Investigation of a Novel Salt Stress-Responsive Pathway Mediated by Arabidopsis DEAD-Box RNA Helicase Gene AtRH17 Using RNA-Seq Analysis
Abstract
1. Introduction
2. Results
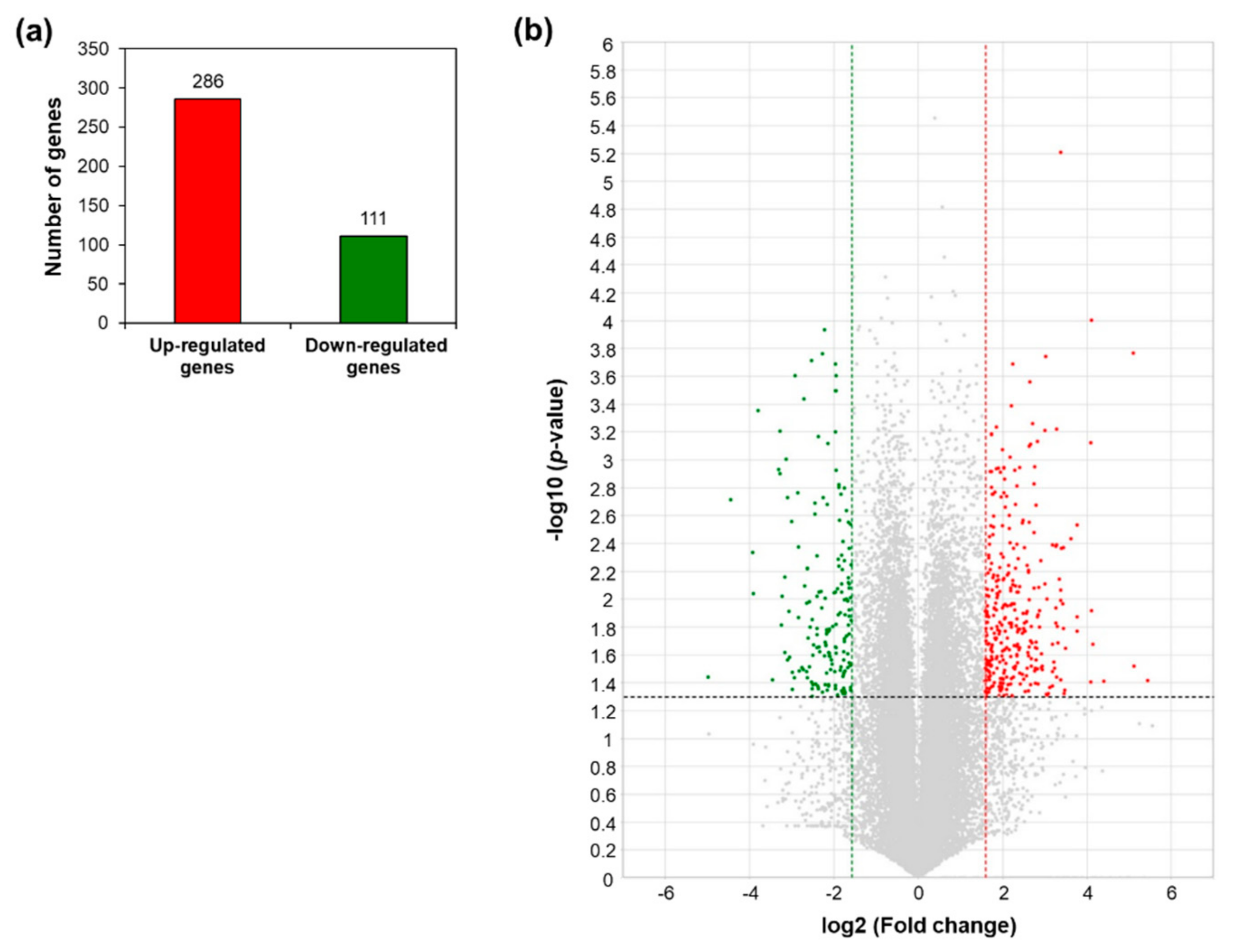
2.1. Transcriptomic Profiling of AtRH17 OXs
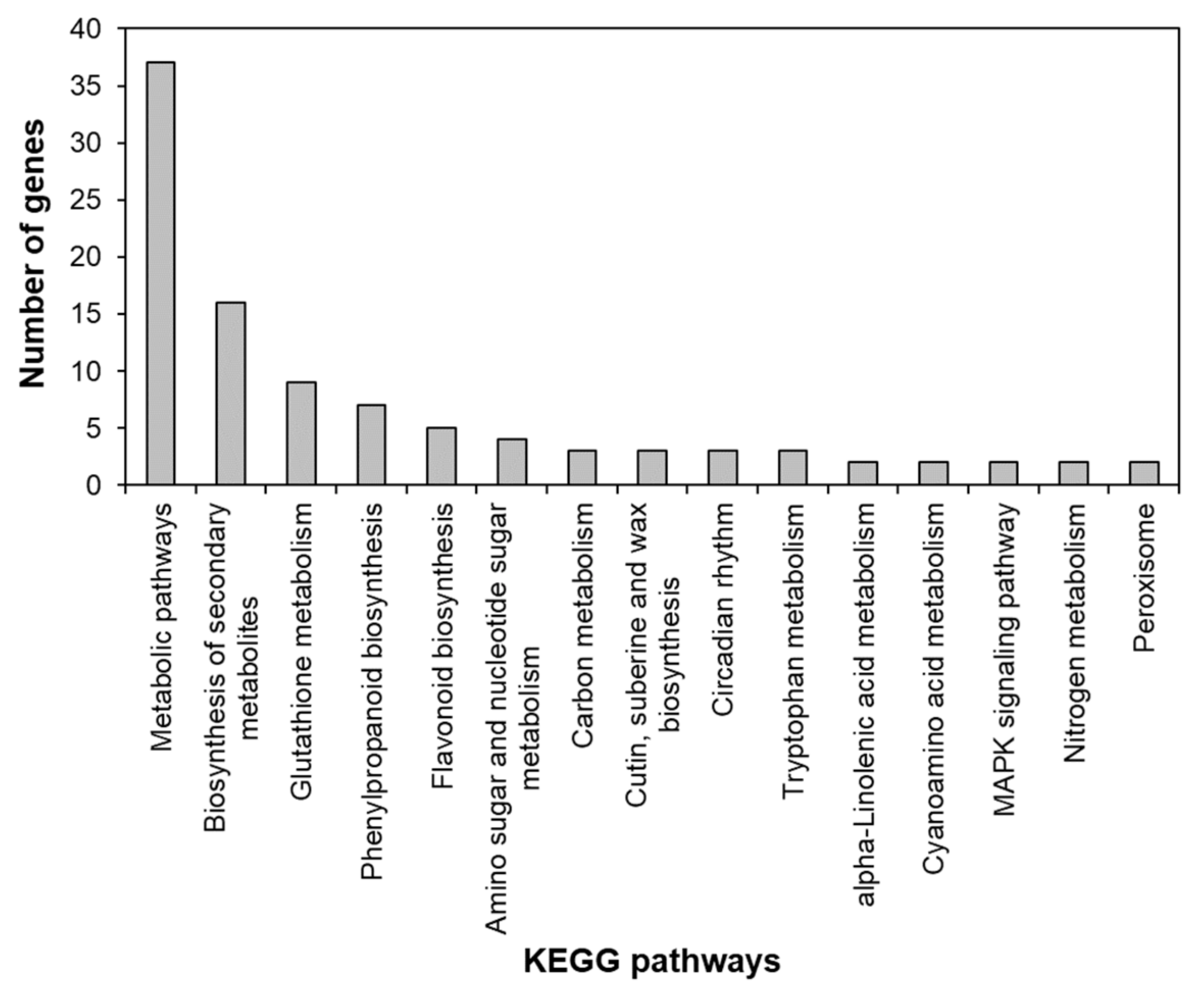
2.2. Analysis of Up- and Downregulated Genes in AtRH17 OXs
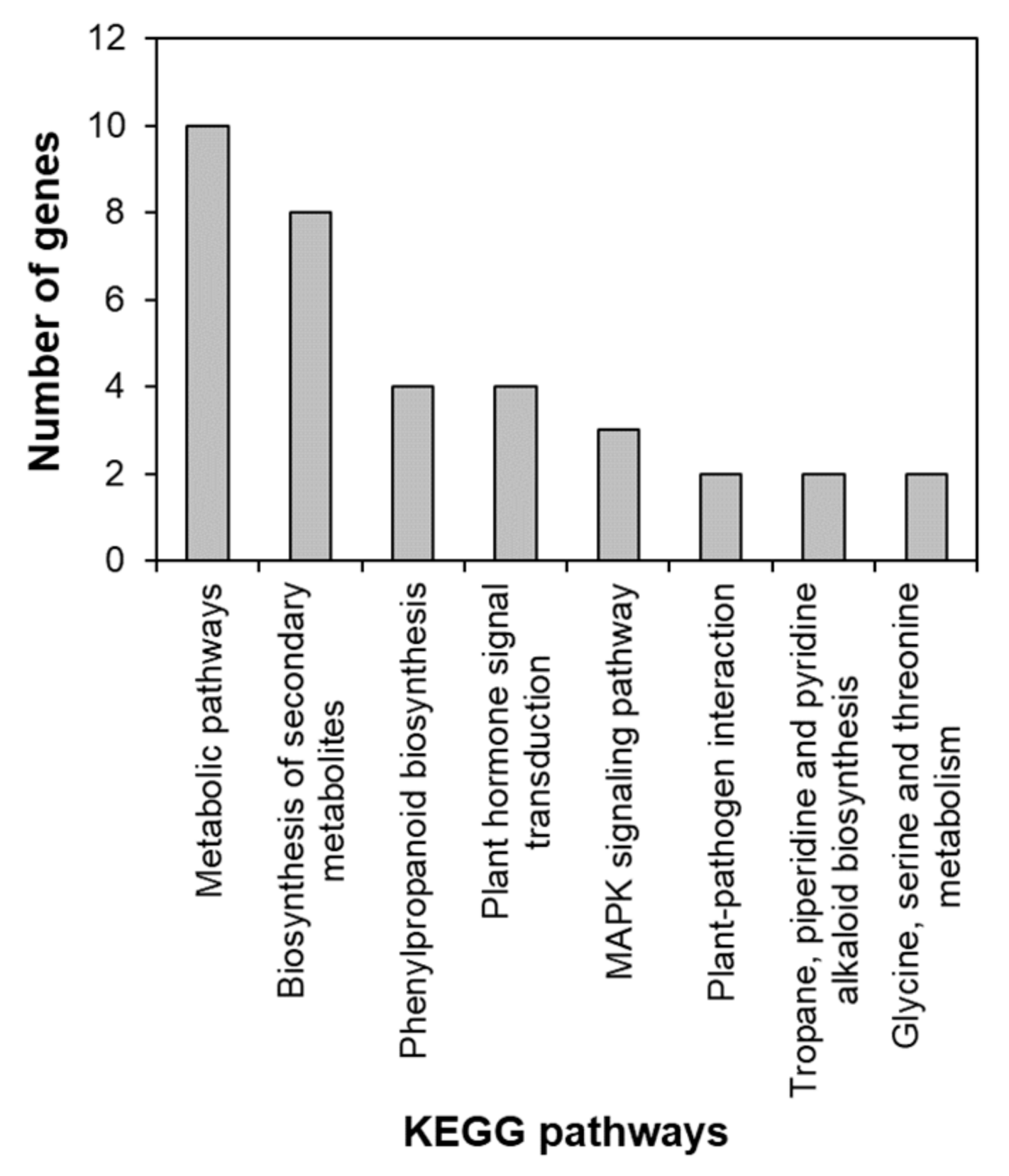
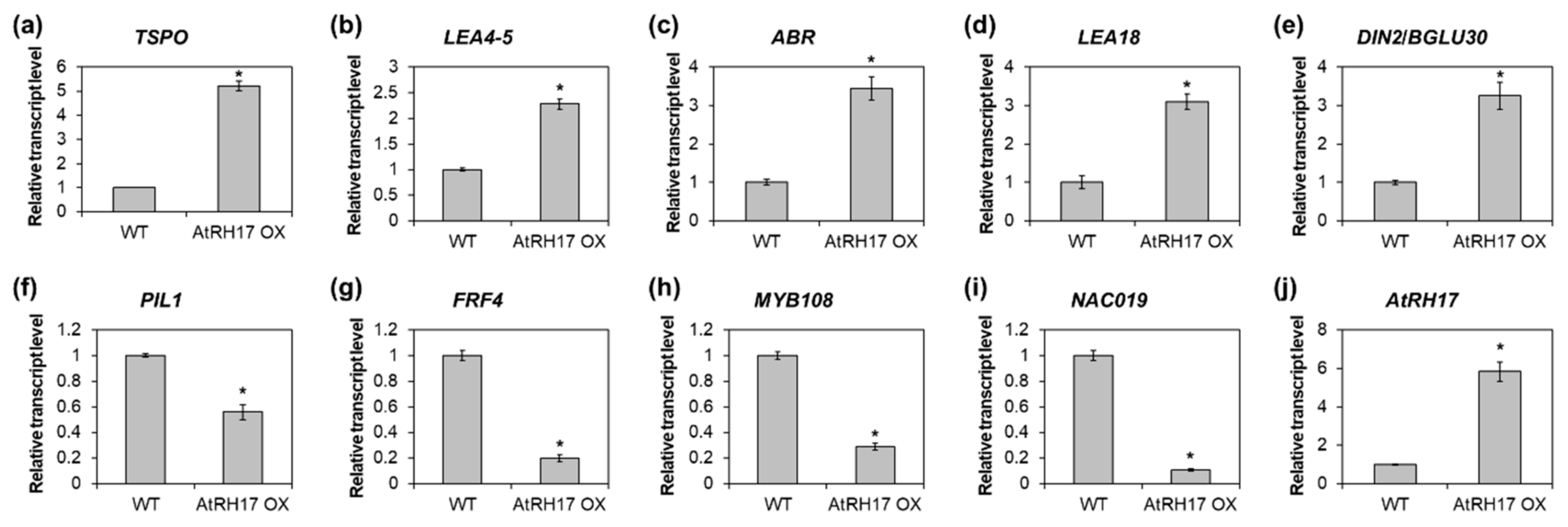
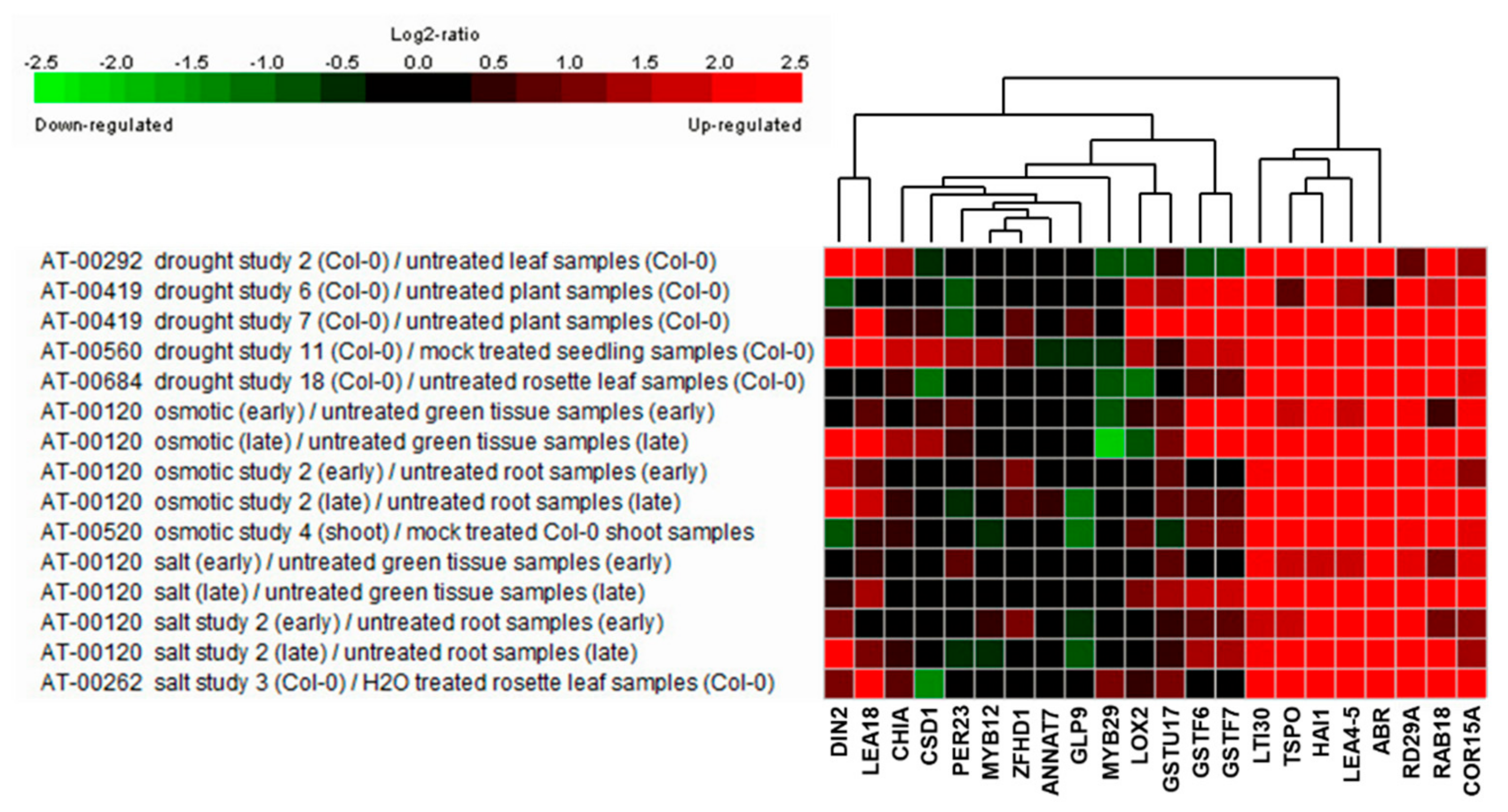
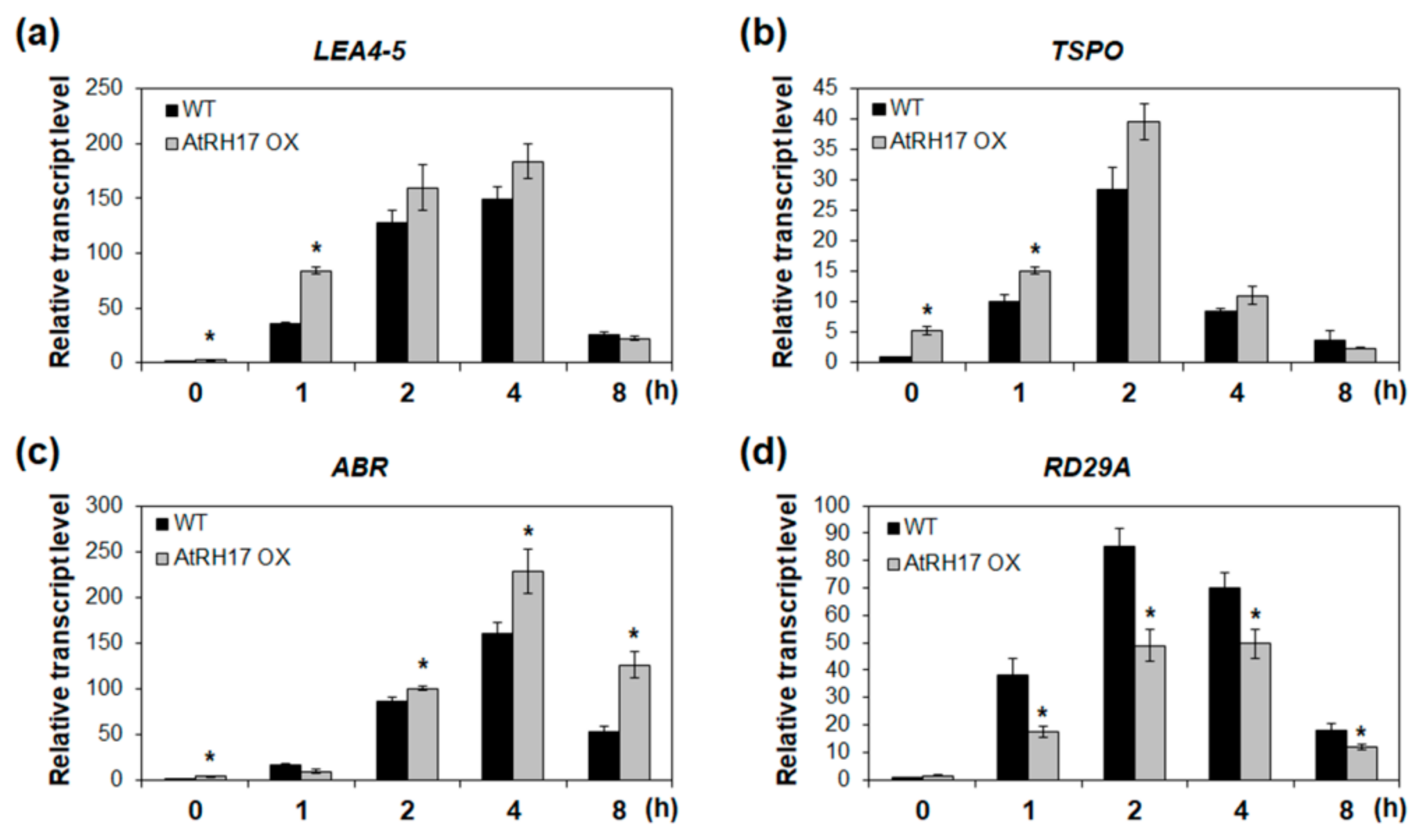
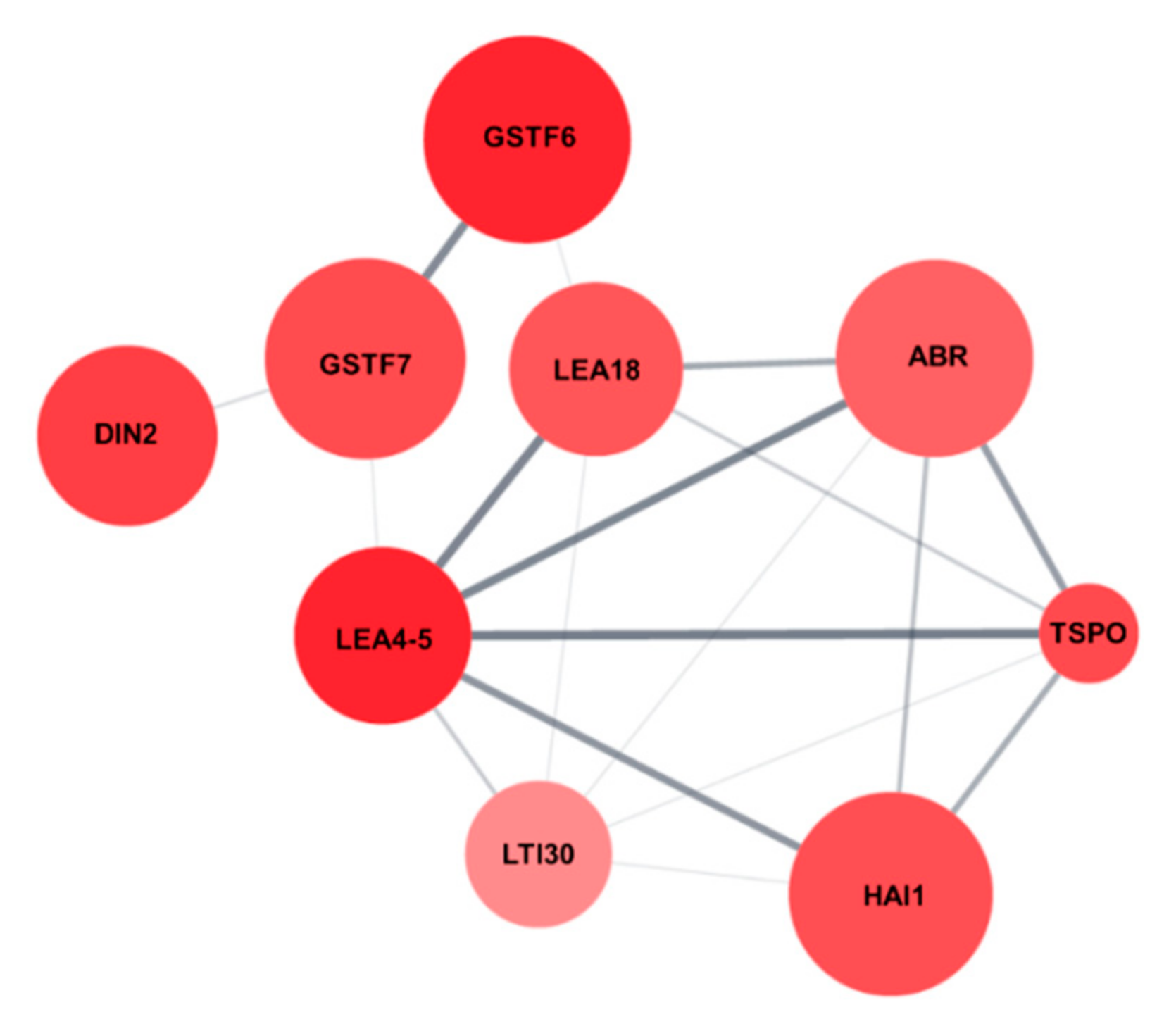
2.3. Identification of the Salt Stress-Tolerance Pathway in AtRH17 OXs
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Plant Stress Treatment for Quantitative RT-PCR
4.3. RNA Isolation and First Strand cDNA Synthesis
4.4. Quantitative RT-PCR
4.5. Library Preparation and RNA-Sequencing
4.6. RNA-Seq Data Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| DAG | Days after germination |
| DEG | Differentially expressed gene |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LD | Long-day |
| MS | Murashige and Skoog |
| OX | Overexpressing transgenic plant |
| RH | RNA helicase |
| SD | Short-day |
References
- Aubourg, S.; Kreis, M.; Lecharny, A. The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res. 1999, 27, 628–636. [Google Scholar] [CrossRef]
- Cordin, O.; Banroques, J.; Tanner, N.K.; Linder, P. The DEAD-box protein family of RNA helicases. Gene 2006, 367, 17–37. [Google Scholar] [CrossRef]
- Linder, P.; Rocak, S. DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004, 5, 232–241. [Google Scholar]
- Linder, P.; Jankowsky, E. From unwinding to clamping—The DEAD box RNA helicase family. Nat. Rev. Mol. Cell. Biol. 2011, 12, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Fuller-Pace, F.V. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006, 34, 4206–4215. [Google Scholar] [CrossRef]
- Nawaz, G.; Kang, H. Chloroplast- or mitochondria-targeted DEAD-box RNA helicases play essential roles in organellar RNA metabolism and abiotic stress responses. Front. Plant Sci. 2017, 8, 871. [Google Scholar] [CrossRef] [PubMed]
- Iost, I.; Dreyfus, M. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 2006, 34, 4189–4197. [Google Scholar] [CrossRef]
- Martin, R.; Straub, A.U.; Doebele, C.; Bohnsack, M.T. DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 2013, 10, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Giraud, C.; Hausmann, S.; Lemeille, S.; Prados, J.; Redder, P.; Linder, P. The C-terminal region of the RNA helicase CshA is required for the interaction with the degradosome and turnover of bulk RNA in the opportunistic pathogen Staphylococcus aureus. RNA Biol. 2015, 12, 658–674. [Google Scholar] [CrossRef]
- Liu, Y.; Imai, R. Function of plant DExD/H-box RNA helicases associated with ribosomal RNA biogenesis. Front. Plant Sci. 2018, 9, 125. [Google Scholar] [CrossRef]
- Baruah, I.; Debbarma, J.; Boruah, H.P.D.; Keshavaiah, C. The DEAD-box RNA helicases and multiple abiotic stresses in plants: A systematic review of recent advantages and challenges. Plant Omics J. 2017, 10, 252–262. [Google Scholar] [CrossRef]
- Huang, C.; Shen, Y.; Huang, L.; Wu, S.; Yeh, C.; Lu, C. The DEAD-box RNA helicase AtRH7/PRH75 participates in pre-rRNA processing, plant development and cold tolerance in Arabidopsis. Plant Cell Physiol. 2016, 57, 174–191. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Dong, C.; Lee, H.; Zhu, J.; Xiong, L.; Gong, D.; Stevenson, B.; Zhu, J.K. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 2005, 17, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Wu, J.; Zhang, Y.; Jiang, C.; Liu, R.; Chai, C.; Zhu, J. A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell 2013, 25, 342–356. [Google Scholar] [CrossRef]
- Kant, P.; Kant, S.; Gordon, M.; Shaked, R.; Barak, S. Stress Response Suppressor1 and Stress Response Suppressor2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol. 2007, 145, 814–830. [Google Scholar] [CrossRef]
- Khan, A.; Garbelli, A.; Grossi, S.; Florentin, A.; Batelli, G.; Acuna, T.; Zolla, G.; Kaye, Y.; Paul, L.K.; Zhu, J.K.; et al. The Arabidopsis STRESS RESPONSE SUPPRESSOR DEAD-box RNA helicases are nucleolar- and chromocenter-localized proteins that undergo stress-mediated relocalization and are involved in epigenetic gene silencing. Plant J. 2014, 79, 28–43. [Google Scholar] [CrossRef]
- Nguyen, V.L.; Seok, H.Y.; Woo, D.H.; Lee, S.Y.; Moon, Y.H. Overexpression of the DEAD-box RNA helicase gene AtRH17 confers tolerance to salt stress in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 3777. [Google Scholar] [CrossRef]
- Candat, A.; Paszkiewicz, G.; Neveu, M.; Gautier, R.; Logan, D.C.; Avelange-Macherel, M.H.; Macherel, D. The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell 2014, 26, 3148–3166. [Google Scholar] [CrossRef]
- Cuevas-Velazquez, C.L.; Saab-Rincón, G.; Reyes, J.L.; Covarrubias, A.A. The unstructured N-terminal region of Arabidopsis group 4 Late Embryogenesis Abundant (LEA) proteins is required for folding and for chaperone-like activity under water deficit. J. Biol. Chem. 2016, 291, 10893–10903. [Google Scholar] [CrossRef]
- Hachez, C.; Veljanovski, V.; Reinhardt, H.; Guillaumot, D.; Vanhee, C.; Chaumont, F.; Batoko, H. The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the aquaporin PIP2;7 through protein-protein interactions and autophagic degradation. Plant Cell 2014, 26, 4974–4990. [Google Scholar] [CrossRef]
- Jurkiewicz, P.; Melser, S.; Maucourt, M.; Ayeb, H.; Veljanovski, V.; Maneta-Peyret, L.; Hooks, M.; Rolin, D.; Moreau, P.; Batoko, H. The multistress-induced Translocator protein (TSPO) differentially modulates storage lipids metabolism in seeds and seedlings. Plant J. 2018, 96, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kang, H. Emerging roles of RNA-binding proteins in plant growth, development, and stress responses. Mol. Cells 2016, 39, 179–185. [Google Scholar] [PubMed]
- Stonebloom, S.; Burch-Smith, T.; Kim, I.; Meinke, D.; Mindrinoss, M.; Zambryski, P. Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc. Natl. Acad. Sci. USA 2009, 106, 17229–17234. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Huang, L.; Huang, J.; Wu, S.; Yeh, C.; Lu, C. A DEAD-box protein, AtRH36, is essential for female gametophyte development and is involved in rRNA biogenesis in Arabidopsis. Plant Cell Physiol. 2010, 51, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.; Chen, Y.; Hsiao, Y.; Wang, B.; Lin, S.; Cheng, W.; Jauh, G.; Harada, J.J.; Wang, C. AtRH57, a DEAD-box RNA helicase, is involved in feedback inhibition of glucose-mediated abscisic acid accumulation during seedling development and additively affects pre-ribosomal RNA processing with high glucose. Plant J. 2014, 77, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Wold, B.; Myers, R.M. Sequence census methods for functional genomics. Nat. Methods 2008, 5, 19–21. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Miao, X.; Luo, Q. Genome-wide transcriptome analysis between small-tail Han sheep and the Surabaya fur sheep using high-throughput RNA sequencing. Reproduction 2013, 145, 587–596. [Google Scholar] [CrossRef]
- French-Pacheco, L.; Cuevas-Velazquez, C.L.; Rivillas-Acevedo, L.; Covarrubias, A.A.; Amero, C. Metal-binding polymorphism in late embryogenesis abundant protein AtLEA4-5, an intrinsically disordered protein. Peer J. 2018, 6, e4930. [Google Scholar] [CrossRef]
- Wang, P.; Shen, L.; Guo, J.; Jing, W.; Qu, Y.; Li, W.; Bi, R.; Xuan, W.; Zhang, Q.; Zhang, W. Phosphatidic acid directly regulates PINOID-dependent phosphorylation and activation of the PIN-FORMED2 auxin efflux transporter in response to salt stress. Plant Cell 2019, 31, 250–271. [Google Scholar] [CrossRef] [PubMed]
- Saini, K.; AbdElgawad, H.; Markakis, M.N.; Schoenaers, S.; Asard, H.; Prinsen, E.; Beemster, G.T.S.; Vissenberg, K. Perturbation of auxin homeostasis and signaling by PINOID overexpression induces stress responses in Arabidopsis. Front. Plant Sci. 2017, 8, 1308. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Georges, N.T.; Walter, Z.; Siméon, F. Frequentist model averaging and applications to Bernoulli trials. Open J. Stat. 2016, 6, 545–553. [Google Scholar]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
| Sample | Total Read | Processed Read | Mapped Read | Mapping Rate |
|---|---|---|---|---|
| WT-1 | 31,584,235 | 30,119,902 | 29,518,240 | 98.01% |
| WT-2 | 32,020,154 | 30,339,538 | 29,616,323 | 97.62% |
| AtRH17 OX-1 | 32,659,954 | 30,992,415 | 30,299,369 | 97.76% |
| AtRH17 OX-2 | 28,802,597 | 27,060,251 | 26,375,818 | 97.47% |
| GO Term | Description | Number in Input List | p-Value |
|---|---|---|---|
| GO:0055144 | oxidation–reduction process | 34 | 0.00000 |
| GO:0050832 | defense response to fungus | 14 | 0.00095 |
| GO:0006952 | defense response | 14 | 0.01700 |
| GO:0006979 | response to oxidative stress | 12 | 0.00020 |
| GO:0009651 | response to salt stress | 11 | 0.00600 |
| GO:0009636 | response to toxic substance | 10 | 0.00000 |
| GO:0009414 | response to water deprivation | 10 | 0.00230 |
| GO:0009407 | toxin catabolic process | 9 | 0.00000 |
| GO:0006749 | glutathione metabolic process | 9 | 0.00000 |
| GO:0006869 | lipid transport | 9 | 0.00015 |
| GO:0009611 | response to wounding | 9 | 0.00100 |
| GO:0009617 | response to bacterium | 8 | 0.00008 |
| GO:0009813 | flavonoid biosynthetic process | 8 | 0.00071 |
| GO:0042742 | defense response to bacterium | 8 | 0.02300 |
| GO:0051707 | response to other organism | 7 | 0.00002 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 7 | 0.02200 |
| GO:0071456 | cellular response to hypoxia | 6 | 0.00001 |
| GO:0010150 | leaf senescence | 6 | 0.00250 |
| GO:0080167 | response to karrikin | 6 | 0.01000 |
| GO:0045454 | cell redox homeostasis | 6 | 0.02100 |
| GO:0042744 | hydrogen peroxide catabolic process | 5 | 0.01300 |
| GO:0009826 | unidimensional cell growth | 5 | 0.02900 |
| GO:0002213 | defense response to insect | 4 | 0.00063 |
| GO:0009682 | induced systemic resistance | 4 | 0.00075 |
| GO:0006032 | chitin catabolic process | 4 | 0.00200 |
| GO:0006040 | amino sugar metabolic process | 3 | 0.00990 |
| GO:0000272 | polysaccharide catabolic process | 3 | 0.01700 |
| GO:0071732 | cellular response to nitric oxide | 3 | 0.01700 |
| GO:0042343 | indole glucosinolate metabolic process | 3 | 0.01900 |
| GO:0016998 | cell wall macromolecule catabolic process | 3 | 0.02800 |
| GO:0055072 | iron ion homeostasis | 3 | 0.04200 |
| GO:0045168 | cell-cell signaling involved in cell fate commitment | 3 | 0.04200 |
| GO:0046256 | 2,4,6-trinitrotoluene catabolic process | 2 | 0.02000 |
| GO:0006801 | superoxide metabolic process | 2 | 0.02000 |
| GO:0009790 | embryo development | 2 | 0.03000 |
| GO:0071457 | cellular response to ozone | 2 | 0.03000 |
| GO Term | Description | Number in Input List | p-Value |
|---|---|---|---|
| GO:0055114 | oxidation-reduction process | 11 | 0.0330 |
| GO:0006952 | defense response | 7 | 0.0430 |
| GO:0009664 | plant-type cell wall organization | 6 | 0.0000 |
| GO:0006979 | response to oxidative stress | 5 | 0.0270 |
| GO:0042744 | hydrogen peroxide catabolic process | 4 | 0.0051 |
| GO:00009741 | response to brassinosteroid | 3 | 0.0077 |
| GO:0009739 | response to gibberellin | 3 | 0.0570 |
| GO:0009826 | unidimensional cell growth | 3 | 0.0730 |
| GO:0009723 | response to ethylene | 3 | 0.0840 |
| GO:0009641 | shade avoidance | 2 | 0.0570 |
| GO:0010017 | red or far-red light signaling pathway | 2 | 0.0930 |
| Locus ID | Gene Symbol | Fold Change | p-Value | Stress Responses |
|---|---|---|---|---|
| At1g02930 | GSTF6 | 10.377 | 0.000 | salt, water deprivation |
| At5g10230 | ANNAT7 | 3.366 | 0.009 | salt, water deprivation |
| At1g10370 | GSTU17 | 9.761 | 0.004 | salt |
| At3g60140 | DIN2/BGLU30 | 8.008 | 0.008 | salt |
| At2g47770 | TSPO | 7.078 | 0.032 | salt |
| At1g02920 | GSTF7 | 6.927 | 0.002 | salt |
| At2g47460 | MYB12 | 4.418 | 0.006 | salt |
| At4g14630 | GLP9 | 4.116 | 0.001 | salt |
| At5g24090 | CHIA | 3.627 | 0.007 | salt |
| At2g38390 | PER23 | 3.387 | 0.017 | salt |
| At1g08830 | CSD1 | 3.087 | 0.013 | salt |
| At5g06760 | LEA4-5 | 10.426 | 0.009 | water deprivation |
| At5g59220 | HAI1 | 6.705 | 0.001 | water deprivation |
| At2g35300 | LEA18 | 6.182 | 0.010 | water deprivation |
| At3g02480 | ABR | 5.526 | 0.003 | water deprivation |
| At1g69600 | ZFHD1 | 4.178 | 0.014 | water deprivation |
| At3g45140 | LOX2 | 4.061 | 0.027 | water deprivation |
| At3g50970 | LTI30 | 3.502 | 0.018 | water deprivation |
| At5g07690 | MYB29 | 3.325 | 0.022 | water deprivation |
| Locus ID | Gene Symbol | Fold Change | p-Value | Description |
|---|---|---|---|---|
| At5g06760 | LEA4-5 | 10.426 | 0.009 | Member of the Late Embryogenesis Abundant (LEA) protein |
| At1g02930 | GSTF6 | 10.377 | 0.000 | Glutathione transferase belonging to the phi class of GSTs |
| At3g60140 | DIN2/BGLU30 | 8.008 | 0.008 | Glycoside hydrolase superfamily protein |
| At2g47770 | TSPO | 7.078 | 0.032 | Membrane-bound protein, TSPO-related protein |
| At1g02920 | GSTF7 | 6.927 | 0.002 | Glutathione transferase belonging to the phi class of GSTs |
| At2g35300 | LEA18 | 6.182 | 0.010 | Member of the Late Embryogenesis Abundant (LEA) protein |
| At5g59220 | HAI1 | 6.705 | 0.001 | Member of the PP2C family (Clade A protein phosphatases type 2C) |
| At3g02480 | ABR | 5.526 | 0.003 | Late embryogenesis abundant protein (LEA) family protein |
| At3g50970 | LTI30 | 3.502 | 0.018 | Dehydrin protein family |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seok, H.-Y.; Nguyen, L.V.; Nguyen, D.V.; Lee, S.-Y.; Moon, Y.-H. Investigation of a Novel Salt Stress-Responsive Pathway Mediated by Arabidopsis DEAD-Box RNA Helicase Gene AtRH17 Using RNA-Seq Analysis. Int. J. Mol. Sci. 2020, 21, 1595. https://doi.org/10.3390/ijms21051595
Seok H-Y, Nguyen LV, Nguyen DV, Lee S-Y, Moon Y-H. Investigation of a Novel Salt Stress-Responsive Pathway Mediated by Arabidopsis DEAD-Box RNA Helicase Gene AtRH17 Using RNA-Seq Analysis. International Journal of Molecular Sciences. 2020; 21(5):1595. https://doi.org/10.3390/ijms21051595
Chicago/Turabian StyleSeok, Hye-Yeon, Linh Vu Nguyen, Doai Van Nguyen, Sun-Young Lee, and Yong-Hwan Moon. 2020. "Investigation of a Novel Salt Stress-Responsive Pathway Mediated by Arabidopsis DEAD-Box RNA Helicase Gene AtRH17 Using RNA-Seq Analysis" International Journal of Molecular Sciences 21, no. 5: 1595. https://doi.org/10.3390/ijms21051595
APA StyleSeok, H.-Y., Nguyen, L. V., Nguyen, D. V., Lee, S.-Y., & Moon, Y.-H. (2020). Investigation of a Novel Salt Stress-Responsive Pathway Mediated by Arabidopsis DEAD-Box RNA Helicase Gene AtRH17 Using RNA-Seq Analysis. International Journal of Molecular Sciences, 21(5), 1595. https://doi.org/10.3390/ijms21051595







