Protein Expression Profile in Rat Silicosis Model Reveals Upregulation of PTPN2 and Its Inhibitory Effect on Epithelial-Mesenchymal Transition by Dephosphorylation of STAT3
Abstract
1. Introduction
2. Results
2.1. Protein Expression Profiles Undersilica Exposure
2.2. Gene Ontology, KEGG Pathway Analysis and Protein-Protein Network Analysis of Differentially Abundant Proteins
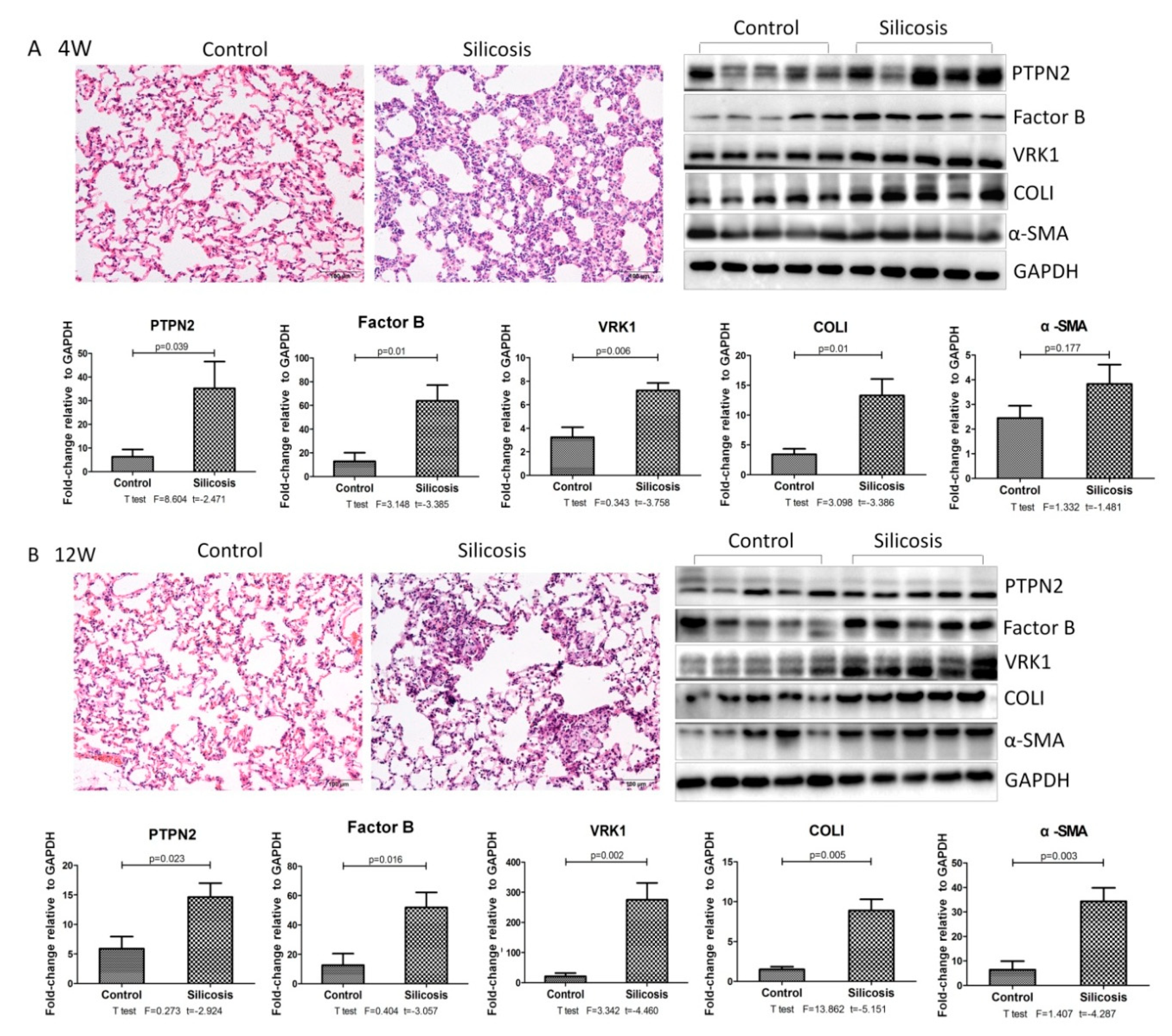
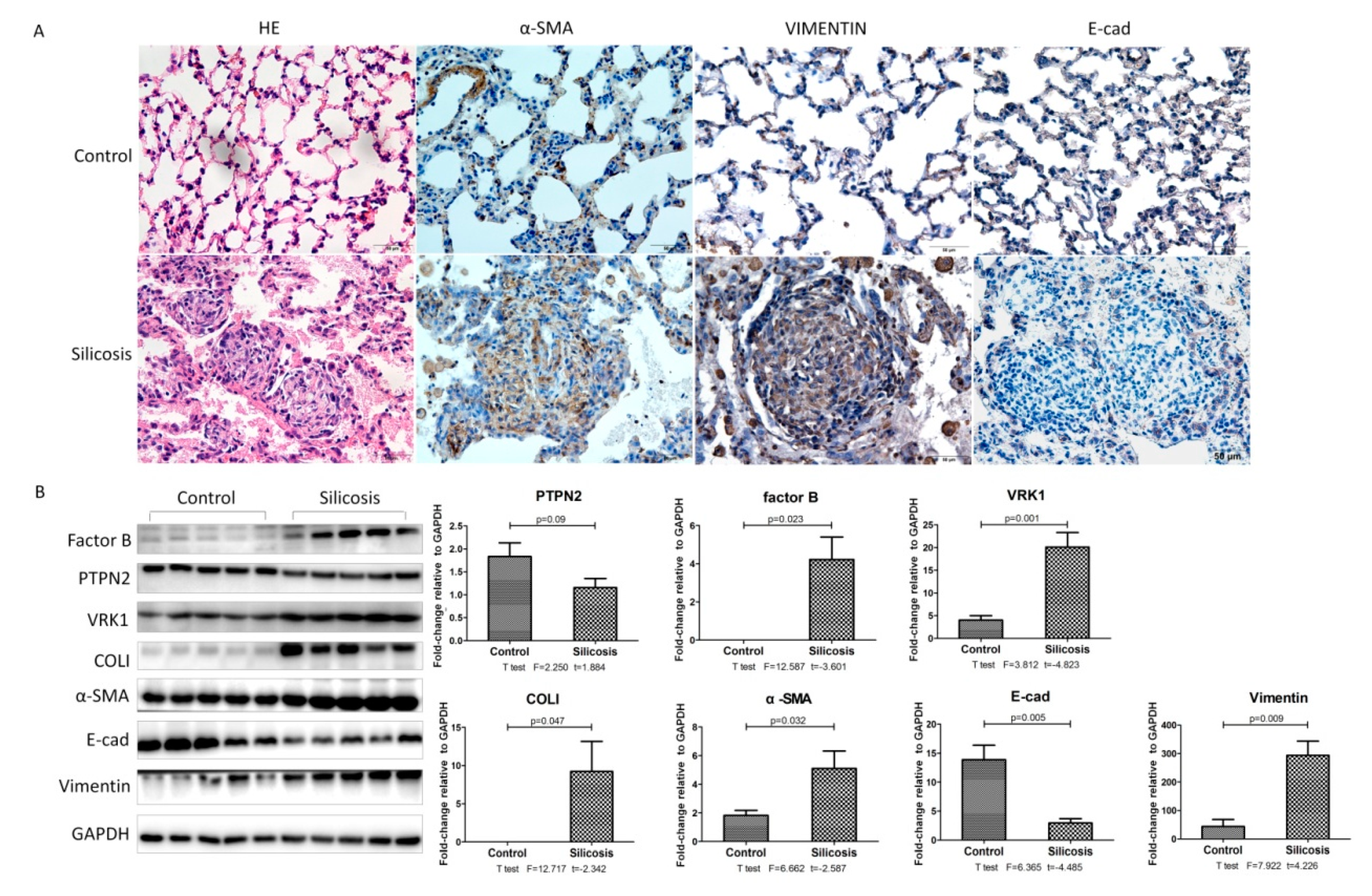
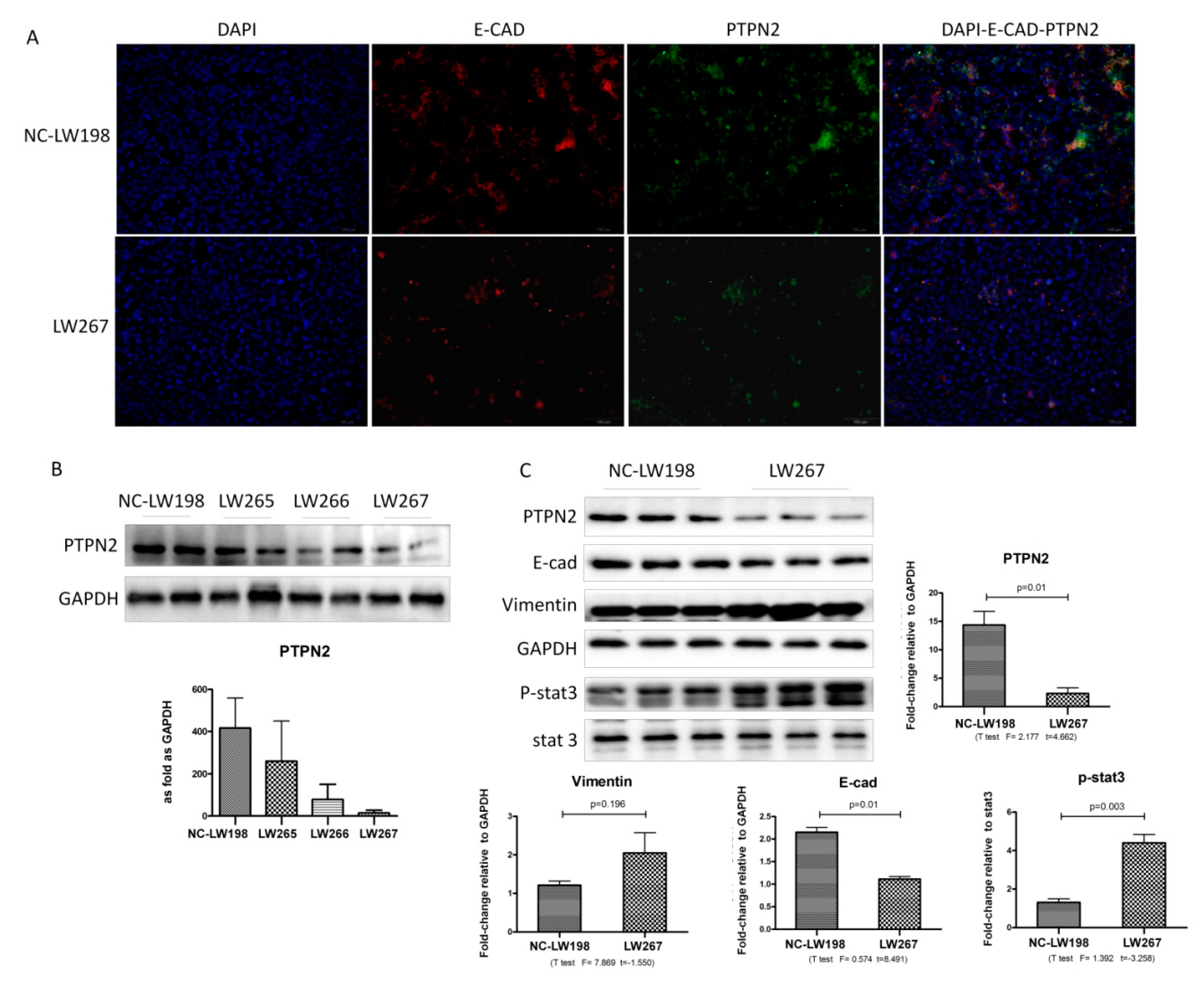
2.3. Validation of iTRAQ-Screened Proteins Exhibiting Increased Expression Levels in the Different Experimental Groups of Silicotic Rats
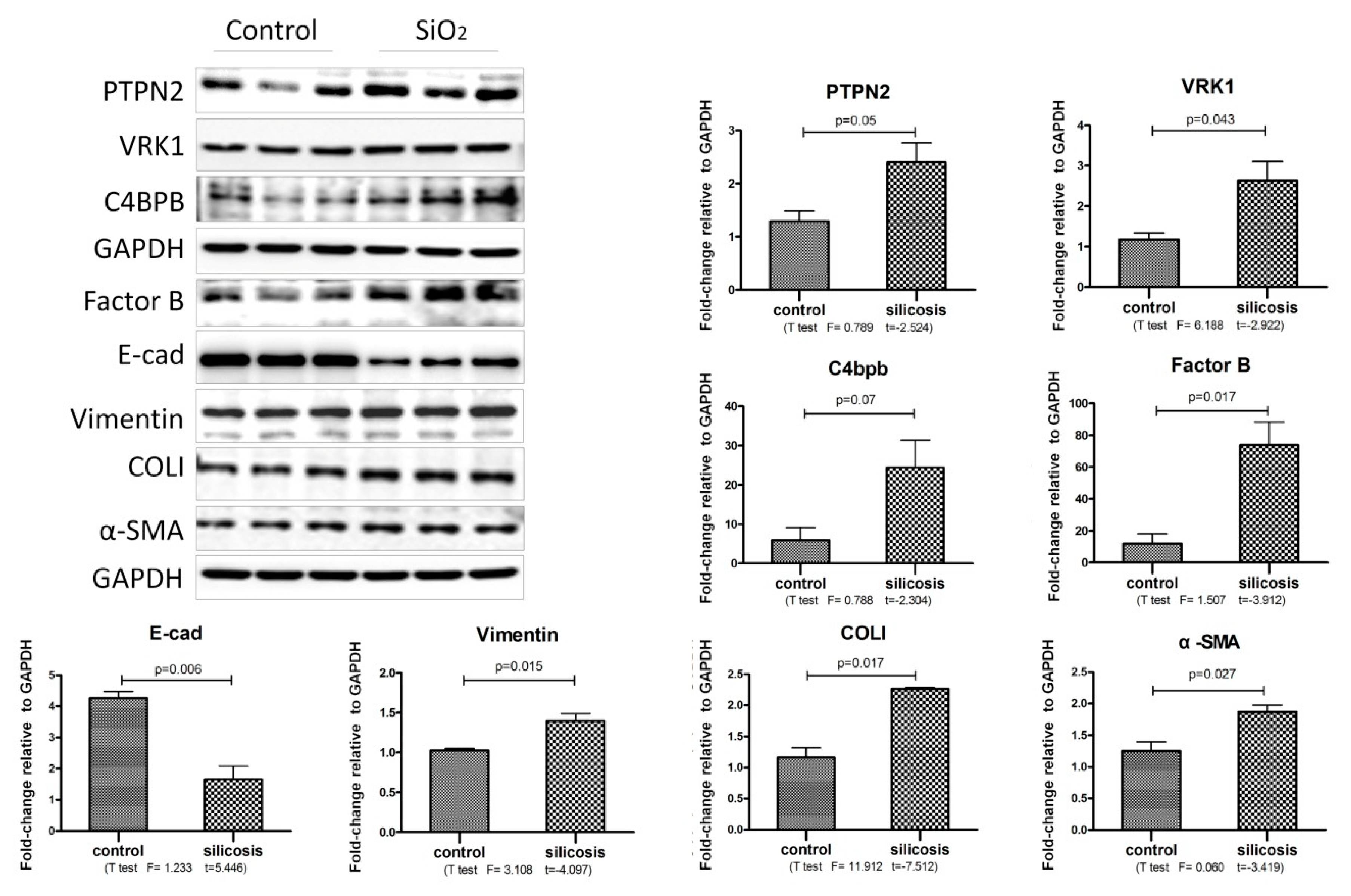
2.4. Validation of iTRAQ-Screened Proteins Exhibiting Increased Expression Levels in SiO2-Stimulated MLE-12 Cells
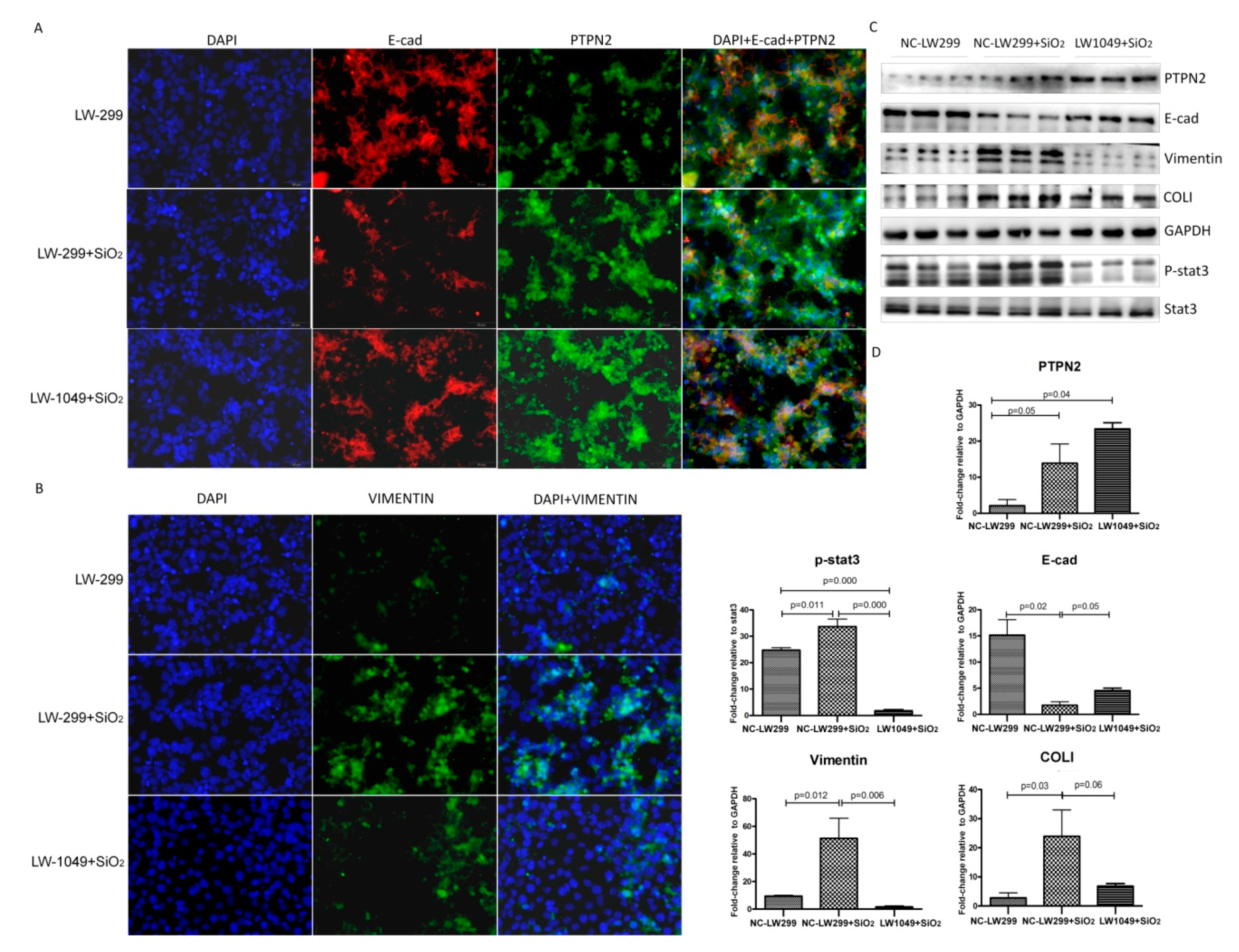
2.5. PTPN2 Overexpression Inhibited the SiO2-Stimulated Epithelial-Mesenchymal Transition of MLE-12 Cells by Dephosphorylating STAT3
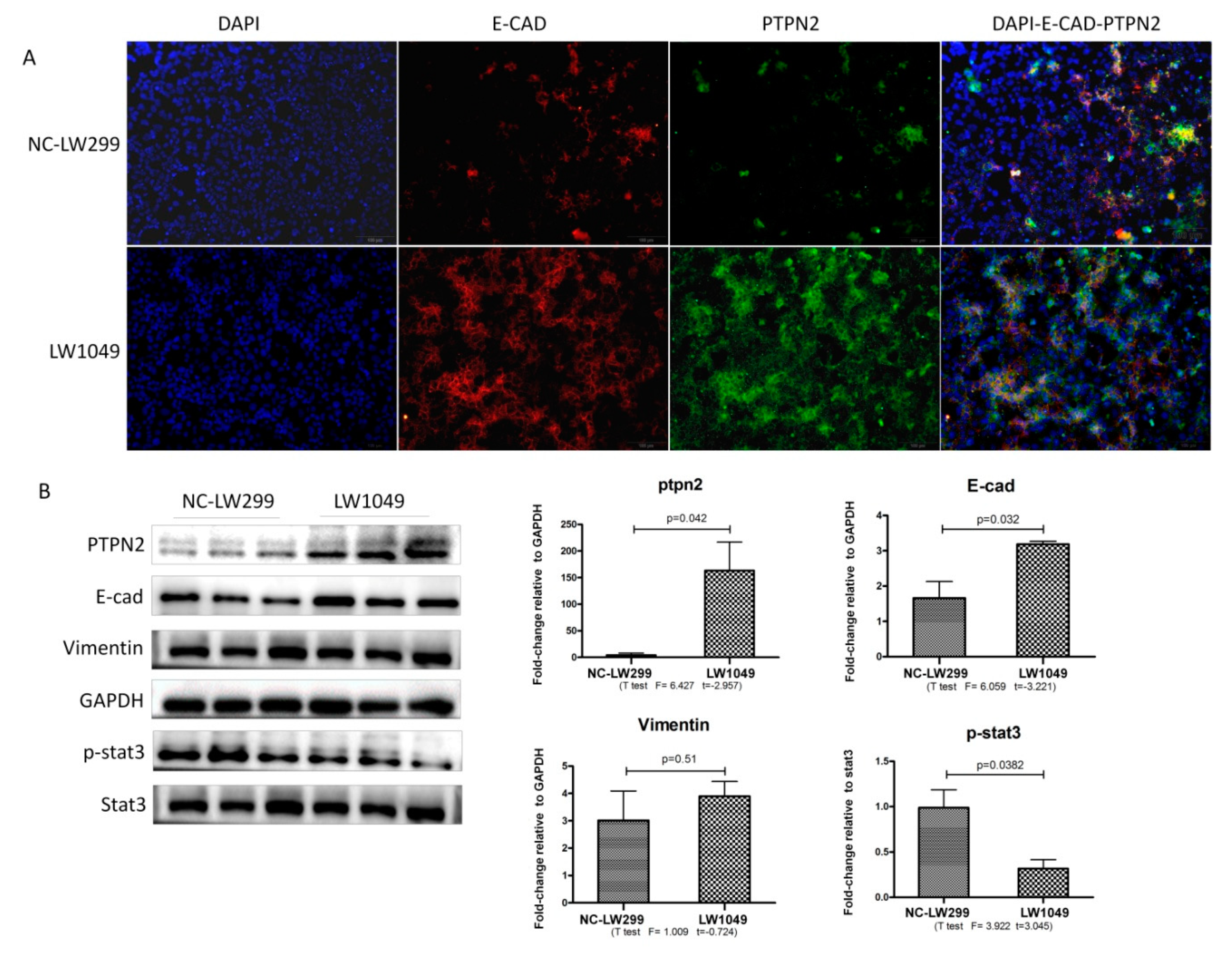
2.6. PTPN2-Gene Silencing Promoted the SiO2-Stimulated EMT of MLE-12 Cells by Phosphorylating STAT3
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Rat Models
4.3. Protein Extraction, Digestion, and iTRAQ Labeling
4.4. Separation of Peptides and LC Mass Spectrometric Analysis
4.5. Bioinformatic Analysis
4.6. Cell Culture and Treatment
4.7. Design and Construction of Lentivirus Vector
4.7.1. Construction of Plasmid Vector for PTPN2 Gene Knockdown Lentivirus
4.7.2. Construction of Plasmid Vector for PTPN2 Gene Overexpress Lentivirus
4.7.3. The Process of Lentivirus Packaging Plasmids
4.8. Lentiviral Vector Infection
4.9. Western Blot Analysis
4.10. Immunofluorescence Staining
4.11. Immunohistochemistry Staining
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethics Approval
References
- Zhao, J.Q.; Li, J.G.; Zhao, C.X. Prevalence of pneumoconiosis among young adults aged 24–44 years in a heavily industrialized province of China. J. Occup. Health 2019, 61, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Komai, M.; Mihira, K.; Shimada, A.; Miyamoto, I.; Ogihara, K.; Naya, Y.; Morita, T.; Inoue, K.; Takano, H. Pathological Study on Epithelial- Mesenchymal Transition in Silicotic Lung Lesions in Rat. Vet. Sci. 2019, 6, 70. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.C.; Yu, I.T.; Chen, W. Silicosis. Lancet 2012, 379, 2008–2018. [Google Scholar] [CrossRef]
- Yang, M.; Wang, N.; Li, W.; Li, H.; Zhao, Y.; Yao, S.; Chen, W. Therapeutic effects of scavenger receptor MARCO ligand on silica-induced pulmonary fibrosis in rats. Toxicol. Lett. 2019, 311, 1–10. [Google Scholar] [CrossRef]
- Yen, C.M.; Lin, C.L.; Lin, M.C.; Chen, H.Y.; Lu, N.H.; Kao, C.H. Pneumoconiosis increases the risk of congestive heart failure: A nationwide population-based cohort study. Medicine (Baltimore) 2016, 95. [Google Scholar] [CrossRef]
- Wang, K.; Wang, X.; Zheng, S.; Niu, Y.; Zheng, W.; Qin, X.; Li, Z.; Luo, J.; Jiang, W.; Zhou, X. iTRAQ-based quantitative analysis of age-specific variations in salivary proteome of caries-susceptible individuals. J. Transl. Med. 2018, 16, 293. [Google Scholar] [CrossRef]
- Yin, H.R.; Zhang, L.; Xie, L.Q.; Huang, L.Y.; Xu, Y.; Cai, S.J.; Yang, P.Y.; Lu, H.J. Hyperplex-MRM: A Hybrid Multiple Reaction Monitoring Method Using mTRAQ/iTRAQ Labeling for Multiplex Absolute Quantification of Human Colorectal Cancer Biomarker. J. Proteome Res. 2013, 12, 3912–3919. [Google Scholar] [CrossRef]
- McIntosh, M.; Fitzgibbon, M. Biomarker validation by targeted mass spectrometry. Nat. Biotechnol. 2009, 27, 622–623. [Google Scholar] [CrossRef]
- Fu, R.; Li, Q.; Fan, R.; Zhou, Q.; Jin, X.; Cao, J.; Wang, J.; Ma, Y.; Yi, T.; Zhou, M. iTRAQ-based secretome reveals that SiO2 induces the polarization of RAW264.7 macrophages by activation of the NOD-RIP2-NF-kappaB signaling pathway. Environ. Toxicol. Pharmacol. 2018, 63, 92–102. [Google Scholar] [CrossRef]
- Na, M.; Hong, X.; Fuyu, J.; Dingjie, X.; Sales, D.; Hui, Z.; Zhongqiu, W.; Shifeng, L.; Xuemin, G.; Wenchen, C.; et al. Proteomic profile of TGF-beta1 treated lung fibroblasts identifies novel markers of activated fibroblasts in the silica exposed rat lung. Exp. Cell Res. 2019, 375, 1–9. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, W.; Zhang, D.; Wang, M.; Aprecio, R.; Ji, N.; Mohamed, O.; Li, Y.; Ding, Y.; Wang, Q. 25-Hydroxyvitamin D-3-enhanced PTPN2 positively regulates periodontal inflammation through the JAK/STAT pathway in human oral keratinocytes and a mouse model of type 2 diabetes mellitus. J. Periodontal Res. 2018, 53, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.C.; Huang, C.M.; Bamodu, O.A.; Lin, C.S.; Liu, B.L.; Tzeng, Y.M.; Tsai, J.T.; Lee, W.H.; Chen, T.M. Ovatodiolide suppresses nasopharyngeal cancer by targeting stem cell-like population, inducing apoptosis, inhibiting EMT and dysregulating JAK/STAT signaling pathway. Phytomedicine 2019, 56, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Pedroza, M.; Le, T.T.; Lewis, K.; Karmouty-Quintana, H.; To, S.; George, A.T.; Blackburn, M.R.; Tweardy, D.J.; Agarwal, S.K. STAT-3 contributes to pulmonary fibrosis through epithelial injury and fibroblast-myofibroblast differentiation. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2016, 30, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Morales, L.D.; Jang, I.S.; Cho, Y.Y.; Kim, D.J. Protein Tyrosine Phosphatases as Potential Regulators of STAT3 Signaling. Int. J. Mol. Sci. 2018, 19, 2708. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Yan, N.; Du, Y.; Liu, X.; Chu, M.; Shi, J.; Zhang, H.; Liu, Y.; Zhang, Z. iTRAQ-based proteomic analysis reveals the accumulation of bioactive compounds in Chinese wild rice (Zizania latifolia) during germination. Food Chem. 2019, 289, 635–644. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhang, C.; Gan, L.; Jia, Y.; Xiong, Y.; Chen, Y.; Wang, Z.; Wang, L.; Luo, H.; Li, J. iTRAQ-Based Quantitative Proteomics Approach Identifies Novel Diagnostic Biomarkers that were Essential for Glutamine Metabolism and Redox Homeostasis for Gastric Cancer. Proteom. Clin. Appl. 2018, 13. [Google Scholar] [CrossRef]
- Du, C.; Weng, Y.; Lou, J.; Zeng, G.; Liu, X.; Jin, H.; Lin, S.; Tang, L. Isobaric tags for relative and absolute quantitationbased proteomics reveals potential novel biomarkers for the early diagnosis of acute myocardial infarction within 3 h. Int. J. Mol. Med. 2019, 43, 1991–2004. [Google Scholar]
- Zhou, F.; Chen, X.; Chen, G.; Yan, J.; Xiao, Y. Identification of SAA and ACTB as potential biomarker of patients with severe HFMD using iTRAQ quantitative proteomics. Clin. Biochem. 2019, 67, 1–6. [Google Scholar] [CrossRef]
- Arora, A.; Patil, V.; Kundu, P.; Kondaiah, P.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Pal, D.; Somasundaram, K. Serum biomarkers identification by iTRAQ and verification by MRM: S100A8/S100A9 levels predict tumor-stroma involvement and prognosis in Glioblastoma. Sci. Rep. 2019, 9, 2749. [Google Scholar] [CrossRef]
- Ricklin, D.; Lambris, J.D. Complement in Immune and Inflammatory Disorders: Pathophysiological Mechanisms. J. Immunol. 2013, 190, 3831–3838. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Chen, M.H.; Jing, Y.; Gu, J.H.; Mei, S.Q.; Yao, Q.; Zhou, J.; Yang, M.; Sun, L.J.; Wang, W.T. The C-terminal tail of polycystin-1 regulates complement factor B expression by signal transducer and activator of transcription 1. Am. J. Physiol. Ren. Physiol. 2016, 310, F1284–F1294. [Google Scholar] [CrossRef] [PubMed]
- Kaczorowski, D.J.; Afrazi, A.; Scott, M.J.; Kwak, J.H.; Gill, R.; Edmonds, R.D.; Liu, Y.J.; Fan, J.; Billiar, T.R. Pivotal Advance: The pattern recognition receptor ligands lipopolysaccharide and polyinosine-polycytidylic acid stimulate factor B synthesis by the macrophage through distinct but overlapping mechanisms. J. Leukoc. Biol. 2010, 88, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Lake, F.R.; Noble, P.W.; Henson, P.M.; Riches, D.W.H. Functional Switching of Macrophage Responses to Tumor-Necrosis-Factor-Alpha (Tnf-Alpha) by Interferons—Implications for the Pleiotropic Activities of Tnf-Alpha. J. Clin. Investig. 1994, 93, 1661–1669. [Google Scholar] [CrossRef] [PubMed]
- Dobrzynska, A.; Askjaer, P. Vaccinia-related kinase 1 is required for early uterine development in Caenorhabditis elegans. Dev. Biol. 2016, 411, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Salzano, M.; Sanz-Garcia, M.; Monsalve, D.M.; Moura, D.S.; Lazo, P.A. VRK1 chromatin kinase phosphorylates H2AX and is required for foci formation induced by DNA damage. Epigenetics 2015, 10, 373–383. [Google Scholar] [CrossRef]
- Molitor, T.P.; Traktman, P. Molecular genetic analysis of VRK1 in mammary epithelial cells: Depletion slows proliferation in vitro and tumor growth and metastasis in vivo. Oncogenesis 2013, 2, e48. [Google Scholar] [CrossRef]
- Mon, A.M.; MacKinnon, A.C.; Traktman, P. Overexpression of the VRK1 kinase, which is associated with breast cancer, induces a mesenchymal to epithelial transition in mammary epithelial cells. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Cockrell, A.S.; Kafri, T. Gene delivery by lentivirus vectors. Mol. Biotechnol. 2007, 36, 184–204. [Google Scholar] [CrossRef]
- Zhang, L.J.; Xu, D.J.; Li, Q.; Yang, Y.; Xu, H.; Wei, Z.Q.; Wang, R.M.; Zhang, W.L.; Liu, Y.; Geng, Y.C.; et al. N-acetyl-seryl-aspartyl-lysyl-proline (Ac-SDKP) attenuates silicotic fibrosis by suppressing apoptosis of alveolar type II epithelial cells via mediation of endoplasmic reticulum stress. Toxicol. Appl. Pharmacol. 2018, 350, 1–10. [Google Scholar] [CrossRef]
- Wang, X.J.; Liu, Y.; Xu, H.; Zhang, X.H.; Li, S.F.; Xu, D.J.; Gao, X.M.; Zhang, L.J.; Zhang, B.N.; Wei, Z.Q.; et al. Acetylated alpha-Tubulin Regulated by N-Acetyl-Seryl-Aspartyl-Lysyl-Proline(Ac-SDKP) Exerts the Antifibrotic Effect in Rat Lung Fibrosis Induced by Silica. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Jin, X.; Zhu, L.; Tao, C.; Xie, Q.; Xu, X.; Chang, L.; Tan, Y.; Ding, G.; Li, H.; Wang, X. An improved protein extraction method applied to cotton leaves is compatible with 2-DE and LC-MS. BMC Genom. 2019, 20, 285. [Google Scholar] [CrossRef] [PubMed]
- Zor, T.; Selinger, Z. Linearization of the Bradford protein assay increases its sensitivity: Theoretical and experimental studies. Anal. Biochem. 1996, 236, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, A.J. Java Treeview--extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Rodriguez, M.; Aladowicz, E.; Lanfrancone, L.; Goding, C.R. Tbx3 represses E-cadherin expression and enhances melanoma invasiveness. Cancer Res. 2008, 68, 7872–7881. [Google Scholar] [CrossRef]


| No. | Protein Name | Accession Number | Fold Change |
|---|---|---|---|
| 1 | Acidic mammalian chitinase | [CHIA_RAT] | 4.277554 |
| 2 | Tyrosine-protein phosphatase non-receptor type 2 | [PTN2_RAT] | 2.292036 |
| 3 | Protein Vrk1 | [VRK1_RAT] | 2.126730 |
| 4 | B-factor, properdin | [Q6MG74_RAT] | 1.970988 |
| 5 | Complement component 4, gene 2 | [Q6MG90_RAT] | 1.822588 |
| 6 | Cd68 molecule | [Q4FZY1_RAT] | 1.773507 |
| 7 | Protein Spcs1 | [D3ZFK5_RAT] | 1.749611 |
| 8 | Equilibrative nucleoside transporter 3 | [ENT3_RAT] | 1.731603 |
| 9 | BPI fold-containing family B member 1 | [BPIB1_RAT] | 1.716078 |
| 10 | Monocarboxylate transporter 4 | [MOT4_RAT] | 1.680098 |
| 11 | Transmembrane glycoprotein NMB | [GPNMB_RAT] | 1.676391 |
| 12 | A-kinase anchor protein SPHKAP | [SPKAP_RAT] | 1.672275 |
| 13 | Complement C4 | [CO4_RAT] | 1.652953 |
| 14 | Olfactory receptor | [D3ZRJ8_RAT] | 1.652572 |
| 15 | Phospholipase B-like 1 | [PLBL1_RAT] | 1.585530 |
| 16 | Chitinase 3-like 1 protein (Fragment) | [A4LA56_RAT] | 1.558782 |
| 17 | Cell division cycle protein 123 homolog | [CD123_RAT] | 1.546649 |
| 18 | Protein Stk33 | [D4A165_RAT] | 1.532094 |
| 19 | Beta-defensin 4 | [DEFB4_RAT] | 1.519443 |
| 20 | UDP-glucuronosyltransferase | [Q6T5E8_RAT] | 1.506820 |
| 21 | Protein Slc16a2 | [G3V9C2_RAT] | −1.877088 |
| 22 | Carboxylic ester hydrolase | [A0A0G2JV37_RAT] | −1.842240 |
| 23 | Protein Spns2 | [D3ZPS4_RAT] | −1.796205 |
| 24 | PDZ domain-containing protein 2 | [F1M785_RAT] | −1.666518 |
| 25 | Protein Dcp1b | [D3ZZJ7_RAT] | −1.571295 |
| 26 | CD34 antigen isoform 2 | [B1PLB2_RAT] | −1.566583 |
| 27 | Protein Shroom2 | [SHRM2_RAT] | −1.541996 |
| 28 | Periaxin | [PRAX_RAT] | −1.528930 |
| Vector Number | Lenticirus Titer |
|---|---|
| pHS-ASR-LW265 | 1.08 × 108 TU/mL |
| pHS-ASR-LW266 | 1.02 × 108 TU/mL |
| pHS-ASR-LW267 | 1.37 × 108 TU/mL |
| pHS-ASR-LW198 | 1.09 × 108 TU/mL |
| pHS-AVC-LW1049 | 1.03 × 108 TU/mL |
| pHS-BVC-LW299 | 1.83 × 108 TU/mL |
| Vector Number | Inserted Content | shRNA Sequence |
|---|---|---|
| pHS-ASR-LW265 | PTPN2 shRNA01 | 5′-GCATTCTACGGAAACGTATTC CGAAGAATACGTTTCCGTAGAATGC-3′ |
| pHS-ASR-LW266 | PTPN2 shRNA02 | 5′-GCATTAATCTCAAGGGTTTGT CGAAACAAACCCTTGAGATTAATGC-3′ |
| pHS-ASR-LW267 | PTPN2 shRNA03 | 5′-GCAGGTAAGAAGGATGTTTCC CGAAGGAAACATCCTTCTTACCTGC-3′ |
| pHS-ASR-LW198 | Neg control shRNA | 5′-CCTAAGGTTAAGTCGCCCTCGC CGAAGCGAGGGCGACTTAACCTTAGG-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Yao, J.; Duan, Y.; Xu, H.; Cheng, Q.; Gao, X.; Li, S.; Yang, F.; Liu, H.; Yuan, J. Protein Expression Profile in Rat Silicosis Model Reveals Upregulation of PTPN2 and Its Inhibitory Effect on Epithelial-Mesenchymal Transition by Dephosphorylation of STAT3. Int. J. Mol. Sci. 2020, 21, 1189. https://doi.org/10.3390/ijms21041189
Zhu Y, Yao J, Duan Y, Xu H, Cheng Q, Gao X, Li S, Yang F, Liu H, Yuan J. Protein Expression Profile in Rat Silicosis Model Reveals Upregulation of PTPN2 and Its Inhibitory Effect on Epithelial-Mesenchymal Transition by Dephosphorylation of STAT3. International Journal of Molecular Sciences. 2020; 21(4):1189. https://doi.org/10.3390/ijms21041189
Chicago/Turabian StyleZhu, Ying, Jingxin Yao, Yuxia Duan, Hong Xu, Qiyun Cheng, Xuemin Gao, Shumin Li, Fang Yang, Heliang Liu, and Juxiang Yuan. 2020. "Protein Expression Profile in Rat Silicosis Model Reveals Upregulation of PTPN2 and Its Inhibitory Effect on Epithelial-Mesenchymal Transition by Dephosphorylation of STAT3" International Journal of Molecular Sciences 21, no. 4: 1189. https://doi.org/10.3390/ijms21041189
APA StyleZhu, Y., Yao, J., Duan, Y., Xu, H., Cheng, Q., Gao, X., Li, S., Yang, F., Liu, H., & Yuan, J. (2020). Protein Expression Profile in Rat Silicosis Model Reveals Upregulation of PTPN2 and Its Inhibitory Effect on Epithelial-Mesenchymal Transition by Dephosphorylation of STAT3. International Journal of Molecular Sciences, 21(4), 1189. https://doi.org/10.3390/ijms21041189





