Conversion of Glycosylated Platycoside E to Deapiose-Xylosylated Platycodin D by Cytolase PCL5
Abstract
1. Introduction
2. Results and Discussion
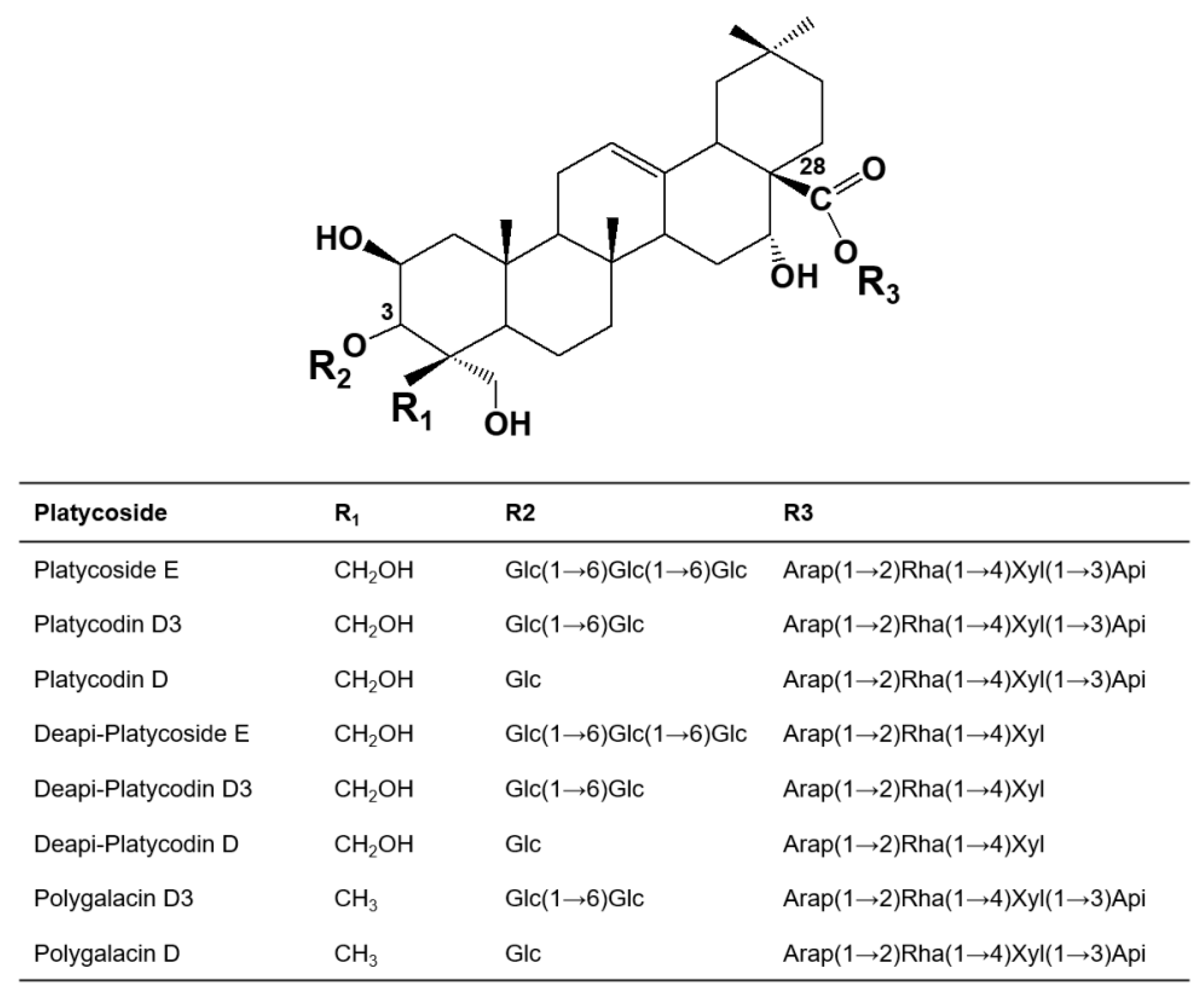
2.1. Identification of Products Obtained by the Action of Cytolase PCL5 on Platycoside E and Polygalacin D3 by
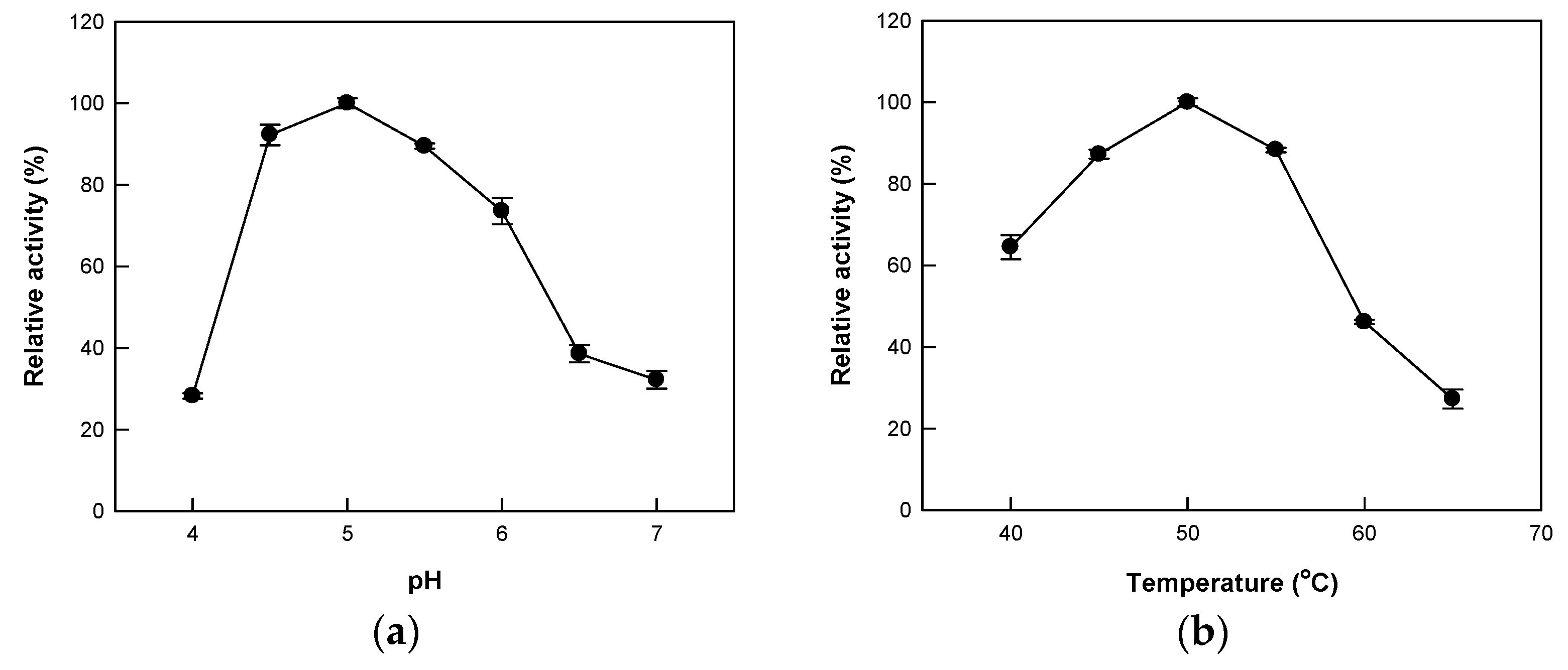
2.2. Effects of pH and Temperature on the Hydrolytic Activity of Cytolase PCL5
2.3. Substrate Specificity of Cytolase PCL5 for Platycosides
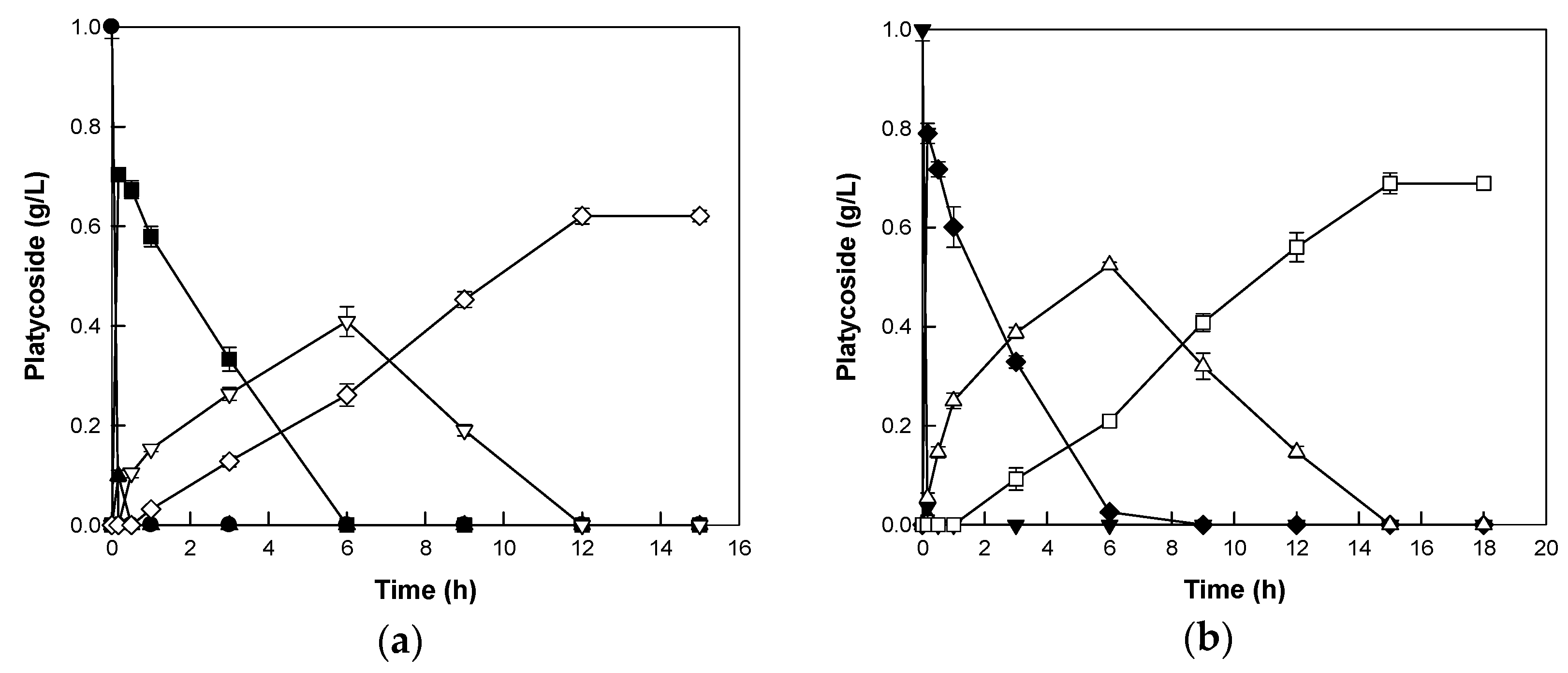
2.4. Biotransformations of Platycoside E and Polygalacin D3 into Deapiose-Xylosylated Platycosides by Cytolase PCL5
3. Materials and Methods
3.1. Materials
3.2. Hydrolytic Activity Assay
3.3. Biotransformation
3.4. HPLC Analysis
3.5. Liquid Chromatography-Mass Spectrometry Analysis of Platycosides
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nyakudya, E.; Jeong, J.H.; Lee, N.K.; Jeong, Y.S. Platycosides from the Roots of Platycodon grandiflorum and Their Health Benefits. Prev. Nutr. Food Sci. 2014, 19, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Y.; Yang, D.; Zhang, C.; Zhang, N.; Li, M.; Liu, Y. Platycodon Grandiflorus-an Ethnopharmacological, Phytochemical and Pharmacological Review. J. Ethnopharmacol. 2015, 164, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Ryu, C.S.; Kim, C.H.; Lee, S.Y.; Lee, K.S.; Choung, K.J.; Song, G.Y.; Kim, B.H.; Ryu, S.Y.; Lee, H.S.; Kim, S.K. Evaluation of the Total Oxidant Scavenging Capacity of Saponins Isolated from Platycodon grandiflorum. Food Chem. 2012, 132, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Lin, S.; Ren, F.; Wu, L.; Chen, L.; Sun, Y. Antioxidant and Antimicrobial Capacity of Chinese Medicinal Herb Extracts in Raw Sheep Meat. J. Food Prot. 2007, 70, 1440–1445. [Google Scholar] [CrossRef]
- Kim, J.Y.; Hwang, Y.P.; Kim, D.H.; HAN, E.H.; Chung, Y.C.; Roh, S.H.; Jeong, H.G. Inhibitory Effect of the Saponins Derived from Roots of Platycodon grandiflorum on Carrageenan-Induced Inflammation. Biosci. Biotechnol. Biochem. 2006, 70, 858–864. [Google Scholar] [CrossRef]
- Kim, M.; Hwang, I.G.; Kim, S.B.; Choi, A.J. Chemical Characterization of Balloon Flower (Platycodon grandiflorum) Sprout Extracts and Their Regulation of Inflammatory Activity in Lipopolysaccharide-Stimulated Raw 264.7 Murine Macrophage Cells. Food Sci. Nutr. 2020, 8, 246–256. [Google Scholar] [CrossRef]
- Yim, N.H.; Hwang, Y.H.; Liang, C.; Ma, J.Y. A Platycoside-Rich Fraction from the Root of Platycodon grandiflorum Enhances Cell Death in A549 Human Lung Carcinoma Cells Via Mainly Ampk/Mtor/Akt Signal-Mediated Autophagy Induction. J. Ethnopharmacol. 2016, 194, 1060–1068. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, J.S.; Choi, S.U.; Kim, J.S.; Lee, H.S.; Roh, S.H.; Jeong, Y.C.; Kim, Y.K.; Ryu, S.Y. Isolation of a New Saponin and Cytotoxic Effect of Saponins from the Root of Platycodon grandiflorum on Human Tumor Cell Lines. Planta. Med. 2005, 71, 566–568. [Google Scholar] [CrossRef]
- Khan, M.; Maryam, A.; Zhang, H.; Mehmood, T.; Ma, T. Killing Cancer with Platycodin D through Multiple Mechanisms. J. Cell Mol. Med. 2016, 20, 389–402. [Google Scholar] [CrossRef]
- Halliwell, B. Oxidative Stress and Neurodegeneration: Where Are We Now? J. Neurochem. 2006, 97, 1634–1658. [Google Scholar] [CrossRef]
- Oh, Y.C.; Kang, O.H.; Choi, J.G.; Lee, Y.S.; Brice, O.O.; Jung, H.J.; Hong, S.H.; Lee, Y.M.; Shin, D.W.; Kim, Y.S.; et al. Anti-Allergic Activity of a Platycodon Root Ethanol Extract. Int. J. Mol. Sci 2010, 11, 2746–2758. [Google Scholar] [CrossRef]
- Han, L.K.; Zheng, Y.N.; Xu, B.J.; Okuda, H.; Kimura, Y. Saponins from Platycodi Radix Ameliorate High Fat Diet-Induced Obesity in Mice. J. Nutr. 2002, 132, 2241–2245. [Google Scholar] [CrossRef]
- Zhao, H.L.; Harding, S.V.; Marinangeli, C.P.F.; Kim, Y.S.; Jones, P.J.H. Hypocholesterolemic and Anti-Obesity Effects of Saponins from Platycodon grandiflorum in Hamsters Fed Atherogenic Diets. J. Food Sci. 2008, 73, H195–H200. [Google Scholar] [CrossRef]
- Hwang, K.A.; Hwang, Y.J.; Im, P.R.; Hwang, H.J.; Song, J.; Kim, Y.J. Platycodon grandiflorum Extract Reduces High-Fat Diet-Induced Obesity through Regulation of Adipogenesis and Lipogenesis Pathways in Mice. J. Med. Food 2019, 22, 993–999. [Google Scholar] [CrossRef]
- Xie, Y.; Ye, Y.P.; Sun, H.X.; Li, D. Contribution of the Glycidic Moieties to the Haemolytic and Adjuvant Activity of Platycodigenin-Type Saponins from the Root of Platycodon grandiflorum. Vaccine 2008, 26, 3452–3460. [Google Scholar] [CrossRef] [PubMed]
- Noh, E.M.; Kim, J.M.; Lee, H.Y.; Song, H.K.; Joung, S.O.; Yang, H.J.; Kim, M.J.; Kim, K.S.; Lee, Y.R. Immuno-Enhancement Effects of Platycodon grandiflorum Extracts in Splenocytes and a Cyclophosphamide-Induced Immunosuppressed Rat Model. BMC Complement. Altern. Med. 2019, 19, 322. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, Y.; Yan, P.; Cheng, G.; Wang, C.; Geng, N.; Wang, X.; Liu, J. Effects of Polysaccharides from Platycodon grandiflorum on Immunity-Enhancing Activity in Vitro. Molecules 2017, 22, 1918. [Google Scholar] [CrossRef]
- Ha, Y.W.; Na, Y.C.; Seo, J.J.; Kim, S.N.; Linhardt, R.J.; Kim, Y.S. Qualitative and Quantitative Determination of Ten Major Saponins in Platycodi Radix by High Performance Liquid Chromatography with Evaporative Light Scattering Detection and Mass Spectrometry. J. Chromatogr. A 2006, 1135, 27–35. [Google Scholar] [CrossRef]
- Yoo, D.S.; Choi, Y.H.; Cha, M.R.; Lee, B.H.; Kim, S.J.; Yon, G.H.; Hong, K.S.; Jang, Y.S.; Lee, H.S.; Kim, Y.S.; et al. Hplc-Elsd Analysis of 18 Platycosides from Balloon Flower Roots (Platycodi Radix) Sourced from Various Regions in Korea and Geographical Clustering of the Cultivation Areas. Food Chem. 2011, 129, 645–651. [Google Scholar] [CrossRef]
- Park, C.S.; Yoo, M.H.; Noh, K.H.; Oh, D.K. Biotransformation of Ginsenosides by Hydrolyzing the Sugar Moieties of Ginsenosides Using Microbial Glycosidases. Appl. Microbiol. Biotechnol. 2010, 87, 9–19. [Google Scholar] [CrossRef]
- Shin, K.C.; Oh, D.K. Classification of Glycosidases That Hydrolyze the Specific Positions and Types of Sugar Moieties in Ginsenosides. Crit. Rev. Biotechnol. 2016, 36, 1036–1049. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.Y.; Kim, J.M.; Han, S.B.; Lee, S.K.; Kim, N.D.; Park, M.K.; Kim, C.K.; Park, J.H. Steaming of Ginseng at High Temperature Enhances Biological Activity. J. Nat. Prod. 2000, 63, 1702–1704. [Google Scholar] [CrossRef] [PubMed]
- Bae, E.A.; Han, M.J.; Kim, E.J.; Kim, D.H. Transformation of Ginseng Saponins to Ginsenoside Rh2 by Acids and Human Intestinal Bacteria and Biological Activities of Their Transformants. Arch. Pharm. Res. 2004, 27, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yan, Q.; Li, J.Y.; Zhang, X.C.; Zhou, P. Biotransformation of Panax Notoginseng Saponins into Ginsenoside Compound K Production by Paecilomyces bainier Sp. 229. J. Appl. Microbiol. 2008, 104, 699–706. [Google Scholar] [CrossRef]
- Cui, C.H.; Choi, T.E.; Yu, H.; Jin, F.; Lee, S.T.; Kim, S.C.; Im, W.T. Mucilaginibacter composti Sp. Nov., with Ginsenoside Converting Activity, Isolated from Compost. J. Microbiol. 2011, 49, 393–398. [Google Scholar] [CrossRef]
- Shin, K.C.; Lee, H.J.; Oh, D.K. Substrate Specificity of Beta-Glucosidase from Gordonia Terrae for Ginsenosides and Its Application in the Production of Ginsenosides Rg(3), Rg(2), and Rh(1) from Ginseng Root Extract. J. Biosci. Bioeng. 2015, 119, 497–504. [Google Scholar] [CrossRef]
- Kil, T.G.; Kang, S.H.; Kim, T.H.; Shin, K.C.; Oh, D.K. Enzymatic Biotransformation of Balloon Flower Root Saponins into Bioactive Platycodin D by Deglucosylation with Caldicellulosiruptor Bescii Beta-Glucosidase. Int. J. Mol. Sci. 2019, 20, 3854. [Google Scholar] [CrossRef]
- Shin, K.C.; Seo, M.J.; Kim, D.W.; Yeom, S.J.; Kim, Y.S. Characterization of Β-Glycosidase from Caldicellulosiruptor Owensensis and Its Application in the Production of Platycodin D from Balloon Flower Leaf. Catalysts 2019, 9, 1025. [Google Scholar] [CrossRef]
- Ahn, H.J.; You, H.J.; Park, M.S.; Johnston, T.V.; Ku, S.; Ji, G.E. Biocatalysis of Platycoside E and Platycodin D3 Using Fungal Extracellular Beta-Glucosidase Responsible for Rapid Platycodin D Production. Int. J. Mol. Sci. 2018, 19, 2671. [Google Scholar] [CrossRef]
- Li, W.; Zhao, L.C.; Wang, Z.; Zheng, Y.N.; Liang, J.; Wang, H. Response Surface Methodology to Optimize Enzymatic Preparation of Deapio-Platycodin D and Platycodin D from Radix Platycodi. Int. J. Mol. Sci. 2012, 13, 4089–4100. [Google Scholar] [CrossRef]
- Jeong, E.K.; Ha, I.J.; Kim, Y.S.; Na, Y.C. Glycosylated Platycosides: Identification by Enzymatic Hydrolysis and Structural Determination by Lc-Ms/Ms. J. Sep. Sci. 2014, 37, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Ha, I.J.; Ha, Y.W.; Kang, M.; Lee, J.; Park, D.; Kim, Y.S. Enzymatic Transformation of Platycosides and One-Step Separation of Platycodin D by High-Speed Countercurrent Chromatography. J. Sep. Sci. 2010, 33, 1916–1922. [Google Scholar] [CrossRef]
- Kang, S.H.; Kim, T.H.; Shin, K.C.; Ko, Y.J.; Oh, D.K. Biotransformation of Food-Derived Saponins, Platycosides, into Deglucosylated Saponins Including Deglucosylated Platycodin D and Their Anti-Inflammatory Activities. J. Agric. Food. Chem. 2019, 67, 1470–1477. [Google Scholar] [CrossRef] [PubMed]
- Wie, H.J.; Zhao, H.L.; Chang, J.H.; Kim, Y.S.; Hwang, I.K.; Ji, G.E. Enzymatic Modification of Saponins from Platycodon grandiflorum with Aspergillus Niger. J. Agric. Food Chem. 2007, 55, 8908–8913. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Yeo, E.; Song, E.; Chang, Y.H.; Han, B.K.; Choi, H.J.; Hwang, J. Bioconversion of Citrus Unshiu Peel Extracts with Cytolase Suppresses Adipogenic Activity in 3t3-L1 Cells. Nutr. Res. Pract. 2015, 9, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Lim, S.; Kim, S.O.; Ahn, S.H.; Choi, Y.J. Optimization of Enzymatic Treatment for Compound K Production from White Ginseng Extract by Response Surface Methodology. Biosci. Biotechnol. Biochem. 2013, 77, 1138–1140. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Kim, S.Y.; Park, Y.; Jung, E.Y.; Suh, H.J. Enzymatic Transformation of Ginsenosides in Korean Red Ginseng (Panax ginseng Meyer) Extract Prepared by Spezyme and Optidex. J. Ginseng. Res. 2014, 38, 264–269. [Google Scholar] [CrossRef]
- Kim, H.W.; Han, S.H.; Lee, S.W.; Choi, H.S.; Suh, H.J.; Hong, K.B. Enzymatic Hydrolysis Increases Ginsenoside Content in Korean Red Ginseng (Panax ginseng Ca Meyer) and Its Biotransformation under Hydrostatic Pressure. J. Sci. Food Agric. 2019, 99, 6806–6813. [Google Scholar] [CrossRef]
- Chapman, J.; Ismail, A.E.; Dinu, C.Z. Industrial Applications of Enzymes: Recent Advances, Techniques, and Outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef]
- Yushkova, E.D.; Nazarova, E.A.; Matyuhina, A.V.; Noskova, A.O.; Shavronskaya, D.O.; Vinogradov, V.V.; Skvortsova, N.N.; Krivoshapkina, E.F. Application of Immobilized Enzymes in Food Industry. J. Agric. Food Chem. 2019, 67, 11553–11567. [Google Scholar] [CrossRef]
- Basso, A.; Serban, S. Overview of Immobilized Enzymes’ Applications in Pharmaceutical, Chemical, and Food Industry. Methods Mol. Biol. 2020, 2100, 27–63. [Google Scholar] [PubMed]
- Ricardi, N.C.; de Menezes, E.W. Valmir Benvenutti, E.; da Natividade Schoffer, J.; Hackenhaar, C.R.; Hertz, P.F.; Costa, T.M.H. Highly Stable Novel Silica/Chitosan Support for Beta-Galactosidase Immobilization for Application in Dairy Technology. Food Chem. 2018, 246, 343–350. [Google Scholar] [CrossRef] [PubMed]
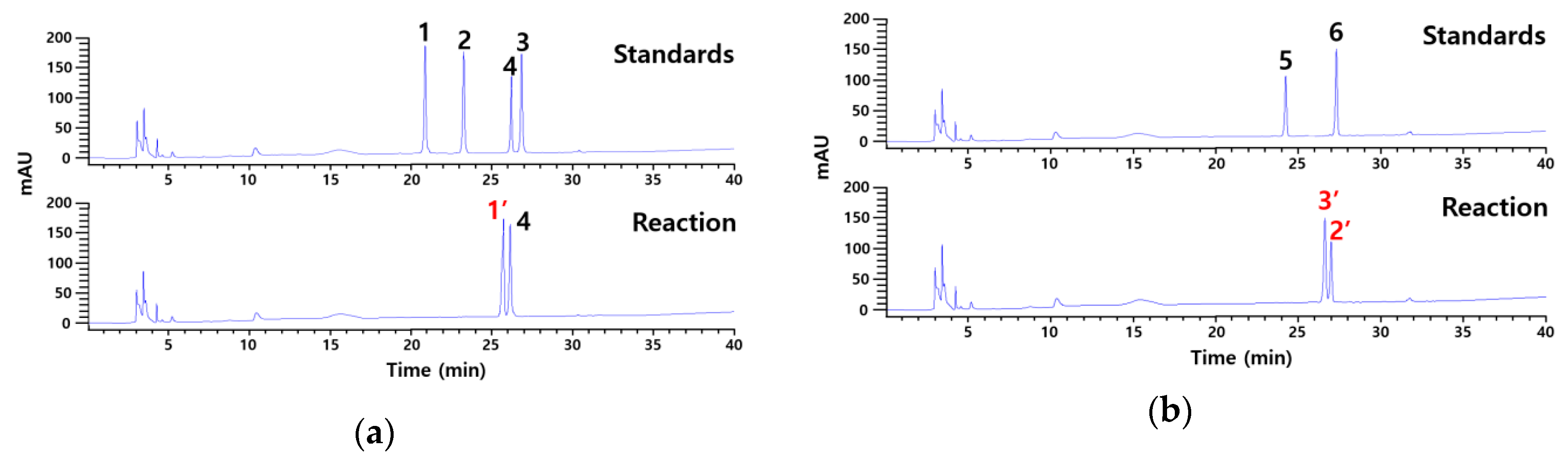

| Compound | Formula | Selected Ion | m/zexperimetnal | m/zcalculated | Error | |
|---|---|---|---|---|---|---|
| mDa | ppm | |||||
| Deapiose-xylosylated platycodin D (1′) | C47H76O20 | [M+H]+ | 961.4966 | 961.5003 | −3.7 | 3.8 |
| Deapiose-xylosylated polygalacin D (2′) | C48H77O21 | [M–H + HCO2H]− | 989.4985 | 989.4957 | 2.8 | 2.8 |
| Deapiosylated polygalacin D (3′) | C53H85O25 | [M–H + HCO2H]− | 1121.5436 | 1124.5380 | 5.6 | 5.0 |
| Substrate | Product | Specific Activity (nmol/min/mg) |
|---|---|---|
| PE | PD3 | 15481.2 ± 27.5 |
| PD3 | PD | 270.9 ± 11.0 |
| PD | Deapi-PD | 30.8 ± 3.2 |
| Deapi-PD | Deapi-Dexyl-PD | 13.3 ± 1.5 |
| Deapi-xyl-PD | − | ND |
| PGD3 | PGD | 844.4 ± 10.2 |
| PGD | Deapi-PGD | 38.6 ± 1.6 |
| Deapi-PGD | Deapi-Dexyl-PGD | 10.7 ± 0.5 |
| Deapi-xyl-PGD | − | ND |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, K.-C.; Kim, D.W.; Woo, H.S.; Oh, D.-K.; Kim, Y.-S. Conversion of Glycosylated Platycoside E to Deapiose-Xylosylated Platycodin D by Cytolase PCL5. Int. J. Mol. Sci. 2020, 21, 1207. https://doi.org/10.3390/ijms21041207
Shin K-C, Kim DW, Woo HS, Oh D-K, Kim Y-S. Conversion of Glycosylated Platycoside E to Deapiose-Xylosylated Platycodin D by Cytolase PCL5. International Journal of Molecular Sciences. 2020; 21(4):1207. https://doi.org/10.3390/ijms21041207
Chicago/Turabian StyleShin, Kyung-Chul, Dae Wook Kim, Hyun Sim Woo, Deok-Kun Oh, and Yeong-Su Kim. 2020. "Conversion of Glycosylated Platycoside E to Deapiose-Xylosylated Platycodin D by Cytolase PCL5" International Journal of Molecular Sciences 21, no. 4: 1207. https://doi.org/10.3390/ijms21041207
APA StyleShin, K.-C., Kim, D. W., Woo, H. S., Oh, D.-K., & Kim, Y.-S. (2020). Conversion of Glycosylated Platycoside E to Deapiose-Xylosylated Platycodin D by Cytolase PCL5. International Journal of Molecular Sciences, 21(4), 1207. https://doi.org/10.3390/ijms21041207





