p in different liver stages. β-actin was used as an internal reference gene to estimate the relative expression of mRNA. U6 was used as an internal reference gene to estimate the relative expression of miRNA. Data are represented as mean ± SD (n = 6). * means 0.01 < p < 0.05.
2.3. Screening of gga-miR-221-5p Candidate Target Genes
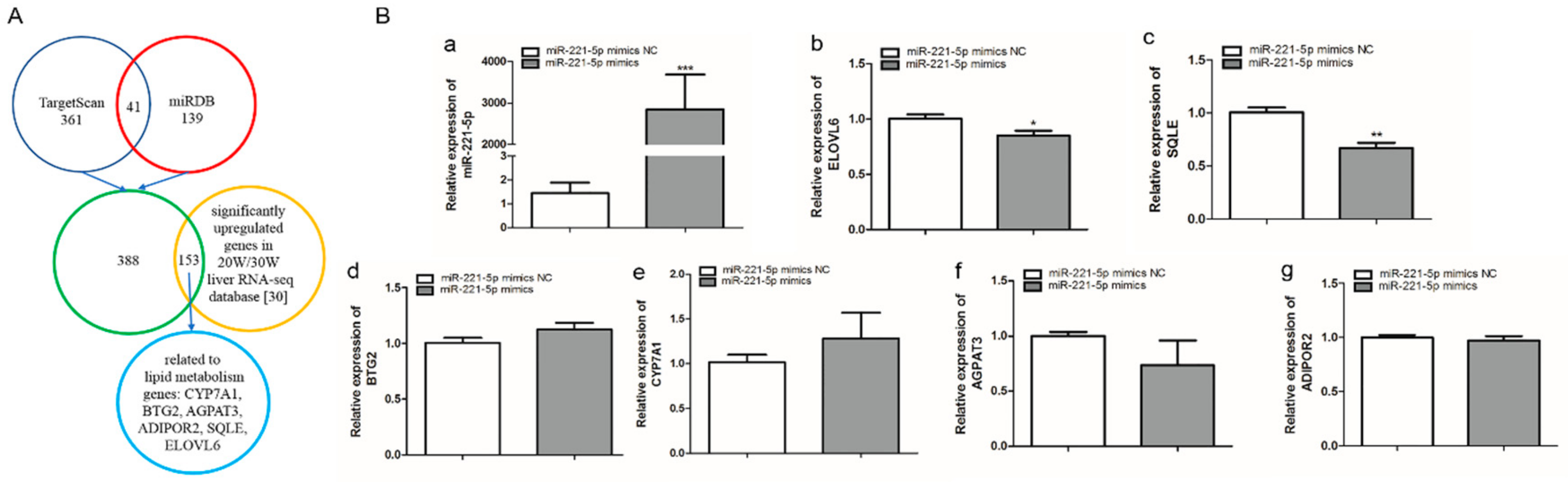
402 and 180 target genes of gga-miR-221-5p were predicted by TargetScan and miRDB online software, respectively, taking the union set of the two online software predicted target genes to obtain 541 potential target genes; these genes were intersected with the significantly upregulated genes in the 20-week and 30-week liver RNA-seq database [30], and 153 potential target genes were obtained, of which BTG anti-proliferation factor 2 (BTG2), cytochrome P450 family 7 subfamily A member 1 (CYP7A1), 1-acylglycerol-3-phosphate O-acyltransferase 3 (AGPAT3), ADIPOR2, SQLE, and ELOVL6 genes related to lipid metabolism were selected as potential target genes for further validation (Figure 3A). To further screen the possible targets, gga-miR-221-5p mimics was used to transfect LMH chicken hepatoma cells. According to the qRT-PCR analysis result, compared with the miR-221-5p mimics negative control (miR-221-5p mimic NC) treatment group, the relative expression of gga-miR-221-5p was significantly increased in the miR-221-5p mimics treated group (p < 0.0001). Only the mRNA relative expression levels of SQLE and ELOVL6 were significantly downregulated in the miR-221-5p mimics treated group (p < 0.05 or p < 0.01), and there was no significant alteration in BTG2, CYP7A1, AGPAT3, and ADIPOR2 compared to the miR-221-5p mimic NC group (p > 0.05; Figure 3B).
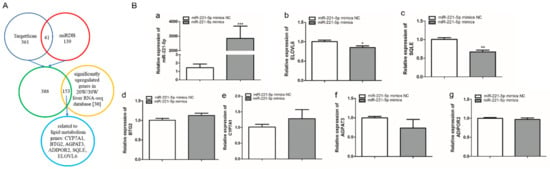
Figure 3.
Screening of gga-miR-221-5p candidate target genes. (A) The selection process of the gga-miR-221-5p target genes. (B) Relative expression levels of gga-miR-221-5p and candidate target genes after gga-miR-221-5p mimics treatment for 48 h. * means 0.01 < p < 0.05, ** means 0.001 < p < 0.01, *** means p < 0.0001.
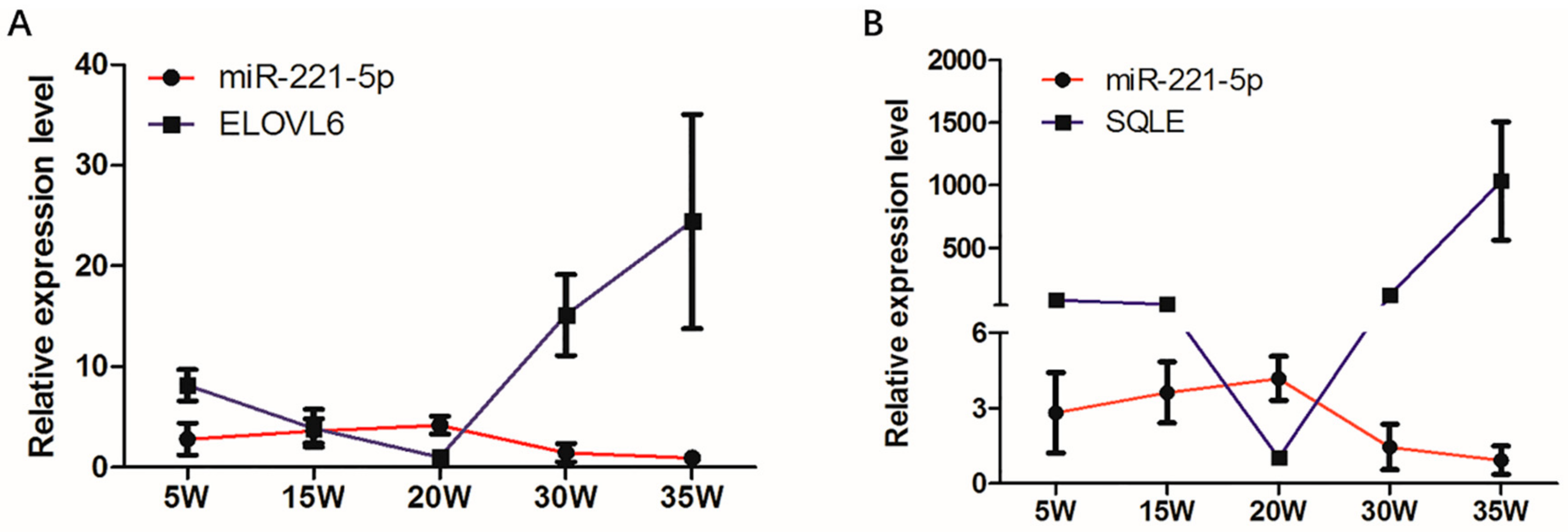
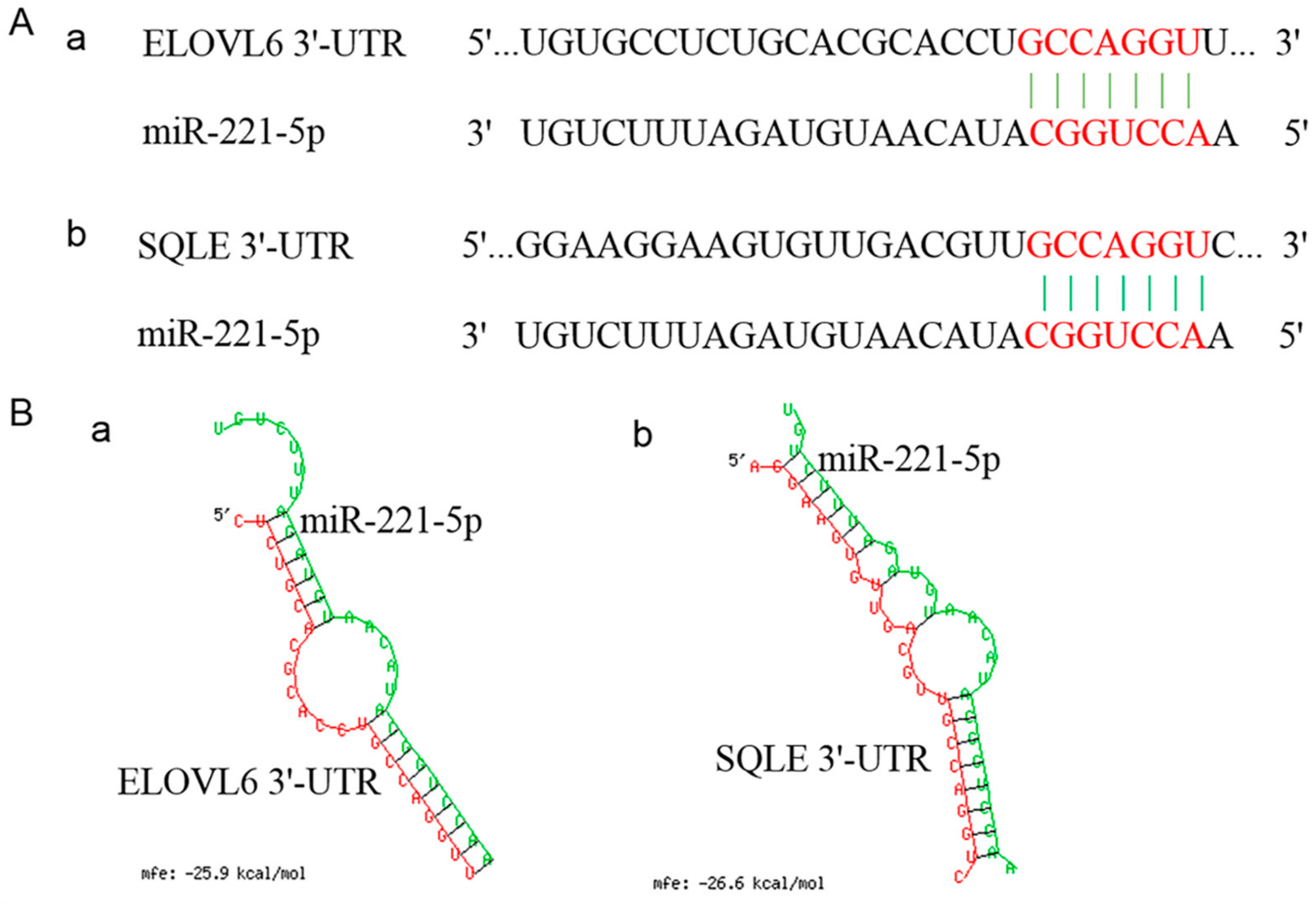
The spatio-temporal expression analysis displayed that the expression modes of ELOVL6 and SQLE in the chicken livers at different stages were completely opposite to that of gga-miR-221-5p (Figure 4). In addition, the sequences located in the 3′UTR of ELOVL6 and SQLE genes where the gga-miR-221-5p seed region was recognized are shown in Figure 5A. The MFE of gga-miR-221-5p binding to the 3′UTR of ELOVL6 and SQLE was −25.9 kcal/mol and −26.6 kcal/mol, respectively (Figure 5B), indicating that the complexes of gga-miR-221-5p binding with ELOVL6 and SQLE occurred with high stability. It was suggested that SQLE and ELOVL6 might be the possible direct target genes of gga-miR-221-5p.
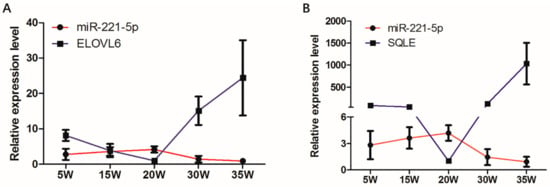
Figure 4.
Expression of gga-miR-221-5p and candidate target genes in different liver stages. (A) Expression of gga-miR-221-5p and ELOVL6 in different liver stages. (B) Expression of gga-miR-221-5p and SQLE in different liver stages. β-actin was used as an internal reference gene to estimate the relative expression of mRNA. U6 was used as an internal reference gene to estimate the relative expression of miRNA. Data are represented as mean ± SD (n = 6).
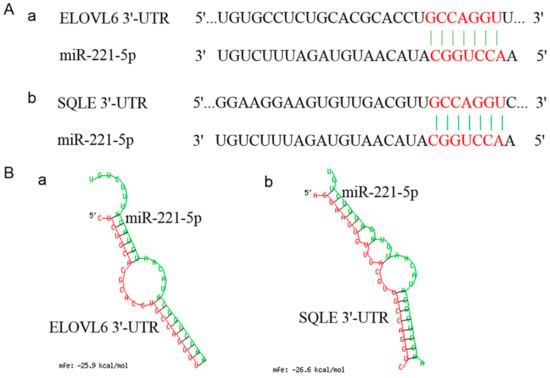
Figure 5.
Sequencing information of ELOVL6’s and SQLE’s 3′ untranslated regions (3′UTR) binding to the gga-miR-221-5p seed region and the prediction of its free energy. (A) Sequencing information of ELOVL6’s and SQLE’s 3′UTR binding to the gga-miR-221-5p seed region. (B) Free energy information of ELOVL6’s and SQLE’s 3′UTR binding to gga-miR-221-5p.
2.4. Verification of the Interaction Relationship between gga-miR-221-5p and Potential Target Genes
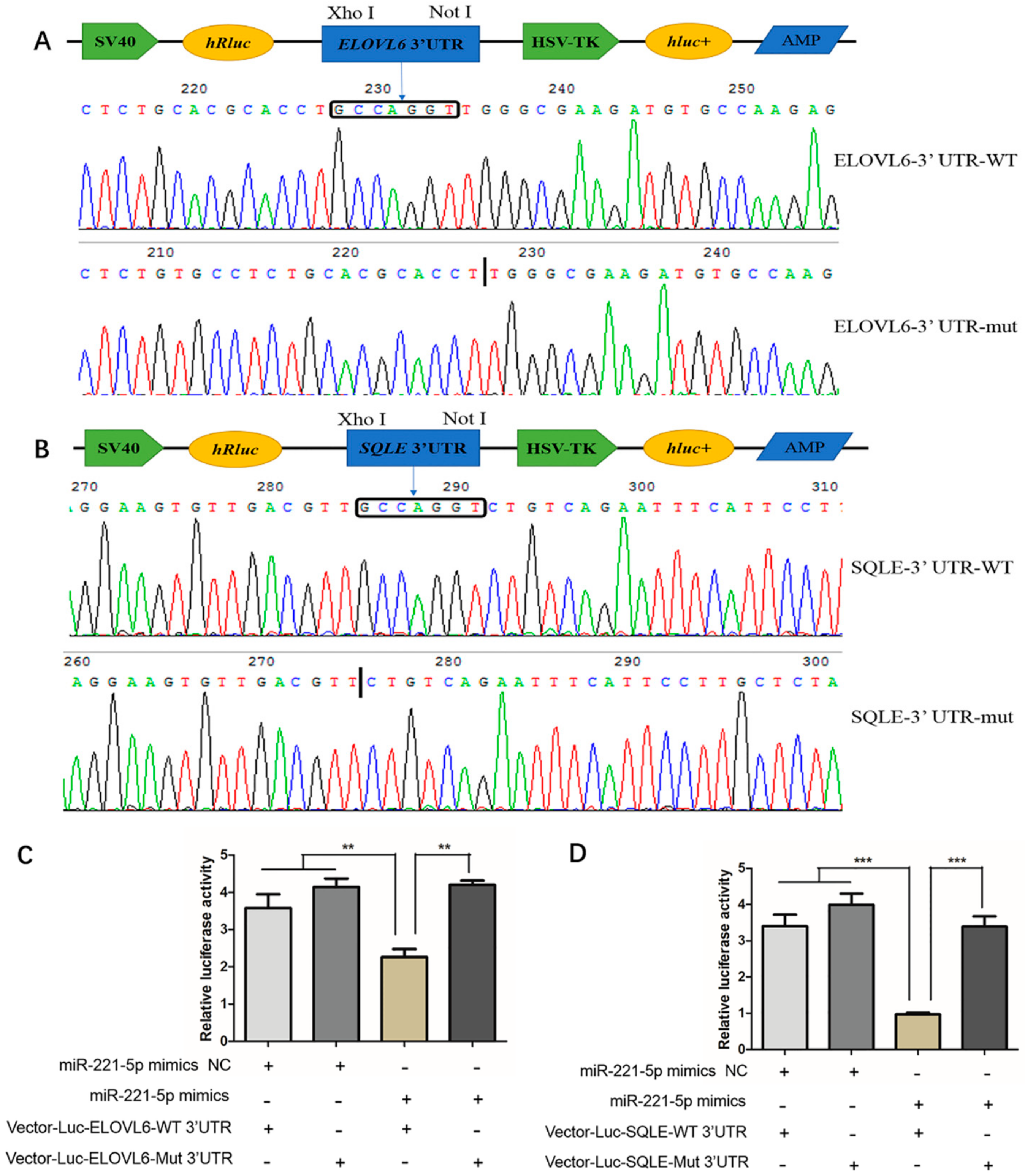
To verify whether gga-miR-221-5p interacts with ELOVL6 and SQLE genes, we constructed Vector-Luc-ELOVL6-WT 3′UTR, Vector-Luc-ELOVL6-Mut 3′UTR, Vector-Luc-SQLE-WT 3′UTR, and Vector-Luc-SQLE-Mut 3′UTR for the verification of the interaction between gga-miR-221-5p and the two potential target genes (Figure 6A,B), and the dual luciferase reporter assay was conducted in LMH chicken hepatoma cells. The results showed that, when co-transfected with gga-miR-221-5p mimics and Vector-Luc-ELOVL6-WT 3′UTR, the relative fluorescence activity was significantly inhibited (p < 0.01), and the inhibition was abolished in the gga-miR-221-5p mimics and Vector-Luc-ELOVL6-Mut 3′UTR co-treated group. No significant difference was observed among gga-miR-221-5p mimics and Vector-Luc-ELOVL6-Mut 3′UTR co-treated, gga-miR-221-5p mimics NC and Vector-Luc-ELOVL6-WT 3’UTR co-treated, gga-miR-221-5p mimics NC and Vector-Luc-ELOVL6-Mut 3’UTR co-treated groups (p > 0.05). Furthermore, the same interaction relationship was also found between gga-miR-221-5p and the SQLE gene (Figure 6C,D).
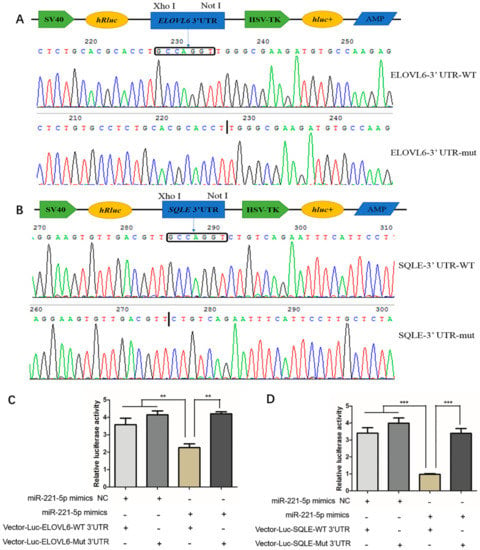
Figure 6.
Validation of gga-miR-221-5p and the ELOVL6 and SQLE targeting relationship. (A,B) Recombinant plasmid sequencing. (C,D) Dual luciferase reporter assay analysis of the target relationship of gga-miR-221-5p with ELOVL6 and SQLE in LMH chicken hepatoma cells. The long box □ indicates the position of the gga-miR-221-5p seed region in combination with the ELOVL6 and the SQLE gene 3′UTR, the black vertical line indicates the position of the ELOVL6 and SQLE gene 3′UTR deletion binding to the gga-miR-221-5p seed region. WT indicates wild, Mut indicates mutation, + indicates added, - indicates not added. Data are represented as mean ± SD (n = 6). ** means 0.001 < p < 0.01, *** means p < 0.0001.
2.5. Effect of gga-miR-221-5p on Intracellular Triglyceride and Total Cholesterol Content
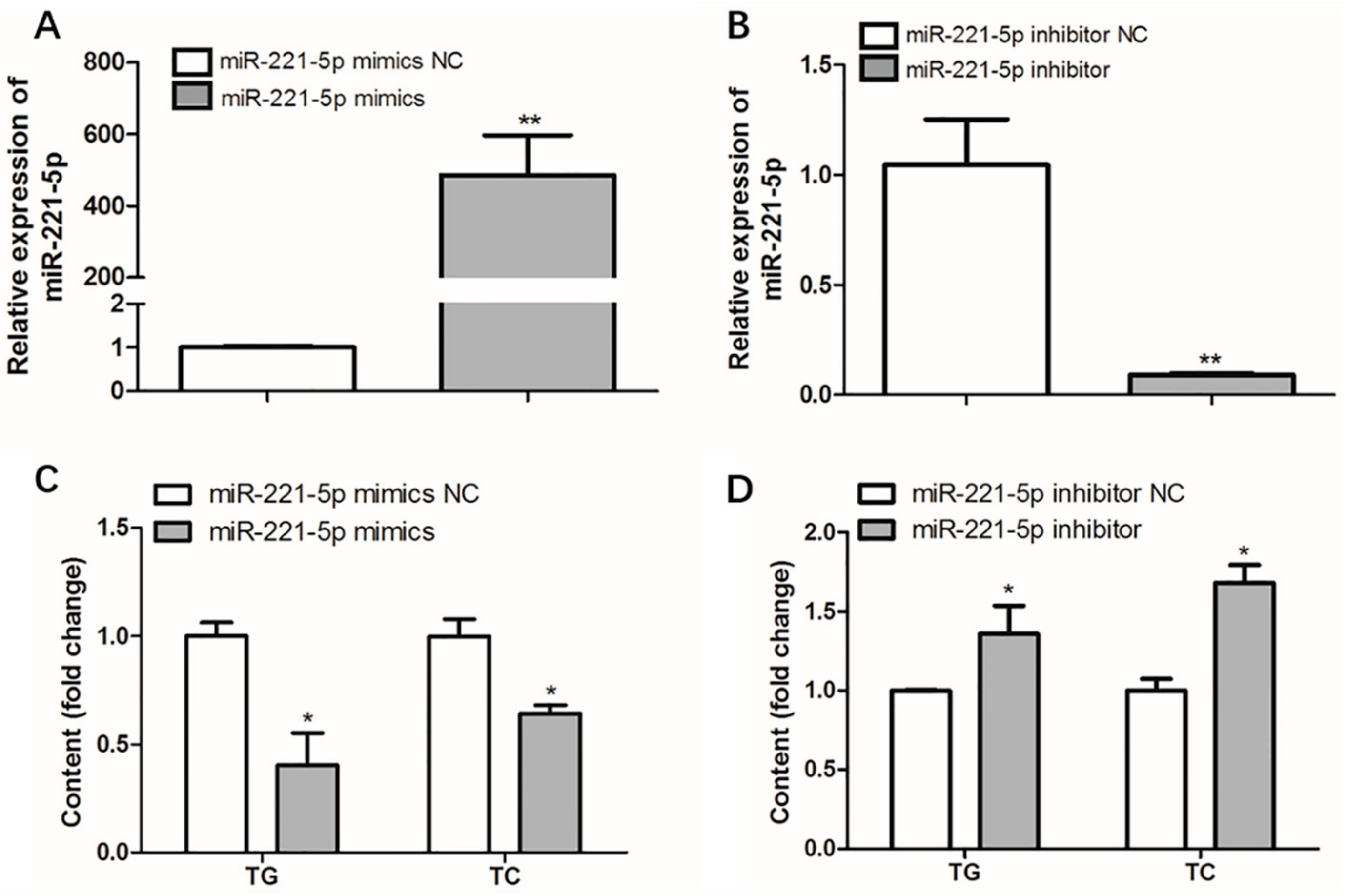
To measure the effect of gga-miR-221-5p on intracellular triglyceride (TG) and total cholesterol (TC) levels, over-expression and interference assays of gga-miR-221-5p were carried out, respectively. The over-expression experiment result showed that the expression level of gga-miR-221-5p was upregulated by approximately 500-fold compared with the control group (miR-221-5p mimics NC group). Interestingly, the levels of intracellular TG and TC were significantly downregulated in the miR-221-5p mimics group compared with the miR-221-5p mimics NC group (p < 0.05). The interference experiment showed that the expression level of gga-miR-221-5p was downregulated by approximately 90%, and the contents of intracellular TG and TC were significantly upregulated in the miR-221-5p inhibitor group than the miR-221-5p inhibitor NC group (p < 0.05; Figure 7).
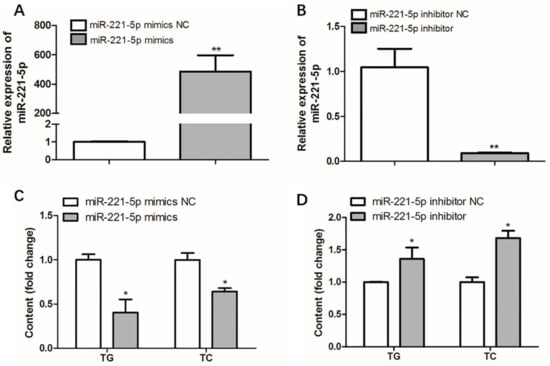
Figure 7.
Effect of gga-miR-221-5p overexpression and inhibition on intracellular triglycerides (TG) and cholesterol (TC). (A,B) The expression of gga-miR-221-5p under the gga-miR-221-5p overexpression and inhibition in LMH chicken hepatoma cells, respectively. (C,D) Changes of TG and TC contents under gga-miR-221-5p overexpression and inhibition in LMH chicken hepatoma cells, respectively. Data are represented as mean ± SD (n = 6). * means 0.01 < p < 0.05, ** means 0.001 < p < 0.01.
2.6. 17β-Estradiol Repressed the Expression of gga-miR-221-5p and Promoted the Expression of Target Genes in Chicken Primary Hepatocytes
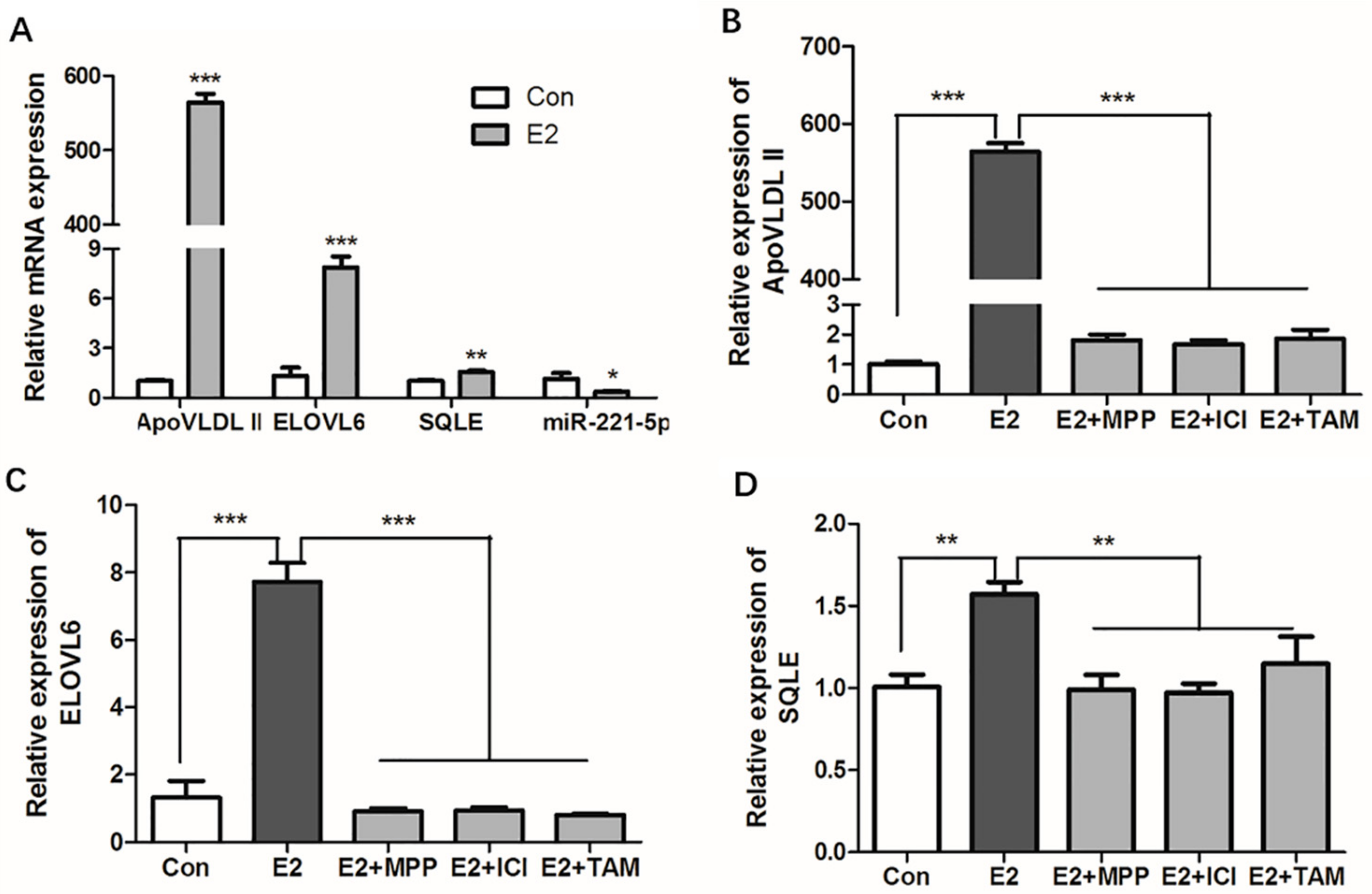
Estrogen is an important regulator in lipid metabolism, and 17β-estradiol was used to stimulate the primary hepatocytes. The results showed that the estrogen response marker gene, apovitellenin very low density lipoprotein Ⅱ (ApoVLDL II) [31], was significantly upregulated by 17β-estradiol (p < 0.0001) in chicken primary hepatocytes, which indicated the estrogen exerted effects in primary hepatocytes, and an in vitro estrogen induction model was successfully constructed. Compared with the control group, gga-miR-221-5p was significantly repressed (p < 0.05); in contrast, the expression of ELOVL6 and SQLE was significantly promoted by 17β-estradiol (p < 0.0001 or p < 0.001) in chicken primary hepatocytes (Figure 8A).
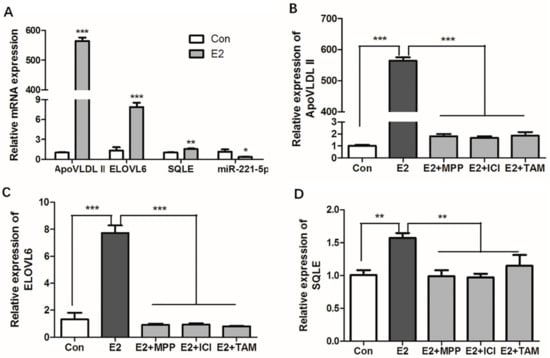
Figure 8.
Effects of estrogen and estrogen antagonists on the expression of ApoVLDL II, gga-miR-21-5p, ELOVL6, and SQLE gene in vitro. (A) Effects of estrogen on the expression of ApoVLDL II, gga-miR-21-5p ELOVL6, and SQLE gene in vitro. (B–D) Effects of estrogen and estrogen antagonists co-treated on the expression of ApoVLDL II, ELOVL6, and SQLE gene in vitro. Con indicates control group, E2 indicates 17β-estradiol (100nM) treated group, E2 + MPP indicates 17β-estradiol (100 nM) and MPP co- treated group, E2 + ICI indicates 17β-estradiol (100 nM), and ICI 182,780 co-treated group, E2 +TAM indicates 17β-estradiol (100 nM), and tamoxifen co-treated group, respectively. β-actin was used as an internal reference gene. Data are represented as mean ± SD (n = 6). * means 0.01 < p < 0.05, ** means 0.001 < p < 0.01, *** means p < 0.0001.
The classic model of estrogen action requires that the estrogen receptor (ER) activates gene expression. To know which ER subtype estrogen acts through, chicken primary hepatocytes were treated with different ER antagonists. The literature states that ApoVLDL II gene is regulated by ERα [31], and the expression of the ApoVLDL II gene under the estrogen antagonist treatment results indicated that estrogen antagonist treatment experiment was successful (Figure 8B). The results showed that 17β-estradiol could significantly increase the mRNA expression levels of ELOVL6 and SQLE in primary hepatocytes (p < 0.0001 or p < 0.001). Compared with the 17β-estradiol treatment group, the expressions of ELOVL6 and SQLE were significantly decreased in the ER antagonist treatment groups (p < 0.0001 or p < 0.001). The expression levels of the genes returned to the control level in the ER subtype antagonist-treated groups (p > 0.05; Figure 8C,D). Online software predicted results showed that atypical estrogen responsive elements (ERE) binding sites (GGGCACTAAGACC and GCTCACATTGTCC) were found in the -5000 bp promoter region of ELOVL6 and SQLE genes, respectively, and the SQLE gene was also found in our chip database established by estrogen receptor alpha chromatin immunoprecipitation experiment (unpublished data) [20]. Since methyl-piperidino-pyrazole (MPP) is the antagonist of estrogen receptor alpha (ERα), the ICI 182,780 and tamoxifen (TAM) are the antagonists of ERα and estrogen receptor beta (ERβ). The expression of ELOVL6 and SQLE in 17β-estradiol and ICI 182,780 co-treated, 17β-estradiol and tamoxifen co-treated groups did not decrease further compared with the 17β-estradiol and MPP co-treated group. This revealed that estrogen could activate the expression of SQLE via ERα, while estrogen might activate the expression of ELOVL6 via other estrogen receptors or intermediates.
2.7. Effect of 17β-Estradiol on gga-miR-221-5p and the Target Genes in Chicken Liver
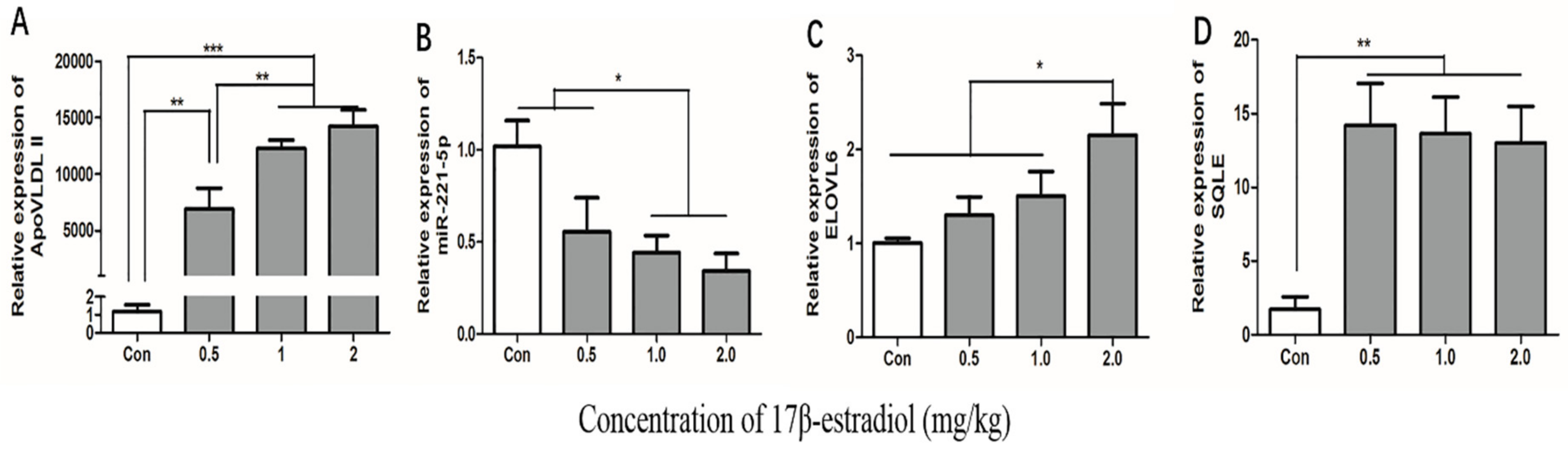
To estimate whether the 17β-estradiol acted similarly on the expression of gga-miR-221-5p and the target genes in vivo as in vitro, chickens were treated with different doses of 17β-estradiol through intramuscular injection. The effect of 17β-estradiol on the expressions of gga-miR-221-5p and the target genes in the liver of chickens were detected by using qRT-PCR (Figure 9). The ApoVLDL II expression level, a marker gene [31], was significantly increased in a dose-dependent manner under the action of estrogen, indicating that we successfully constructed an estrogen-treated biological model in vivo. The expression of gga-miR-221-5p was significantly decreased in the 17β-estradiol 1.0 mg/kg and 2.0 mg/kg treated groups (p < 0.05). Conversely, the relative expression levels of ELOVL6 were significantly upregulated in the 2.0 kg/mg 17β-estradiol group (p < 0.05), and the relative expression levels of SQLE were significantly upregulated in the 17β-estradiol group (p < 0.01).
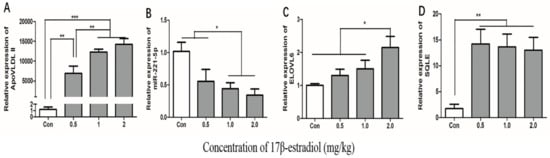
Figure 9.
Effects of estrogen on the expression of ApoVLDL II, gga-miR-221-5p, ELOVL6, and SQLE gene in vivo. Con indicates control group. β-actin was used as an internal reference gene. U6 was used as an internal reference gene to estimate the relative expression of miRNA. Data are represented as mean ± SD (n = 8). * means 0.01 < p < 0.05, ** means 0.001 < p <0.01, *** means p < 0.0001.
3. Discussion
Numerous studies have shown that miRNAs, as a conserved endogenous regulator in vivo, participate in various metabolic processes of organisms [32,33]. In this study, it was found that the mature sequence of miR-221-5p was relatively highly conserved among species, suggesting that miR-221-5p played an important biological function among species. It has been reported that hsa-miR-221-5p regulates the proliferation and metastasis of prostate cancer cells by targeting the expression of the suppressor of cytokine signaling 1 (SOCS1) gene [34]. The analysis of miRNA transcriptomes in Marek’s disease (MD) lymphoma showed that gga-miR-221-5p was significantly downregulated in MD lymphoma, and gga-miR-221-5p inhibited the proliferation of MDV-transformed lymphoid cell line (MSB1), suggesting that gga-miR-221-5p had an anti-cancer effect [27]. The expression profiles of gga-miR-221-5p in different tissues of chicken at the age of 30 weeks showed that it was expressed in a broad-spectrum in various tissues, indicating that gga-miR-221-5p might exert certain biological effects in various tissues of chicken as in other species. The expressions of the liver at different stages showed that the expression level of gga-miR-221-5p at the peak-laying stage was significantly lower than that of the pre-laying stages. The most physiological difference between the Lushi chickens at the age of 20 weeks and 30 weeks was in whether they laid or not; therefore, this further suggests that gga-miR-221-5p might be involved in the chicken egg-laying related processes.
As is well known, miRNAs are involved in different biological processes by regulating the expression of target genes at the post-transcriptional level by binding to the target gene 3′UTR [5,6]. One miRNA may regulate the expression of multiple target genes, and one gene may also be regulated by multiple miRNAs [35]. Gga-miR-221-5p was demonstrated to directly interact with ELOVL6 and SQLE genes by binding to the 3′UTR of mRNAs with high stability, and controlling the expression of ELOVL6 and SQLE at the post-transcriptional level. The ELOVL6 gene, a member of the fatty acyl elongase (ELOVL) gene family, is the only enzyme that participated in de novo lipogenesis and is responsible for the final step of endogenous saturated fatty acid synthesis, which plays an important role in the fatty acid synthesis pathway [36]. It was found that the ELOVL6 promoter region has a QTL polymorphism site that can regulate the composition of porcine fatty acid [37]. Studies in poultry have found that single nucleotide polymorphisms in the ELOVL6 gene are associated with the deposition of subcutaneous fat [38]. SQLE located on the endoplasmic reticulum is the first oxygenation step in the biosynthesis of sterols and is considered to be one of the key enzymes in this pathway, which has an important impact on the flux synthesis of cholesterol [39]. An abnormal expression of the SQLE gene in abnormal cholesterol metabolism cells has been reported, such as prostate cancer, breast cancer, and lung cancer [40,41,42]. The function loss or gain assays of gga-miR-221-5p showed a promotion or inhibition role on the content of intracellular TG and TC, respectively. Our result demonstrated that gga-miR-221-5p participated in the lipid metabolism pathway by regulating the expression of the ELOVL6 and SQLE genes.
The relative expression between gga-miR-221-5p and ELOVL6 or SQLE in the liver at different stages showed a negative correlation, which revealed that gga-miR-221-5p might be involved in different physiological periods by targeting the ELOVL6 and SQLE genes in a chicken. It was interested to found that gga-miR-221-5p was significantly downregulated at the egg-laying stages compared with pre-laying stages. Studies report that, with the arrival of sexual maturity, estrogen levels gradually rise and reach the highest level at sexual maturity (i.e., when the first egg is laid) [43,44]. During the egg-laying stage, under the promotion of estrogen, a large number of neutral lipids including triacylglycerols, cholesteryl esters, and free fatty acids are synthesized in the liver, then assembled into very-low-density lipoprotein (VLDL) and vitellogenin particles and transported to ovary to generate the egg-yolk precursors through blood circulation [45,46]. Genes involved in this lipid synthesis process, including ELOVL6 related to fatty acids and SQLE related to cholesterol, were upregulated by estrogen at the genome level, while estrogen repressed the expression of gga-miR-221-5p, which was proved by in vitro and in vivo trial. Moreover, we found that the TG and TC contents were also dramatically increased under the estrogen treatment in chicken [47], and our results were consistent with previous studies [48]. Meanwhile, it is well known that liver X receptor alpha (LXR-α) is also an important regulators of liver lipid metabolism. LXRα is functional when heterodimerized with retinoid x receptors (RXRs), and regulated the transcription of their target genes by binding to specific response elements (LXREs) that contain a hexametric nucleotide direct repeat spaced by four bases (DR4) [49]. A previous study showed that LXR gene knockout mice blocked cholesterol metabolism by downregulating ATP-binding cassette A1 (ABCA1) gene expression [50]. In many species, the activation of LXR-α can promote the biosynthesis of cholesterol and fatty acids, key genes for lipid metabolism such as SREBP-1c, Acetyl-CoA carboxylase (ACC) and fatty acid synthase (FASN) gene expression levels are significantly upregulated [51,52,53]. While, LXR-α was not significantly changed in our databases from the liver transcriptome data of the pre- and peak-laying hens [30], and the liver transcriptome data of estrogen-treated hens and untreated hens (unpublished data) [20]. It was suggested that LXR-α might be not involve in the estrogen signal pathway.
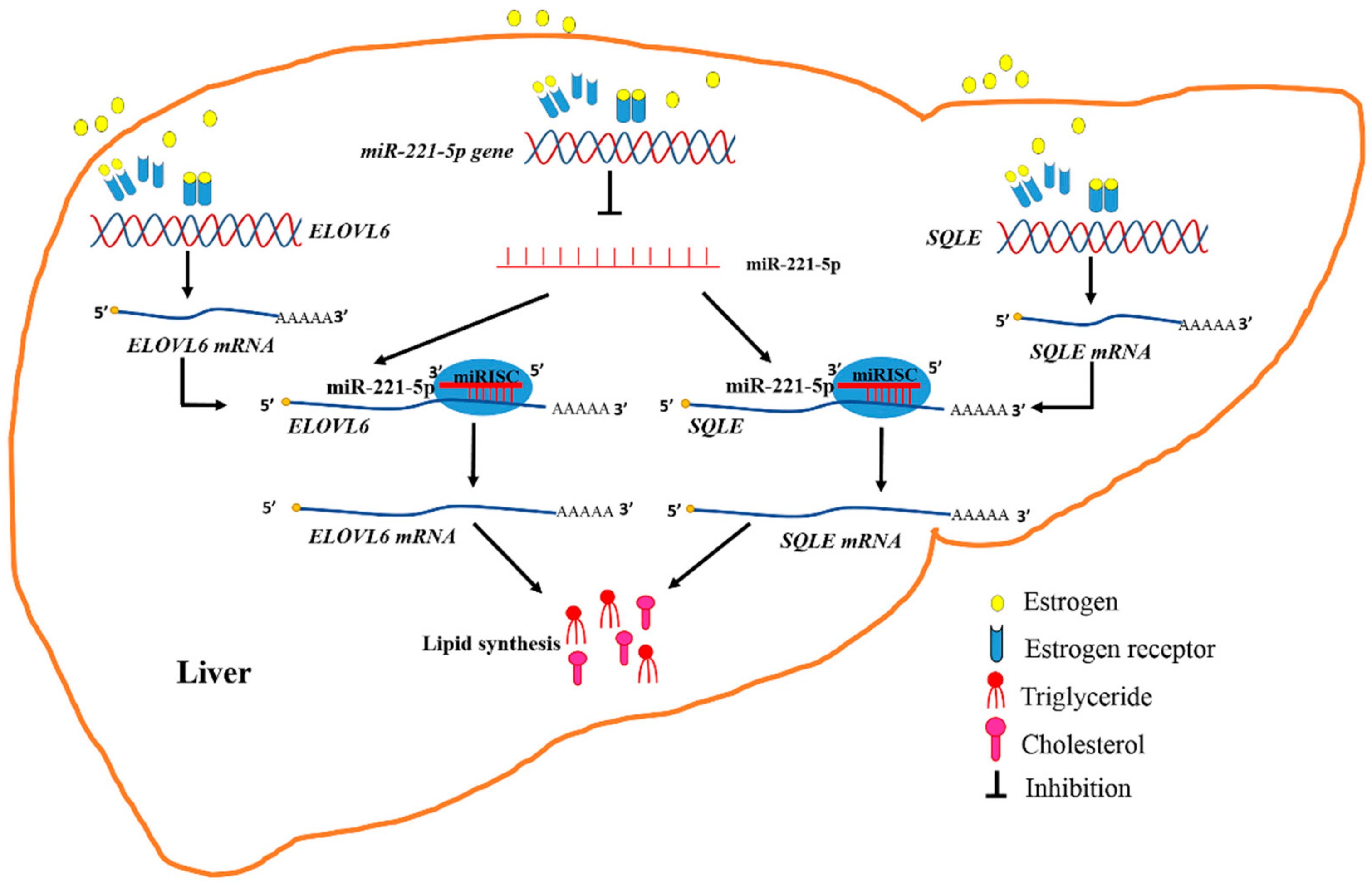
Therefore, it was determined that estrogen abolished the inhibitory effect of gga-miR-221-5p on targeting ELOVL6 and SQLE at post-transcriptional levels during the egg-laying period; meanwhile, estrogen promoted the expression of ELOVL6 and SQLE, which resulted in a dramatic increase in TG and TC content in chicken (Figure 10). Generally, estrogen regulates gene expression by signaling through intracellular hormone-specific estrogen receptors (ERα and ERβ). The canonical model for ER-mediated regulation of gene expression involves the direct binding of dimeric ER to DNA sequences known as estrogen response elements (EREs) or the AP1 enhancer element [54,55,56]. If estrogen signaling at a classical ERE, both ERα and ERβ showed the same transactivation profiles. However, if estrogen signaling at the AP1 element, ERα and ERβ would respond differently. The ligand-induced responses with ERβ at an AP1 site showed a negative transcriptional regulation by the natural hormone. Our research found that estrogen promoted the expression of the SQLE gene through ERα, but estrogen might promote the expression of the ELOVL6 gene through the mediation of intermediates. The expression of gga-miR-221-5p was downregulated by estrogen, and an AP1-binding site existed in the −5000 bp region upstream of pri-miR-221-5p (TGAGTCA). It implied that estrogen-binding ERβ might form a complex with Jun/Fos, and binding to the AP1 site, thereby ultimately inhibiting the formation of gga-miR-221-5p. In addition, some other regulation mechanism would be also possible. For example, it was also reported that some other regulators such as long non-coding RNA (lncRNA) controlled by estrogen, and lncRNA could sponge miRNAs and interfere with miRNA expression [57]. Therefore, the regulation mechanisms of estrogens interfere with miR-221-5p expression are complex, and needed to be studied further in the future.
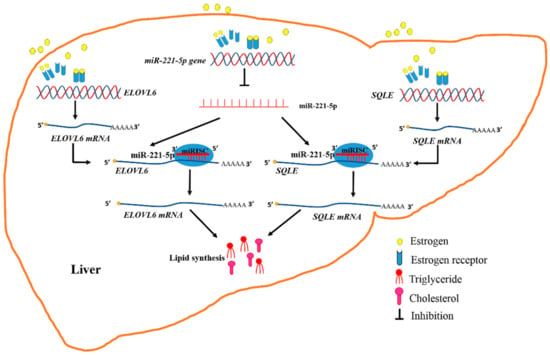
Figure 10.
Diagram illustrating the underlying mechanism by which that estrogen and miR-221-5p target ELOVL6 and SQLE to participate in chicken liver lipid metabolism.
To date, this is the first evidence of the biological function of gga-miR-221-5p controlling the lipid metabolism in the liver of egg-laying chicken. In addition, human lipid metabolism diseases are affected by many factors including lncRNA [58], miRNA [59], transcription factors [60], leucine deficiency [61], etc., and this has always been a hot topic in human research. The literature showed that gga-miR-33 can participate in lipid metabolism by targeting fat mass and obesity-associated (FTO) genes associated with obesity in chicken liver [1], and miR-33 can regulate cholesterol and fatty acid metabolism in mammals (humans and mice), which corresponds to the function of the host gene sterol regulatory element binding transcription factor 2 (SREBP2), which can regulate the synthesis and uptake of triglycerides and cholesterol [62,63], it can be seen that miR-33 functions similarly in chickens and mammals (humans and mice). Other studies have shown that, in human and mouse liver cells, the overexpression of miR-34a can inhibit the expression of hepatocyte nuclear factor 4 alpha (HNF4α) and increase the accumulation of TG. The knockdown of miR-34a can inhibit lipid accumulation and reduce liver cell steatosis [64]. In chicken liver, miR-34a-5p promotes the accumulation of TG by regulating the expression of long-chain acyl-CoA synthetase 1 (ACSL1), and it has a function similar to that of human and mouse hepatocytes, thus achieving the positive regulation of fatty acid synthesis [65]. Interestingly, miR-221-5p was highly conserved among species including mammals, and gga-miR-221-5p was involved in liver lipid metabolism; our study results may also provide a valuable reference for humans in lipid study.
In summary, the highly conserved gga-miR-221-5p was proved to directly target ELOVL6 and SQLE mRNAs to affect the level of intracellular triglyceride and total cholesterol in LMH chicken hepatoma cells. Meanwhile, 17β-estradiol could repress the expression of gga-miR-221-5p but increase the expression of ELOVL6 and SQLE mRNAs, thereby promoting the synthesis of intracellular triglyceride and cholesterol levels in the liver of egg-laying chickens.
4. Materials and Methods
4.1. Animal Ethics
Animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Henan Agricultural University, China (Permit Number: 11-0085; Date: 13 June 2011).
4.2. Treatment and Sample Collection
Chinese, locally breed Lushi green-shell hens were used as the experimental animals. Tissue samples including heart, liver, spleen, lung, kidney, duodenum, pancreas, ovary, and pectoral muscles of eight 30-week-old Lushi green-shell hens were collected at −80 °C after liquid nitrogen freezing. Liver samples of 5, 15, 20, 30, and 35-week-old Lushi green-shell hens were collected and stored at −80 °C after liquid nitrogen freezing.
Thirty-two healthy 10-week-old hens were randomly divided into four groups, with eight birds in each group. They were feed in the same environment with free access to feed and water. 17β-estradiol was dissolved in anhydrous ethanol. The birds in the three experimental groups were intramuscularly injected with 17β-estradiol at the final concentration of 0.5, 1.0, and 2.0 mg·kg−1 body weight, respectively. The birds in the control group were administered an equal volume of anhydrous ethanol. After treatment for 12 hours, chickens in each group were slaughtered, and the liver tissues were collected and stored at −80 °C after liquid nitrogen freezing.
4.3. Target Gene Prediction of gga-miR-221-5p and Bioinformatics Analysis
The miR-221-5p mature sequences of different species including Gallus gallus, Capra hircus, Homo sapiens, Columba livia, Rattus norvegicus, Mus musculus, Sus scrofa, Cricetulus griseus, Macaca mulatta, Danio rerio, Dromaius novaehollandiae, Oryctolagus cuniculus, Crocodylus porosus, Gadus morhua, and Xenopus laevis were downloaded from the miRBase database (http://www.mirbase.org/), and used to analyze the conservatism. The target genes of gga-miR-221-5p were predicted as shown in Figure S1. In detail, the TargetScan 7.1 (http://www.targetscan.org/vert_71/) and miRDB (http://www.mirdb.org/) were used to predict the target genes of gga-miR-221-5p. The potential targets were obtained based on the intersection between the predicted target genes and the differentially upregulated genes in the liver transcriptome of the pre-and peak-laying hens [30]. The genes related to lipid metabolism including BTG2, CYP7A1, ADIPOR2, AGPAT3, SQLE, and ELOVL6 were selected for further experiment.
The minimum free energy (MFE) of the gga-miR-221-5p binding to the candidate target genes was calculated using hybrid online software (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid). The ERE sequence matrix was obtained from JASPAR (http://jaspar.genereg.net/). The promoter sequences of gga-pri-miR-221-5p, ELOVL6, and SQLE were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/gene), then, they were used to analyze the ERE sites via the online software FIMO (http://meme-suite.org/tools/fimo).
4.4. Vector Construction
The 3′UTR fragments of ELOVL6 and SQLE (including Xho I and Not I restriction endonuclease sites) including the binding sites of gga-miR-221-5p were cloned, respectively. Primers used for cloning were designed on the NCBI website (primer information in Table 1). The 3′UTR fragment including the wild-type gga-miR-221-5p binding region was amplified by PCR, and the 3′UTR fragment including the mutant-type gga-miR-221-5p binding region was amplified by overlap-PCR. PCR amplification program: 95 °C for 5 min; 35 cycles at 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s, followed by 72 °C for 10 min. The amplified products were purified and retrieved by using the DNA purification and recovery kit instructions (Tiangen, Beijing, China). Then, the restriction enzymes Xho I and Not I were used to digest the psiCHECK2 plasmid (Promega, Madison, WI, USA) and the retrieved target DNA fragments. Finally, the digested psiCHECK2 vector and target DNA fragments are ligated with T4 DNA ligase (NEB, Beijing, China). The four recombinant vectors were named Vector-Luc-ELOVL6-WT 3′UTR, Vector-Luc-ELOVL6-Mut 3′UTR, Vector-Luc-SQLE-WT 3′UTR and Vector-Luc-SQLE-Mut 3′UTR, respectively. All the recombinant vectors were confirmed by sequencing (BGI, Wuhan, China). The recombinant plasmids were extracted by using the EndoFree Plasmid Maxi Kit (Tiangen, Beijing, China), and stored at −20 °C until use.

Table 1.
Primer sequences used for dual-luciferase reporter vector construction.
4.5. Cell Culture and Treatment
LMH chicken hepatoma cells were cultured in DMEM/F12 medium containing 10% fetal bovine serum (FBS), 1% penicillin G (100 U/mL), and streptomycin (100 μg/mL), and cultured in an incubator containing 5% CO2 at 37 °C. When cells fusion reached 70% in the six-well plate, miR-221-5p mimics and miR-221-5p mimics NC were transfected to the cells using lip2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The cells were collected after 48 h.
When sterilized specific pathogen free (SPF) eggs hatched to 17 embryos, they were removed and placed on a clean bench. Then, we took out chicken embryos and collected liver tissue, washed the liver using D-Hank’s solution and cut it into pieces and digested it with collagenase IV. After digestion was completed, it was filtered through 100-mesh and 500-mesh cell sieves to obtain cell suspension, centrifuged for 5 minutes at 1000 r·min−1 and repeated three times, then, the supernatant was aspirated and resuspended by adding Williams’s medium, and hepatocytes were purified by Percoll gradient centrifugation. The purified hepatocytes were resuspended in Williams’s medium, planted in six-well plates at 5 × 105 cells·mL−1, and placed in a 37 °C, 5% CO2 incubator. We changed the medium after 12 h, when the cell fusion reached 80%, we changed to serum-free medium for 6 hours, and then used 0, 25, 50, and 100 nM 17β-estradiol treatment, respectively. The cells were collected after 12 h.
Estrogen action was mediated by liganded ER. Different estrogen receptor antagonists were used to estimate the type of liganded ER that delivered the estrogen action. The most ERα-selective compound was methyl-piperidino-pyrazole (MPP). The ICI 182,780 and tamoxifen (Sigma-Aldrich, Shanghai, China) were the antagonists of ERα and Erβ, in addition, they could act as agonists on GPR30 due to their high affinity for it. They were dissolved with dimethyl sulfoxide (DMSO). When primary hepatocyte fusion reached to 80%, starvation was performed for 6 h using serum-free medium and treated with MPP (100 mg· L−1), ICI (100 mg· L−1), and TAM (100 mg· L−1) for 3 h, respectively; then, we added 100 nM 17β-estradiol for 12 h. In addition, an equal volume of absolute ethanol treatment for 12 h and 100 nM 17β-estradiol treatment for 12 h were used as the blank control and positive control, respectively. Each group used three replicates. Finally, primary hepatocytes were collected and stored at −80 °C until use. The experiment was repeated independently three times.
4.6. Dual Luciferase Reporter Assay
When LMH chicken hepatoma cells fusion reached 70% in the 24-well plate, the cells were transfected with different combinations, including gga-miR-221-5p mimics combined with Vector-Luc-ELOVL6-WT 3′UTR, gga-miR-221-5p mimics combined with Vector-Luc-ELOVL6-Mut 3′UTR, gga-miR-221-5p mimics NC combined with Vector-Luc-ELOVL6-WT 3′UTR, gga-miR-221-5p mimics NC combined with Vector-Luc-ELOVL6-Mut 3′UTR, gga-miR-221-5p mimics combined with Vector-Luc-SQLE-WT 3′UTR, gga-miR-221-5p mimics combined with Vector-Luc-SQLE-Mut 3′UTR, gga-miR-221-5p mimics NC combined with Vector-Luc-SQLE-WT 3′UTR, and gga-miR-221-5p mimics NC combined with Vector-Luc-SQLE-Mut 3′UTR, respectively. Three biological repeats were performed in each group. Each group was set three replicates. After 48 h, the activity of the double fluorescence was detected according to the kit instructions (Promega, Madison, WI, USA).
4.7. Intracellular Triglyceride and Cholesterol Detection
To determine the effect of gga-miR-221-5p on the contents of intracellular TG and TC, gga-miR-221-5p mimics and inhibitors were transfected into LMH chicken hepatoma cells for 48 hours, respectively. The cells were washed twice with PBS and lysed with lysate according to the kit. Then, the supernatant of cell lysate was incubated at 70 °C for 10 minutes at room temperature and centrifuged at 2000× g for 5 minutes; finally, the supernatant was used to measure intracellular TG and TC contents according to Cell TG and T-CHO ELISA kit instructions (Applygen, Beijing, China), respectively. The total protein content in cells was determined according to the instructions of the Bradford Protein Quantification Kit (Applygen, Beijing, China).
4.8. cDNA Synthesis and Quantitative Real-Time PCR (qRT-PCR)
The total RNA of tissues and cells was extracted with the Trizol reagent according to the manufacturer’s instructions (Takara, Kyoto, Japan). The RNA quality was detected by agarose gel electrophoresis and ultraviolet spectrophotometer and stored at −80 °C According to the instructions of the reverse transcription kit (Takara, Kyoto, Japan), random primers were used for reverse transcription to obtain cDNA for mRNA amplification. The miRNA specific hairpin primers synthesized by RiboBio were used for reverse transcription to obtain cDNA for miRNA amplification.
The sequence information of qRT-PCR primers was listed in Table 2. All primers were synthesized by Shangya except gga-miR-221-5p primers, and gga-miR-221-5p primers were synthesized by RiboBio. The qRT-PCR was carried out according to the instructions of the QuantiFast SYBR Green PCR kit (Takara). The β-actin and U6 genes served as the internal reference genes of mRNA and miRNA quantification, respectively. Each reaction was repeated three times. The 10 μL qRT-PCR reaction system consisted of 2 × QuantiFast SYBR Green Master Mix 5 μL, 0.5 μL each of the upstream and downstream primers (10 μM), 1 μL of cDNA (about ng), and 5 μL RNase-free water. The normal qRT-PCR reaction procedure was as follows: pre-denaturation at 95 °C for 5 min; denaturation at 95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s for 40 cycles, and 72 °C for 10 min. The gga-miR-221-5p qRT-PCR reaction procedure was as follows: pre-denaturation at 95 °C for 3 min, denaturation at 95 °C for 12 s, 60 °C for 40 s, 72 °C for 30 s for 40 cycles, and 72 °C for 10 min.

Table 2.
Primer sequences used for qRT-PCR.
4.9. Statistical Analysis
The gene relative expression level was analyzed using the 2−ΔΔCt method. The data were presented as the mean ± standard error. A one-way analysis of variance (ANOVA) and t-test analysis of SPSS 20.0 software were performed to analyze the difference significance. The Duncan method was used to test the statistical significance of the differences among the groups. p < 0.05 indicated a significant difference. The graphs were drawn by using GraphPad Prism 5.0 software.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/5/1624/s1.
Author Contributions
D.-D.Z. performed the majority of experiments and wrote the manuscript. D.-D.W. and Z.W. helped to collect the samples. Y.-B.W., G.-R.S. and G.-X.L. participated in cell culture. Y.-D.T., R.-L.H., Z.-J.L. and R.-R.J. involved in data analysis. X.-J.L. and X.-T.K. was responsible for the experiment design and provided overall supervision. H.L. participated in the experiment design and modification of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the China Postdoctoral Science Foundation (No. 2017M520301), the National Natural Science Foundation of China (No.31902146, and No. U1704233), the Earmarked Fund for Modern Agro-Industry Technology Research System (No. CARS-41-K04) and the Scientific Studio of Zhongyuan Scholars (No. 30601985).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ELOVL6 | elongase of very long chain fatty acids 6 |
| SQLE | squalene epoxidase |
| TG | Triglyceride |
| TC | Total cholesterol |
| VLDL | Very low density lipoprotein |
| MicroRNAs | miRNAs |
| WT | Wild-type |
| Mut | Mutation |
| ApoVLDL II | apovitellenin very low density lipoprotein Ⅱ |
| ERE | estrogen responsive elements |
| ER | estrogen receptor |
| miR-221-5p mimics NC | miR-221-5p mimics negative control |
| AP1 | activator protein-1 |
References
- Shao, F.; Wang, X.; Yu, J.; Jiang, H.; Zhu, B.; Gu, Z. Expression of miR-33 from an SREBF2 Intron targets the FTO gene in the chicken. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Kang, L.; Cui, X.; Zhang, Y.; Yang, C.; Jiang, Y. Identification of miRNAs associated with sexual maturity in chicken ovary by Illumina small RNA deep sequencing. BMC Genom. 2013, 14. [Google Scholar] [CrossRef]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of MicroRNA-target recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef]
- Li, H.; Sun, G.R.; Lv, S.J.; Wei, Y.; Han, R.L.; Tian, Y.D.; Kang, X.T. Association study of polymorphisms inside the miR-1657 seed region with chicken growth and meat traits. Br. Poult. Sci. 2012, 53, 770–776. [Google Scholar] [CrossRef]
- Pillai, R.S.; Bhattacharyya, S.N.; Biology, W.F. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007, 17, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N. The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012, 19, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Heneghan, H.M.; Miller, N.; Kerin, M.J. Role of microRNAs in obesity and the metabolic syndrome. Obes. Rev. 2010, 11, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, A.Y.; Lee, H.W.; Son, Y.H.; Lee, G.Y.; Lee, J.-W.; Lee, Y.S.; Kim, J.B. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPAR gamma expression. Biochem. Biophy. Res. Commun. 2010, 392, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Xie, H.; Mori, M.A.; Alexander, R.; Yuan, B.; Hattangadi, S.M.; Liu, Q.; Kahn, C.R.; Lodish, H.F. Mir193b-365 is essential for brown fat differentiation. Nat. Cell Biol. 2011, 13, 958–965. [Google Scholar] [CrossRef]
- Moschenross, D.M. Characterization of a Liver Specific microRNA: miR-122. Ph.D. Thesis, University of Connecticut, Mansfield, UK, 2011. [Google Scholar]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef]
- Rayner, K.J.; Suarez, Y.; Davalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernandez-Hernando, C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Li, H.; Zheng, H.; Jiang, K.; Yan, F.; Tian, Y.; Kang, X.; Wang, Y.; Liu, X. Hepatic ELOVL6 mRNA is regulated by the gga-miR-22-3p in egg-laying hen. Gene 2017, 623, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Li, H.; Zheng, H.; Jiang, K.; Jia, L.; Yan, F.; Tian, Y.; Kang, X.; Wang, Y.; Liu, X. MicroRNA-101-2-5p Targets the ApoB gene in the liver of chicken (Gallus gallus). Genome 2017, 60, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ma, Z.; Jia, L.; Li, Y.; Xu, C.; Wang, T.; Han, R.; Jiang, R.; Li, Z.; Sun, G.; et al. Systematic analysis of the regulatory functions of microRNAs in chicken hepatic lipid metabolism. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Fu, T.; Choi, S.-E.; Kim, D.-H.; Seok, S.; Suino-Powell, K.M.; Xu, H.E.; Kemper, J.K. Aberrantly elevated microRNA-34a in obesity attenuates hepatic responses to FGF19 by targeting a membrane coreceptor β-Klotho. Proc. Natl. Acad. Sci. USA. 2012, 109, 16137–16142. [Google Scholar] [CrossRef]
- Ortega, F.J.; Moreno-Navarrete, J.M.; Pardo, G.; Sabater, M.; Hummel, M.; Ferrer, A.; Rodriguez-Hermosa, J.I.; Ruiz, B.; Ricart, W.; Peral, B.; et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- O’Hea, E.K.; Leveille, G.A. Lipid biosynthesis and transport in the domestic chick (Gallus domesticus). Comp. Bioche. Physi. 1969, 30, 149–159. [Google Scholar] [CrossRef]
- Walzem, R.L.; Hansen, R.J.; Williams, D.L.; Hamilton, R.L. Estrogen induction of VLDLy assembly in egg-laying hens. J. Nutr. 1999, 129, 467S–472S. [Google Scholar] [CrossRef]
- Ren, J.X.; Tian, W.H.; Yang, L.Y.; Zheng, H.; Li, Z.J.; Yan, F.B.; Wang, Y.B.; Tian, Y.D.; Wang, H.J.; Kang, X.T.; et al. Global investigation of estrogen-responsive genes regulating lipid metabolism in liver of chicken (Gallus gallus). Int. J. Biol. Sci. 2020, in press. [Google Scholar]
- Li, T.; Li, M.; Hu, S.; Cheng, X.; Gao, Y.; Jiang, S.; Yu, Q.; Zhang, C.; Sun, P.; Xian, W.; et al. MiR-221 mediates the epithelial-mesenchymal transition of hepatocellular carcinoma by targeting AdipoR1. Int. J. Biol. Macromol. 2017, 103, 1054–1061. [Google Scholar] [CrossRef]
- Celic, T.; Metzinger-Le Meuth, V.; Six, I.; Massy, Z.A.; Metzinger, L. The mir-221/222 cluster is a key player in vascular biology via the fine-tuning of endothelial cell physiology. Curr. Vasc. Pharmacol. 2017, 15, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Baxi, S.M.J.P.O. Retraction: miR-221 promotes tumorigenesis in human triple negative breast cancer cells. PLoS ONE 2017, 12, e0175869. [Google Scholar] [CrossRef]
- Jin, X.; Cai, L.; Wang, C.; Deng, X.; Yi, S.; Lei, Z.; Xiao, Q.; Xu, H.; Luo, H.; Sun, J. CASC2/miR-24/miR-221 modulates the TRAIL resistance of hepatocellular carcinoma cell through caspase-8/caspase-3. Cell Death. Dis. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-F.; Huang, J.; Li, H.; Zhang, C.; Huang, X.; Tong, G.; Xu, Y.-Z. MicroRNA-221 regulates endothelial nitric oxide production and inflammatory response by targeting adiponectin receptor 1. Gene 2015, 565, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Lustig, Y.; Barhod, E.; Ashwal-Fluss, R.; Gordin, R.; Shomron, N.; Baruch-Umansky, K.; Hemi, R.; Karasik, A.; Kanety, H. RNA-Binding protein PTB and MicroRNA-221 coregulate AdipoR1 translation and adiponectin signaling. Diabetes 2014, 63, 433–445. [Google Scholar] [CrossRef]
- Lian, L.; Zhang, D.; Wang, Q.; Yang, N.; Qu, L.J. The inhibitory effects of gga-miR-199-3p, gga-miR-140-3p, and gga-miR-221-5p in Marek’s disease tumorigenesis. Poul. Sci. 2015, 94, 2131–2135. [Google Scholar] [CrossRef]
- Li, H.; Ji, J.; Xie, Q.; Shang, H.; Zhang, H.; Xin, X.; Chen, F.; Sun, B.; Xue, C.; Ma, J.; et al. Aberrant expression of liver microRNA in chickens infected with subgroup J avian leukosis virus. Virus Res. 2012, 169, 268–271. [Google Scholar] [CrossRef]
- Dai, Z.; Ji, J.; Yan, Y.; Lin, W.; Li, H.; Chen, F.; Liu, Y.; Chen, W.; Bi, Y.; Xie, Q. Role of gga-miR-221 and gga-miR-222 during tumour formation in chickens infected by subgroup J avian leukosis virus. Viruses 2015, 7, 6538–6551. [Google Scholar] [CrossRef]
- Li, H.; Wang, T.A.; Xu, C.L.; Wang, D.D.; Ren, J.X.; Li, Y.M.; Tian, Y.D.; Wang, Y.B.; Jiao, Y.P.; Kang, X.T.; et al. Transcriptome profile of liver at different physiological stages reveals potential mode for lipid metabolism in laying hens. BMC Genom. 2015, 16, 763. [Google Scholar] [CrossRef]
- Klein-Hitpass, L.; Ryffel, G.U.; Heitlinger, E.; Cato, A.C. A 13 bp palindrome is a functional estrogen responsive element and interacts specifically with estrogen receptor. Nucleic Acids Res. 1988, 16, 647–663. [Google Scholar] [CrossRef]
- Pollock, A.; Bian, S.; Zhang, C.; Chen, Z.; Sun, T. Growth of the developing cerebral cortex is controlled by MicroRNA-7 through the p53 Pathway. Cell Rep. 2014, 8, 1184–1196. [Google Scholar] [CrossRef]
- Ng, R.; Wu, H.; Xiao, H.; Chen, X.; Willenbring, H.; Steer, C.J.; Song, G. Inhibition of MicroRNA-24 expression in liver prevents hepatic lipid accumulation and hyperlipidemia. Hepatology 2014, 60, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Shao, N.; Ma, G.; Zhang, J.; Zhu, W. miR-221-5p enhances cell proliferation and metastasis through post-transcriptional regulation of SOCS1 in human prostate cancer. BMC Urol. 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Krek, A.; Grun, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Shimano, H. Novel qualitative aspects of tissue fatty acids related to metabolic regulation: Lessons from Elovl6 knockout. Prog. Lipid Res. 2012, 51, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Corominas, J.; Marchesi, J.A.P.; Puig-Oliveras, A.; Revilla, M.; Estelle, J.; Alves, E.; Folch, J.M.; Ballester, M. Epigenetic regulation of the ELOVL6 gene is associated with a major QTL effect on fatty acid composition in pigs. Genet. Sel. Evol. 2015, 47. [Google Scholar] [CrossRef]
- D’Andre, H.C.; Paul, W.; Shen, X.; Jia, X.; Zhang, R.; Sun, L.; Zhang, X. Identification and characterization of genes that control fat deposition in chickens. J. Anim. Sci. Biotechnol. 2013, 4. [Google Scholar] [CrossRef]
- Lorbek, G.; Perse, M.; Horvat, S.; Bjorkhem, I.; Rozman, D. Sex Differences in the Hepatic Cholesterol Sensing Mechanisms in Mice. Molecules 2013, 18, 11067–11085. [Google Scholar] [CrossRef]
- Stopsack, K.H.; Gerke, T.A.; Andren, O.; Andersson, S.-O.; Giovannucci, E.L.; Mucci, L.A.; Rider, J.R. Cholesterol uptake and regulation in high-grade and lethal prostate cancers. Carcinogenesis 2017, 38, 806–811. [Google Scholar] [CrossRef]
- Helms, M.W.; Kemming, D.; Pospisil, H.; Vogt, U.; Buerger, H.; Korsching, E.; Liedtke, C.; Schlotter, C.M.; Wang, A.; Chan, S.Y.; et al. Squalene epoxidase, located on chromosome 8q24.1, is upregulated in 8q+ breast cancer and indicates poor clinical outcome in stage I and II disease. Br. J. Cancer 2008, 99, 774–780. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, W.; Zhang, K.; Zheng, H.; Ma, Y.; Lin, D.; Zhang, X.; Feng, L.; Lei, W.; Zhang, Z.; et al. Identification of genes differentially expressed in human primary lung squamous cell carcinoma. Lung Cancer 2007, 56, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, Y.; Nakamura, T.; Tanase, H.; Doi, O. Comparisons of plasma LH, progesterone, testosterone and estradiol concentrations in male and female chickens (Gallus domesticus) from 28 to 1141 days of age. Endocrinol. Jpn. 1981, 28, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.; Harvey, S. Plasma concentrations of luteinizing hormone growth hormone, oestradiol, testosterone and androstenedione in the domestic hen from 6 weeks of age to sexual maturity. Reprod. Nutr. Dev. 1986, 26, 515–522. [Google Scholar] [CrossRef]
- Palmisano, B.T.; Zhu, L.; Stafford, J.M. Role of Estrogens in the regulation of liver lipid metabolism. Adv. Exp. Med. Biol. 2017, 1043, 227–256. [Google Scholar] [CrossRef]
- Pellegrini, M.; Pallottini, V.; Marin, R.; Marino, M. Role of the sex hormone estrogen in the prevention of lipid disorder. Curr. Med. Chem. 2014, 21, 2734–2742. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, Z.; Ou, K.; Wang, T.; Li, Z.; Tian, Y.; Wang, Y.; Kang, X.; Li, H.; Liu, X. Evolution, dynamic expression changes and regulatory characteristics of gene families involved in the glycerophosphate pathway of triglyceride synthesis in chicken (Gallus gallus). Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Ivessa, N.E.; Rehberg, E.; Kienzle, B.; Seif, F.; Hermann, R.; Hermann, M.; Schneider, W.J.; Gordon, D.A. Molecular cloning, expression, and hormonal regulation of the chicken microsomal triglyceride transfer protein. Gene 2013, 523, 1–9. [Google Scholar] [CrossRef]
- Lu, T.T.; Repa, J.J.; Mangelsdorf, D.J. Orphan nuclear receptors as eLiXiRs and FiXeRs of sterol metabolism. J. Biol. Chem. 2001, 276, 37735–37738. [Google Scholar] [CrossRef]
- Peet, D.J.; Turley, S.D.; Ma, W.; Janowski, B.; Lobaccare, J.; Hammer, R.; Mangelsdorf, D. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXRα. Cell 1998, 93, 693–704. [Google Scholar] [CrossRef]
- Mcfadden, J.W.; Corl, B.A. Activation of liver X receptor (LXR) enhances de novo fatty acid synthesis in bovine mammary epithelial cells. J. Dairy Sci. 2010, 93, 4651–4658. [Google Scholar] [CrossRef]
- Darimont, C.; Avanti, O.; Zbinden, I.; Leonevautravers, P.; Mansourian, R.; Giusti, V.; Mace, K. Liver X receptor preferentially activates de novo lipogenesis in human preadipocytes. Biochimie 2006, 88, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Breevoort, S.R.; Angdisen, J.; Mingui, F.; Daniel, R.S.; Sam, R.H.; Steven, A.K.; David, J.M.; Ira, G.S. Liver LXRα expression is crucial for whole body cholesterol homeostasis and reverse cholesterol transport in mice. J. Clin. Investig. 2012, 122, 1688–1699. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Zheng, H.; Yang, L.; Li, H.; Tian, Y.; Wang, Y.; Lyu, S.; Brockmann, G.A.; Kang, X.; Liu, X. Dynamic expression profile, regulatory mechanism and correlation with egg-laying performance of ACSF gene family in chicken (Gallus gallus). Sci. Rep. 2018, 8, 8457–8467. [Google Scholar] [CrossRef] [PubMed]
- Paech, K.; Webb, P.; Kuiper, G.J.M.; Nilsson, S.; Gustafsssn, J.A.; Kushiser, P.J.; Scanlan, T.S. Differential ligand activation of estrogen receptors ER and ER at AP1 sites. Science 1997, 277, 1508–1510. [Google Scholar] [CrossRef]
- Petz, L.N.; Ziegler, Y.S.; Schultz, J.R.; Nardulli, A.M. Fos and Jun inhibit estrogen-induced transcription of the human progesterone receptor gene through an activator protein-1 site. Mol. Endocrinol. 2004, 18, 521–532. [Google Scholar] [CrossRef]
- Xiao, B.; Zhang, W.; Chen, L.; Hang, J.; Wang, L.; Zhang, R.; Liao, Y.; Chen, J.; Ma, Q.; Sun, Z.; et al. Analysis of the miRNA–mRNA–lncRNA network in human estrogen receptor-positive and estrogen receptor-negative breast cancer based on TCGA data. Gene 2018, 658, 28–35. [Google Scholar] [CrossRef]
- Zhao, X.-Y.; Li, S.; Wang, G.-X.; Yu, Q.; Lin, J.D. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol. Cell 2014, 55, 372–382. [Google Scholar] [CrossRef]
- Wagschal, A.; Najafi-Shoushtari, S.H.; Wang, L.; Goedeke, L.; Sinha, S.; deLemos, A.S.; Black, J.C.; Ramirez, C.M.; Li, Y.; Tewhey, R.; et al. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat. Med. 2015, 21, 1290–1297. [Google Scholar] [CrossRef]
- Li, Y.; Song, Y.; Zhao, M.; Guo, Y.; Yu, C.; Chen, W.; Shao, S.; Xu, C.; Zhou, X.; Zhao, L.J.H. A novel role for CRTC2 in hepatic cholesterol synthesis via SREBP-2. Hepatology 2017, 66, 481–497. [Google Scholar] [CrossRef]
- Cheng, Y.; Meng, Q.; Wang, C.; Li, H.; Huang, Z.; Chen, S.; Xiao, F.; Guo, F. Leucine deprivation decreases fat mass by stimulation of lipolysis in white adipose tissue and upregulation of uncoupling protein 1 (UCP1) in brown adipose tissue. Diabetes 2010, 59, 17–25. [Google Scholar] [CrossRef]
- Marquart, T.J.; Allen, R.M.; Ory, D.S.; Baldan, A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc. Natl. Acad. Sci. USA 2010, 107, 12228–12232. [Google Scholar] [CrossRef] [PubMed]
- Gerin, I.; Clerbaux, L.A.; Haumont, O.; Lanthier, N.; Das, A.K.; Burant, C.F.; Leclercq, I.A.; MacDougald, O.A.; Bommer, G.T. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J. Biol. Chem. 2010, 285, 33652–33661. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zalzala, M.; Xu, J.; Li, Y.; Yin, L.; Zhang, Y. A metabolic stress-inducible miR-34a-HNF4α pathway regulates lipid and lipoprotein metabolism. Nat. Commun. 2015, 6, 7466. [Google Scholar] [CrossRef]
- Tian, W.H.; Wang, Z.; Yue, Y.X.; Li, H.; Li, Z.J.; Han, R.L.; Tian, Y.D.; Kang, X.T.; Liu, X.J. miR-34a-5p increases hepatic triglycerides and total cholesterol levels by regulating ACSL1 protein expression in laying hens. Int. J. Mol. Sci. 2019, 20, 4420. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).