Novel Biomarkers of Hepatitis B and Hepatocellular Carcinoma: Clinical Significance of HBcrAg and M2BPGi
Abstract
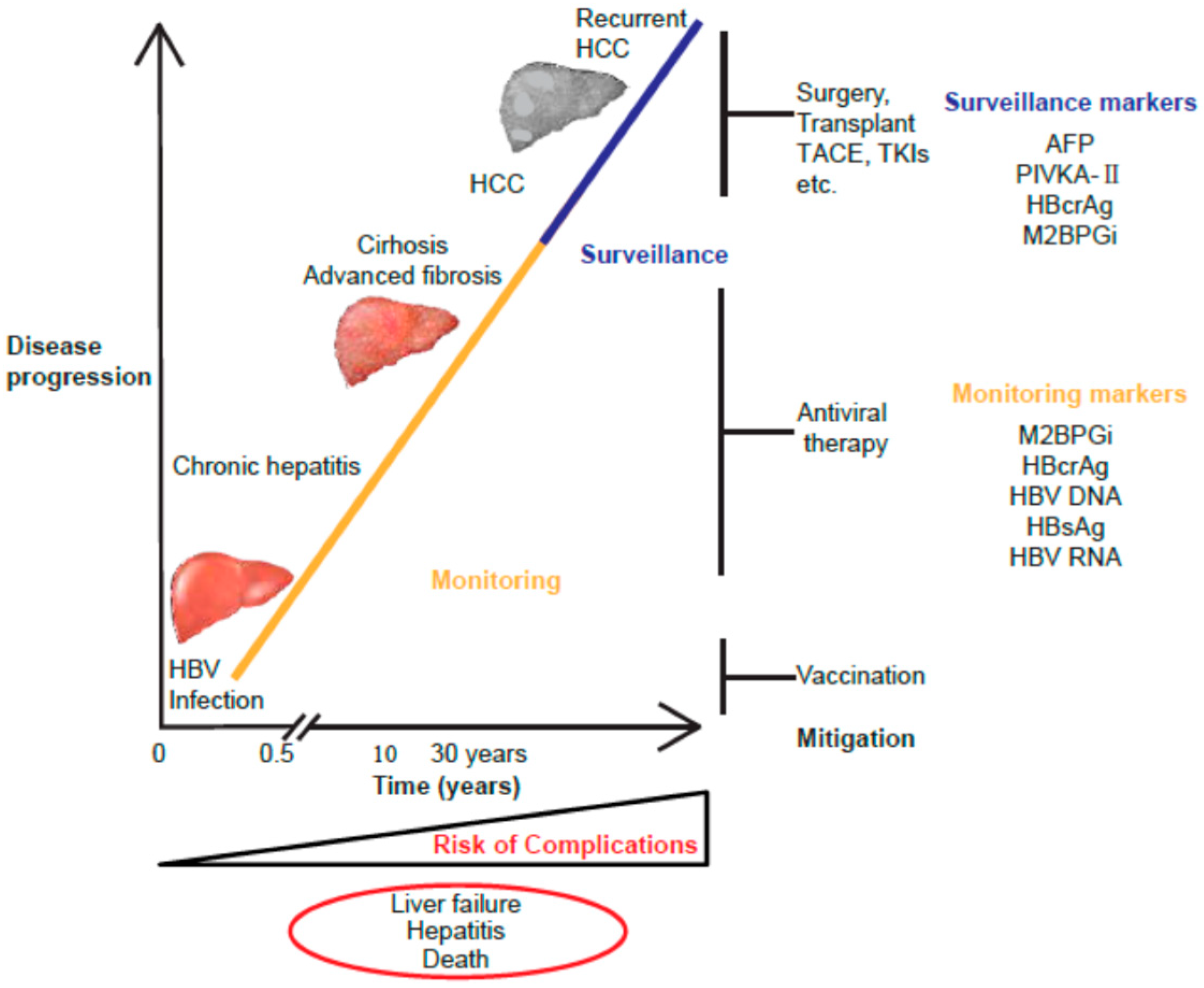
1. Introduction
2. HBV Natural History and Biomarkers
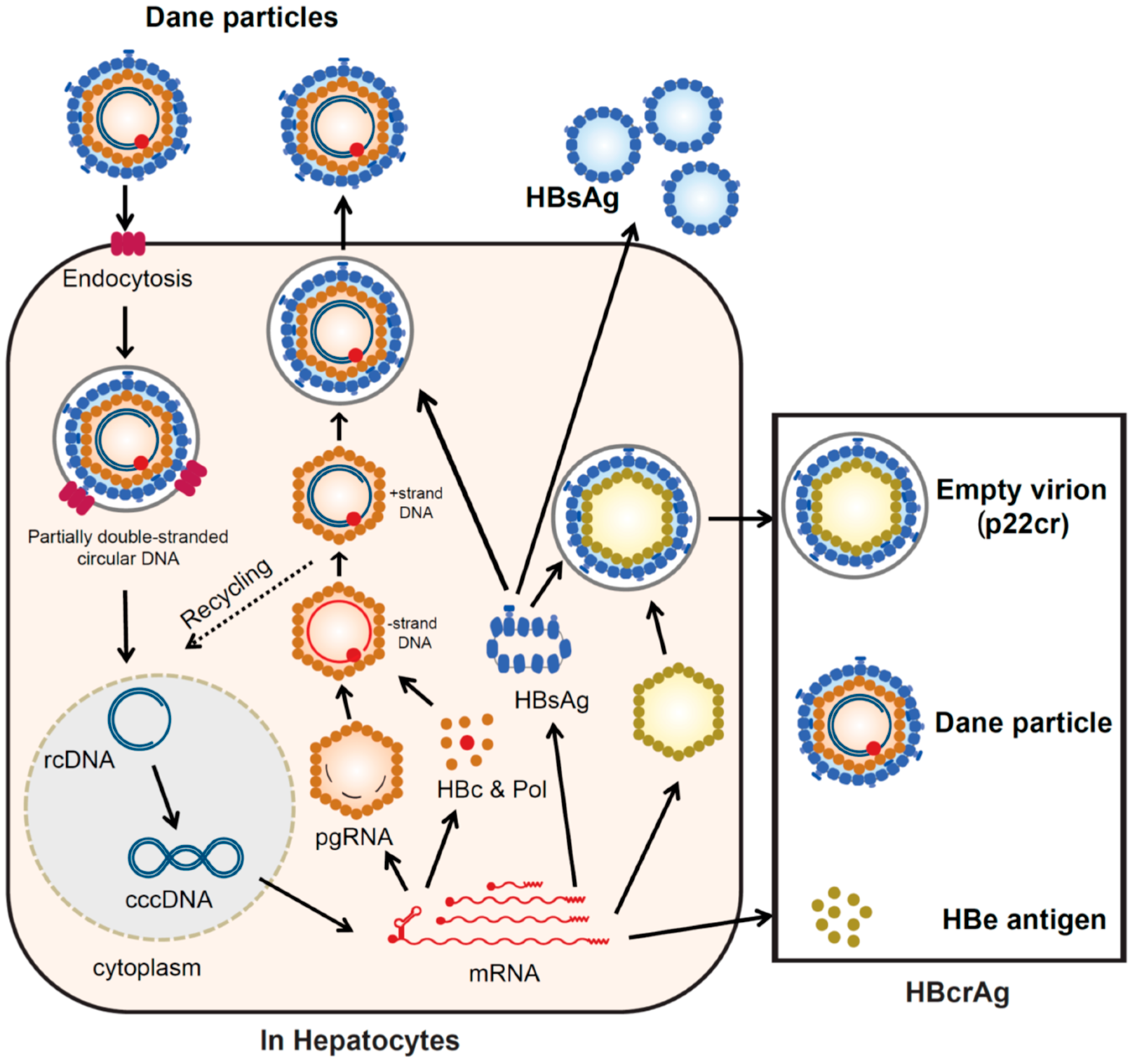
2.1. HBV Replication Cycle
2.2. Screening of Serological Markers Related to HBV Infection
2.3. Monitoring of Serological Markers Related to HBV Infection
3. HBcrAg
3.1. Configuration of HBcrAg
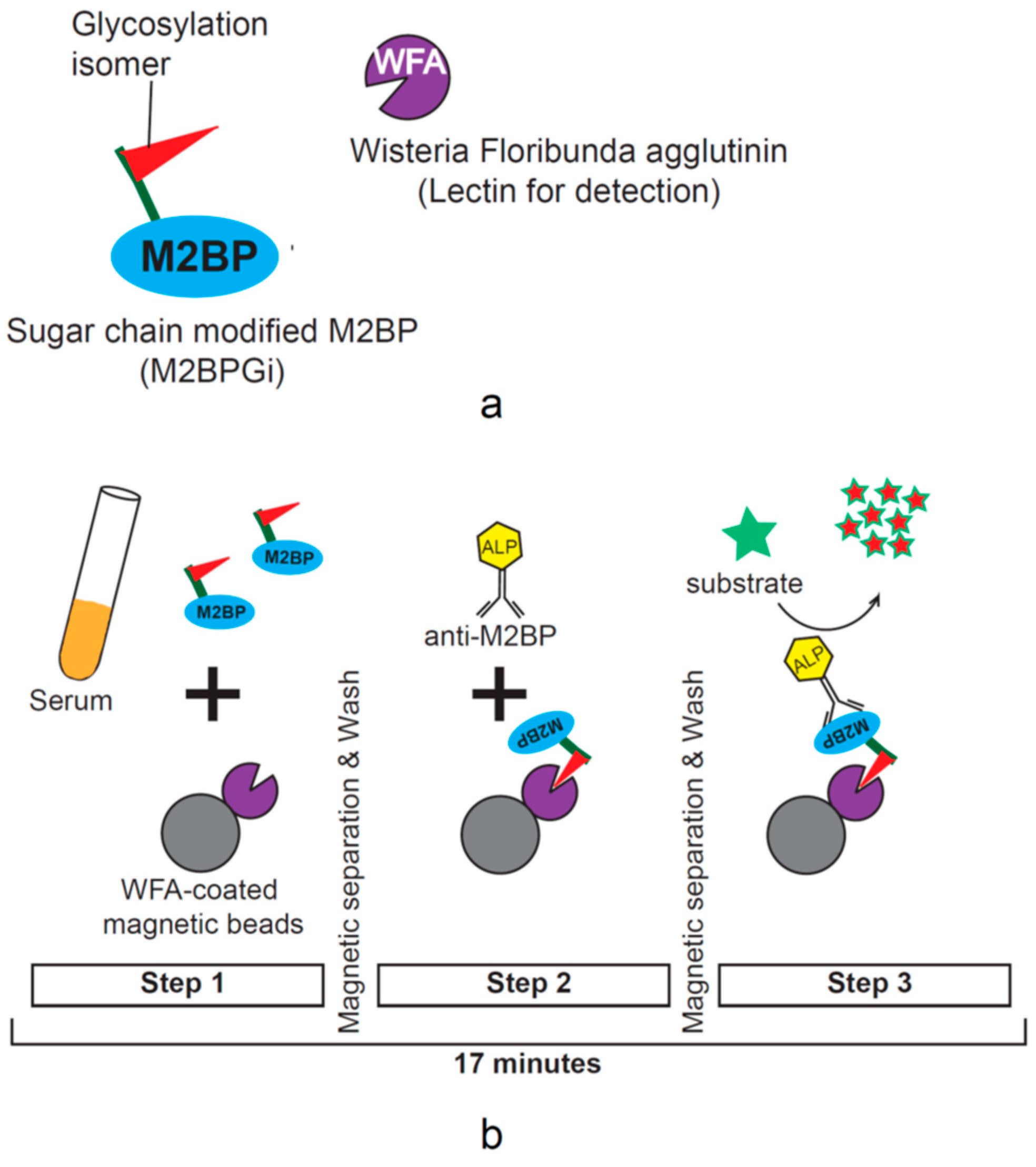
3.2. History of HBcrAg Testing and Development of Quantitative Assays
3.3. Recent Clinical Assessments of HBcrAg
3.4. HBcrAg as a Predictor of HCC Occurrence
3.5. HBcrAg as a Predictor of HCC Recurrence
4. M2BPGi
4.1. M2BPGi and Its Characteristics
4.2. M2BPGi as a Predictor of Hepatocarcinogenesis and Marker of Fibrosis in Chronic Hepatitis C (CHC)
4.3. M2BPGi for HCC Prediction in CHB
5. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HBV | hepatitis B virus |
| HCC | hepatocellular carcinoma |
| CHB | chronic hepatitis B |
| NAs | nucleos(t)ide analogs |
| IFN | interferon |
| cccDNA | covalently closed circular DNA |
| AFP | alpha-fetoprotein |
| PIVKA-II | vitamin K absence or antagonist-II |
| HBcrAg | hepatitis B core-related antigen |
| HBsAg | hepatitis B surface antigen |
| M2BPGi | Mac-2-binding protein glycosylation isomer |
| NTCP | sodium taurocholate co-transporting polypeptide |
| HBeAg | hepatitis B envelope antigen |
| IgM anti-HBc | immunoglobulin M antibody to hepatitis B core antigen |
| IgG anti-HBc | immunoglobulin G antibody to hepatitis B core antigen |
| HBsAg-HQ | high-sensitivity HBsAg assay |
| ICT CLEIA | immune complex transfer chemiluminescence enzyme immunoassay |
| AUROC | the area under the receiver operating characteristic curve |
| BCP | basal core promotor |
| M2BP | Mac-2-binding protein |
| WTA | Wisteria floribunda agglutinin |
| C.O.I. | cutoff index |
| CHC | chronic hepatitis C |
| SVR | sustained virological response |
| BMI | body mass index |
References
- Graber-Stiehl, I. The silent epidemic killing more people than HIV, malaria or TB. Nature 2018, 564, 24–26. [Google Scholar] [CrossRef]
- World Health Organization. Global Hepatitis Report 2017; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Inoue, T.; Tanaka, Y. Hepatitis B virus and its sexually transmitted infection - an update. Microbial Cell 2016, 3, 420–437. [Google Scholar] [CrossRef]
- Pellicoro, A.; Ramachandran, P.; Iredale, J.P.; Fallowfield, J.A. Liver fibrosis and repair: Immune regulation of wound healing in a solid organ. Nat. Rev. Immunol. 2014, 14, 181–194. [Google Scholar] [CrossRef]
- EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017, 67, 370–398. [CrossRef] [PubMed]
- Terrault, N.A.; Lok, A.S.F.; McMahon, B.J.; Chang, K.M.; Hwang, J.P.; Jonas, M.M.; Brown, R.S., Jr.; Bzowej, N.H.; Wong, J.B. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 2018, 67, 1560–1599. [Google Scholar] [CrossRef]
- Lok, A.S.; Zoulim, F.; Dusheiko, G.; Ghany, M.G. Hepatitis B cure: From discovery to regulatory approval. Hepatology 2017, 66, 1296–1313. [Google Scholar] [CrossRef] [PubMed]
- Glebe, D.; Bremer, C.M. The molecular virology of hepatitis B virus. Semin. Liver Dis. 2013, 33, 103–112. [Google Scholar] [PubMed]
- Lopatin, U. Drugs in the Pipeline for HBV. Clin. Liver Dis. 2019, 23, 535–555. [Google Scholar] [CrossRef] [PubMed]
- Gane, E.J. Future anti-HBV strategies. Liver Int. 2017, 37, 40–44. [Google Scholar] [CrossRef]
- Inoue, T.; Tanaka, Y. The Role of Hepatitis B Core-Related Antigen. Genes 2019, 10, 357. [Google Scholar] [CrossRef]
- Bellan, M.; Castello, L.M.; Pirisi, M. Candidate Biomarkers of Liver Fibrosis: A Concise, Pathophysiology-oriented Review. J. Clin. Transl. Hepatol. 2018, 6, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, J.; Verhulst, S.; Mannaerts, I.; Reynaert, H.; van Grunsven, L.A. Prospects in non-invasive assessment of liver fibrosis: Liquid biopsy as the future gold standard? Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1024–1036. [Google Scholar] [CrossRef] [PubMed]
- Parikh, P.; Ryan, J.D.; Tsochatzis, E.A. Fibrosis assessment in patients with chronic hepatitis B virus (HBV) infection. Ann. Transl. Med. 2017, 5, 40. [Google Scholar] [CrossRef] [PubMed]
- Chin, J.L.; Pavlides, M.; Moolla, A.; Ryan, J.D. Non-invasive Markers of Liver Fibrosis: Adjuncts or Alternatives to Liver Biopsy? Front. Pharmacol. 2016, 7, 159. [Google Scholar] [CrossRef] [PubMed]
- JSH Guidelines for the Management of Hepatitis B Virus Infection. Hepatol. Res. 2014, 44 (Suppl S1), 1–58.
- Kawaguchi, K.; Honda, M.; Ohta, H.; Terashima, T.; Shimakami, T.; Arai, K.; Yamashita, T.; Sakai, Y.; Yamashita, T.; Mizukoshi, E.; et al. Serum Wisteria floribunda agglutinin-positive Mac-2 binding protein predicts hepatocellular carcinoma incidence and recurrence in nucleos(t)ide analogue therapy for chronic hepatitis B. J. Gastroenterol. 2018, 53, 740–751. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Q.; Lu, W.; Wang, Y.B.; Weng, Q.C.; Zhang, Z.Y.; Yang, Z.Q.; Feng, Y.L. Measurement of the hepatitis B core-related antigen is valuable for predicting the pathological status of liver tissues in chronic hepatitis B patients. J. Virol. Methods 2016, 235, 92–98. [Google Scholar] [CrossRef]
- Jun, T.; Hsu, Y.C.; Ogawa, S.; Huang, Y.T.; Yeh, M.L.; Tseng, C.H.; Huang, C.F.; Tai, C.M.; Dai, C.Y.; Huang, J.F.; et al. Mac-2 Binding Protein Glycosylation Isomer as a Hepatocellular Carcinoma Marker in Patients With Chronic Hepatitis B or C Infection. Hepatol. Commun. 2019, 3, 493–503. [Google Scholar] [CrossRef]
- Yan, H.; Zhong, G.; Xu, G.; He, W.; Jing, Z.; Gao, Z.; Huang, Y.; Qi, Y.; Peng, B.; Wang, H.; et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 2012, 3. [Google Scholar] [CrossRef]
- Gerlich, W.H. Medical virology of hepatitis B: How it began and where we are now. Virol J. 2013, 10, 239. [Google Scholar] [CrossRef]
- Yang, H.C.; Kao, J.H. Persistence of hepatitis B virus covalently closed circular DNA in hepatocytes: Molecular mechanisms and clinical significance. Emerg. Microbes Infect. 2014, 3, e64. [Google Scholar] [CrossRef] [PubMed]
- Fourati, S.; Pawlotsky, J.M. Recent advances in understanding and diagnosing hepatitis B virus infection. F1000Research 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.L.; Kao, J.H. Perspectives and control of hepatitis B virus infection in Taiwan. J. Formos. Med. Assoc. 2015, 114, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.C.; Kao, J.H. Looking into the crystal ball: Biomarkers for outcomes of HBV infection. Hepatol. Int. 2016, 10, 99–101. [Google Scholar] [CrossRef]
- Iloeje, U.H.; Yang, H.I.; Su, J.; Jen, C.L.; You, S.L.; Chen, C.J. Predicting cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology 2006, 130, 678–686. [Google Scholar] [CrossRef]
- Chen, C.J.; Yang, H.I.; Su, J.; Jen, C.L.; You, S.L.; Lu, S.N.; Huang, G.T.; Iloeje, U.H. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 2006, 295, 65–73. [Google Scholar] [CrossRef]
- Kawanaka, M.; Nishino, K.; Nakamura, J.; Oka, T.; Urata, N.; Goto, D.; Suehiro, M.; Kawamoto, H.; Kudo, M.; Yamada, G. Quantitative Levels of Hepatitis B Virus DNA and Surface Antigen and the Risk of Hepatocellular Carcinoma in Patients with Hepatitis B Receiving Long-Term Nucleos(t)ide Analogue Therapy. Liver Cancer 2014, 3, 41–52. [Google Scholar] [CrossRef]
- Chuaypen, N.; Posuwan, N.; Chittmittraprap, S.; Hirankarn, N.; Treeprasertsuk, S.; Tanaka, Y.; Shinkai, N.; Poovorawan, Y.; Tangkijvanich, P. Predictive role of serum HBsAg and HBcrAg kinetics in patients with HBeAg-negative chronic hepatitis B receiving pegylated interferon-based therapy. Clin. Microbiol. Infect. 2018, 24, 306-e7. [Google Scholar] [CrossRef]
- Brouwer, W.P.; Chan, H.L.; Brunetto, M.R.; Martinot-Peignoux, M.; Arends, P.; Cornberg, M.; Cherubini, B.; Thompson, A.J.; Liaw, Y.F.; Marcellin, P.; et al. Repeated Measurements of Hepatitis B Surface Antigen Identify Carriers of Inactive HBV During Long-term Follow-up. Clin. Gastroenterol. Hepatol. 2016, 14, 1481–1489.e1485. [Google Scholar] [CrossRef]
- Yeo, Y.H.; Ho, H.J.; Yang, H.I.; Tseng, T.C.; Hosaka, T.; Trinh, H.N.; Kwak, M.S.; Park, Y.M.; Fung, J.Y.Y.; Buti, M.; et al. Factors Associated With Rates of HBsAg Seroclearance in Adults With Chronic HBV Infection: A Systematic Review and Meta-analysis. Gastroenterology 2019, 156, 635–646.e639. [Google Scholar] [CrossRef]
- Chen, J.D.; Yang, H.I.; Iloeje, U.H.; You, S.L.; Lu, S.N.; Wang, L.Y.; Su, J.; Sun, C.A.; Liaw, Y.F.; Chen, C.J. Carriers of inactive hepatitis B virus are still at risk for hepatocellular carcinoma and liver-related death. Gastroenterology 2010, 138, 1747–1754. [Google Scholar] [CrossRef]
- Liu, F.; Wang, X.W.; Chen, L.; Hu, P.; Ren, H.; Hu, H.D. Systematic review with meta-analysis: Development of hepatocellular carcinoma in chronic hepatitis B patients with hepatitis B surface antigen seroclearance. Aliment. Pharmacol. Ther. 2016, 43, 1253–1261. [Google Scholar] [CrossRef]
- Kim, G.A.; Lee, H.C.; Kim, M.J.; Ha, Y.; Park, E.J.; An, J.; Lee, D.; Shim, J.H.; Kim, K.M.; Lim, Y.S. Incidence of hepatocellular carcinoma after HBsAg seroclearance in chronic hepatitis B patients: A need for surveillance. J. Hepatol. 2015, 62, 1092–1099. [Google Scholar] [CrossRef] [PubMed]
- Shinkai, N.; Matsuura, K.; Sugauchi, F.; Watanabe, T.; Murakami, S.; Iio, E.; Ogawa, S.; Nojiri, S.; Joh, T.; Tanaka, Y. Application of a newly developed high-sensitivity HBsAg chemiluminescent enzyme immunoassay for hepatitis B patients with HBsAg seroclearance. J. Clin. Microbiol. 2013, 51, 3484–3491. [Google Scholar] [CrossRef] [PubMed]
- Shinkai, N.; Kusumoto, S.; Murakami, S.; Ogawa, S.; Ri, M.; Matsui, T.; Tamori, A.; Toyoda, H.; Ishida, T.; Iida, S.; et al. Novel monitoring of hepatitis B reactivation based on ultra-high sensitive hepatitis B surface antigen assay. Liver Int. 2017, 37, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Seto, W.K.; Wong, D.K.; Fung, J.; Huang, F.Y.; Liu, K.S.; Lai, C.L.; Yuen, M.F. Linearized hepatitis B surface antigen and hepatitis B core-related antigen in the natural history of chronic hepatitis B. Clin. Microbiol. Infect. 2014, 20, 1173–1180. [Google Scholar] [CrossRef]
- Kimura, T.; Ohno, N.; Terada, N.; Rokuhara, A.; Matsumoto, A.; Yagi, S.; Tanaka, E.; Kiyosawa, K.; Ohno, S.; Maki, N. Hepatitis B virus DNA-negative dane particles lack core protein but contain a 22-kDa precore protein without C-terminal arginine-rich domain. J. Biol. Chem. 2005, 280, 21713–21719. [Google Scholar] [CrossRef]
- Locarnini, S.; Zoulim, F. Molecular genetics of HBV infection. Antivir. Ther. 2010, 15 (Suppl 3), 3–14. [Google Scholar] [CrossRef]
- Hadziyannis, E.; Laras, A. Viral Biomarkers in Chronic HBeAg Negative HBV Infection. Genes 2018, 9, 469. [Google Scholar] [CrossRef]
- Lin, C.L.; Kao, J.H. New perspectives of biomarkers for the management of chronic hepatitis B. Clin. Mol. Hepatol. 2016, 22, 423–431. [Google Scholar] [CrossRef]
- Rokuhara, A.; Tanaka, E.; Matsumoto, A.; Kimura, T.; Yamaura, T.; Orii, K.; Sun, X.; Yagi, S.; Maki, N.; Kiyosawa, K. Clinical evaluation of a new enzyme immunoassay for hepatitis B virus core-related antigen; a marker distinct from viral DNA for monitoring lamivudine treatment. J. Viral Hepat. 2003, 10, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Maasoumy, B.; Wiegand, S.B.; Jaroszewicz, J.; Bremer, B.; Lehmann, P.; Deterding, K.; Taranta, A.; Manns, M.P.; Wedemeyer, H.; Glebe, D.; et al. Hepatitis B core-related antigen (HBcrAg) levels in the natural history of hepatitis B virus infection in a large European cohort predominantly infected with genotypes A and D. Clin. Microbiol. Infect. 2015, 21, e601–e606. [Google Scholar] [CrossRef]
- Testoni, B.; Lebosse, F.; Scholtes, C.; Berby, F.; Miaglia, C.; Subic, M.; Loglio, A.; Facchetti, F.; Lampertico, P.; Levrero, M.; et al. Serum hepatitis B core-related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. J. Hepatol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Riveiro-Barciela, M.; Bes, M.; Rodriguez-Frias, F.; Tabernero, D.; Ruiz, A.; Casillas, R.; Vidal-Gonzalez, J.; Homs, M.; Nieto, L.; Sauleda, S.; et al. Serum hepatitis B core-related antigen is more accurate than hepatitis B surface antigen to identify inactive carriers, regardless of hepatitis B virus genotype. Clin. Microbiol. Infect. 2017, 23, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Rokuhara, A.; Sakamoto, Y.; Yagi, S.; Tanaka, E.; Kiyosawa, K.; Maki, N. Sensitive enzyme immunoassay for hepatitis B virus core-related antigens and their correlation to virus load. J. Clin. Microbiol. 2002, 40, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Mak, L.Y.; Wong, D.K.; Cheung, K.S.; Seto, W.K.; Lai, C.L.; Yuen, M.F. Review article: Hepatitis B core-related antigen (HBcrAg): An emerging marker for chronic hepatitis B virus infection. Aliment. Pharmacol. Ther. 2018, 47, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Tada, T.; Kumada, T.; Toyoda, H.; Kiriyama, S.; Tanikawa, M.; Hisanaga, Y.; Kanamori, A.; Kitabatake, S.; Yama, T.; Tanaka, J. HBcrAg predicts hepatocellular carcinoma development: An analysis using time-dependent receiver operating characteristics. J. Hepatol. 2016, 65, 48–56. [Google Scholar] [CrossRef]
- Tseng, T.C.; Liu, C.J.; Hsu, C.Y.; Hong, C.M.; Su, T.H.; Yang, W.T.; Chen, C.L.; Yang, H.C.; Huang, Y.T.; Fang-Tzu Kuo, S.; et al. High Level of Hepatitis B Core-Related Antigen Associated With Increased Risk of Hepatocellular Carcinoma in Patients With Chronic HBV Infection of Intermediate Viral Load. Gastroenterology 2019, 157, 1518–1529. [Google Scholar] [CrossRef]
- Cheung, K.S.; Seto, W.K.; Wong, D.K.; Lai, C.L.; Yuen, M.F. Relationship between HBsAg, HBcrAg and hepatocellular carcinoma in patients with undetectable HBV DNA under nucleos(t)ide therapy. J. Viral Hepat. 2017, 24, 654–661. [Google Scholar] [CrossRef]
- Honda, M.; Shirasaki, T.; Terashima, T.; Kawaguchi, K.; Nakamura, M.; Oishi, N.; Wang, X.; Shimakami, T.; Okada, H.; Arai, K.; et al. Hepatitis B Virus (HBV) Core-Related Antigen During Nucleos(t)ide Analog Therapy Is Related to Intra-hepatic HBV Replication and Development of Hepatocellular Carcinoma. J. Infect. Dis. 2016, 213, 1096–1106. [Google Scholar] [CrossRef]
- Kumada, T.; Toyoda, H.; Tada, T.; Kiriyama, S.; Tanikawa, M.; Hisanaga, Y.; Kanamori, A.; Niinomi, T.; Yasuda, S.; Andou, Y.; et al. Effect of nucleos(t)ide analogue therapy on hepatocarcinogenesis in chronic hepatitis B patients: A propensity score analysis. J. Hepatol. 2013, 58, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Maekawa, S.; Komatsu, N.; Sato, M.; Tatsumi, A.; Miura, M.; Matsuda, S.; Muraoka, M.; Nakakuki, N.; Shindo, H.; et al. Hepatitis B virus (HBV)-infected patients with low hepatitis B surface antigen and high hepatitis B core-related antigen titers have a high risk of HBV-related hepatocellular carcinoma. Hepatol. Res. 2019, 49, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, T.; Suzuki, F.; Kobayashi, M.; Fujiyama, S.; Kawamura, Y.; Sezaki, H.; Akuta, N.; Suzuki, Y.; Saitoh, S.; Arase, Y.; et al. Impact of hepatitis B core-related antigen on the incidence of hepatocellular carcinoma in patients treated with nucleos(t)ide analogues. Aliment. Pharmacol. Ther. 2019, 49, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Liang, X.S. Progression and status of antiviral monitoring in patients with chronic hepatitis B: From HBsAg to HBV RNA. World J. Hepatol. 2018, 10, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Shimakawa, Y.; Ndow, G.; Njie, R.; Njai, H.F.; Takahashi, K.; Akbar, S.M.F.; Cohen, D.; Nayagam, S.; Jeng, A.; Ceesay, A.; et al. Hepatitis B core-related antigen (HBcrAg): An alternative to HBV DNA to assess treatment eligibility in Africa. Clin. Infect. Dis. 2019. [Google Scholar] [CrossRef]
- Suzuki, Y.; Maekawa, S.; Komatsu, N.; Sato, M.; Tatsumi, A.; Miura, M.; Matsuda, S.; Muraoka, M.; Nakakuki, N.; Amemiya, F.; et al. HBV preS deletion mapping using deep sequencing demonstrates a unique association with viral markers. PLoS ONE 2019, 14, e0212559. [Google Scholar] [CrossRef]
- Wong, G.L.; Wong, V.W. Risk prediction of hepatitis B virus-related hepatocellular carcinoma in the era of antiviral therapy. World J. Gastroenterol. 2013, 19, 6515–6522. [Google Scholar] [CrossRef]
- Lee, T.Y.; Lin, J.T.; Zeng, Y.S.; Chen, Y.J.; Wu, M.S.; Wu, C.Y. Association between nucleos(t)ide analog and tumor recurrence in hepatitis B virus-related hepatocellular carcinoma after radiofrequency ablation. Hepatology 2016, 63, 1517–1527. [Google Scholar] [CrossRef]
- Huang, G.; Lau, W.Y.; Zhou, W.P.; Shen, F.; Pan, Z.Y.; Yuan, S.X.; Wu, M.C. Prediction of Hepatocellular Carcinoma Recurrence in Patients with Low Hepatitis B Virus DNA Levels and High Preoperative Hepatitis B Surface Antigen Levels. JAMA Surg. 2014, 149, 519–527. [Google Scholar] [CrossRef]
- Zhou, H.Y.; Luo, Y.; Chen, W.D.; Gong, G.Z. Hepatitis B virus mutation may play a role in hepatocellular carcinoma recurrence: A systematic review and meta-regression analysis. J. Gastroenterol. Hepatol. 2015, 30, 977–983. [Google Scholar] [CrossRef]
- Urabe, A.; Imamura, M.; Tsuge, M.; Kan, H.; Fujino, H.; Fukuhara, T.; Masaki, K.; Kobayashi, T.; Ono, A.; Nakahara, T.; et al. The relationship between HBcrAg and HBV reinfection in HBV related post-liver transplantation patients. J. Gastroenterol. 2017, 52, 366–375. [Google Scholar] [CrossRef]
- Chen, S.; Jia, J.; Gao, Y.; Li, H.; Fang, M.; Feng, H.; Guan, W.; Ji, J.; Gao, Z.; Gao, C. Clinical evaluation of hepatitis B core-related antigen in chronic hepatitis B and hepatocellular carcinoma patients. Clin. Chim. Acta 2018, 486, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Kuno, A.; Ikehara, Y.; Tanaka, Y.; Ito, K.; Matsuda, A.; Sekiya, S.; Hige, S.; Sakamoto, M.; Kage, M.; Mizokami, M.; et al. A serum “sweet-doughnut” protein facilitates fibrosis evaluation and therapy assessment in patients with viral hepatitis. Sci. Rep. 2013, 3, 1065. [Google Scholar] [CrossRef] [PubMed]
- Narimatsu, H. Development of M2BPGi: A novel fibrosis serum glyco-biomarker for chronic hepatitis/cirrhosis diagnostics. Expert Rev. Proteom. 2015, 12, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, R.; Yamasaki, K.; Abiru, S.; Komori, A.; Nagaoka, S.; Saeki, A.; Hashimoto, S.; Bekki, S.; Kugiyama, Y.; Kuno, A.; et al. Serum Wisteria Floribunda Agglutinin-Positive Mac-2 Binding Protein Values Predict the Development of Hepatocellular Carcinoma among Patients with Chronic Hepatitis C after Sustained Virological Response. PLoS ONE 2015, 10, e0129053. [Google Scholar] [CrossRef] [PubMed]
- Shirabe, K.; Bekki, Y.; Gantumur, D.; Araki, K.; Ishii, N.; Kuno, A.; Narimatsu, H.; Mizokami, M. Mac-2 binding protein glycan isomer (M2BPGi) is a new serum biomarker for assessing liver fibrosis: More than a biomarker of liver fibrosis. J. Gastroenterol. 2018, 53, 819–826. [Google Scholar] [CrossRef]
- Toshima, T.; Shirabe, K.; Ikegami, T.; Yoshizumi, T.; Kuno, A.; Togayachi, A.; Gotoh, M.; Narimatsu, H.; Korenaga, M.; Mizokami, M.; et al. A novel serum marker, glycosylated Wisteria floribunda agglutinin-positive Mac-2 binding protein (WFA(+)-M2BP), for assessing liver fibrosis. J. Gastroenterol. 2015, 50, 76–84. [Google Scholar] [CrossRef]
- Ikeda, M.; Fujiyama, S.; Tanaka, M.; Sata, M.; Ide, T.; Yatsuhashi, H.; Watanabe, H. Risk factors for development of hepatocellular carcinoma in patients with chronic hepatitis C after sustained response to interferon. J. Gastroenterol. 2005, 40, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Fattovich, G.; Stroffolini, T.; Zagni, I.; Donato, F. Hepatocellular carcinoma in cirrhosis: Incidence and risk factors. Gastroenterology 2004, 127, S35–S50. [Google Scholar] [CrossRef] [PubMed]
- Morgan, T.R.; Ghany, M.G.; Kim, H.Y.; Snow, K.K.; Shiffman, M.L.; De Santo, J.L.; Lee, W.M.; Di Bisceglie, A.M.; Bonkovsky, H.L.; Dienstag, J.L.; et al. Outcome of sustained virological responders with histologically advanced chronic hepatitis C. Hepatology 2010, 52, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Shiratori, Y.; Moriyama, M.; Arakawa, Y.; Ide, T.; Sata, M.; Inoue, O.; Yano, M.; Tanaka, M.; Fujiyama, S.; et al. Interferon therapy reduces the risk for hepatocellular carcinoma: National surveillance program of cirrhotic and noncirrhotic patients with chronic hepatitis C in Japan. IHIT Study Group. Inhibition of Hepatocarcinogenesis by Interferon Therapy. Ann. Intern. Med. 1999, 131, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Genda, T.; Ichida, T.; Amano, N.; Sato, S.; Murata, A.; Tsuzura, H.; Narita, Y.; Kanemitsu, Y.; Hirano, K.; et al. Prediction of Hepatocellular Carcinoma Development after Hepatitis C Virus Eradication Using Serum Wisteria floribunda Agglutinin-Positive Mac-2-Binding Protein. Int. J. Mol. Sci. 2016, 17, 2143. [Google Scholar] [CrossRef] [PubMed]
- Freeman, A.J.; Law, M.G.; Kaldor, J.M.; Dore, G.J. Predicting progression to cirrhosis in chronic hepatitis C virus infection. J. Viral Hepat. 2003, 10, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.A.; Sheth, S.G.; Chopra, S. Liver biopsy. N. Engl. J. Med. 2001, 344, 495–500. [Google Scholar] [CrossRef]
- Bedossa, P.; Dargere, D.; Paradis, V. Sampling variability of liver fibrosis in chronic hepatitis C. Hepatology 2003, 38, 1449–1457. [Google Scholar] [CrossRef]
- Bruix, J.; Sherman, M. Management of hepatocellular carcinoma. Hepatology 2005, 42, 1208–1236. [Google Scholar] [CrossRef]
- Hanai, T.; Shiraki, M.; Ohnishi, S.; Miyazaki, T.; Ideta, T.; Kochi, T.; Imai, K.; Suetsugu, A.; Takai, K.; Shimizu, M.; et al. Impact of serum glycosylated Wisteria floribunda agglutinin positive Mac-2 binding protein levels on liver functional reserves and mortality in patients with liver cirrhosis. Hepatol. Res. 2015, 45, 1083–1090. [Google Scholar] [CrossRef]
- Zou, X.; Zhu, M.Y.; Yu, D.M.; Li, W.; Zhang, D.H.; Lu, F.J.; Gong, Q.M.; Liu, F.; Jiang, J.H.; Zheng, M.H.; et al. Serum WFA(+) -M2BP levels for evaluation of early stages of liver fibrosis in patients with chronic hepatitis B virus infection. Liver Int. 2017, 37, 35–44. [Google Scholar] [CrossRef]
- Yamasaki, K.; Tateyama, M.; Abiru, S.; Komori, A.; Nagaoka, S.; Saeki, A.; Hashimoto, S.; Sasaki, R.; Bekki, S.; Kugiyama, Y.; et al. Elevated serum levels of Wisteria floribunda agglutinin-positive human Mac-2 binding protein predict the development of hepatocellular carcinoma in hepatitis C patients. Hepatology 2014, 60, 1563–1570. [Google Scholar] [CrossRef]
- Abe, M.; Miyake, T.; Kuno, A.; Imai, Y.; Sawai, Y.; Hino, K.; Hara, Y.; Hige, S.; Sakamoto, M.; Yamada, G.; et al. Association between Wisteria floribunda agglutinin-positive Mac-2 binding protein and the fibrosis stage of non-alcoholic fatty liver disease. J. Gastroenterol. 2015, 50, 776–784. [Google Scholar] [CrossRef]
- Yamada, N.; Sanada, Y.; Tashiro, M.; Hirata, Y.; Okada, N.; Ihara, Y.; Urahashi, T.; Mizuta, K. Serum Mac-2 binding protein glycosylation isomer predicts grade F4 liver fibrosis in patients with biliary atresia. J. Gastroenterol. 2017, 52, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.U.; Heo, J.Y.; Kim, B.K.; Park, J.Y.; Kim, D.Y.; Han, K.H.; Ahn, S.H.; Kim, H.S. Wisteria floribunda agglutinin-positive human Mac-2 binding protein predicts the risk of HBV-related liver cancer development. Liver Int. 2017, 37, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hu, H.-H.; Lee, M.-H.; Korenaga, M.; Jen, C.-L.; Batrla-Utermann, R.; Lu, S.-N.; Wang, L.-Y.; Mizokami, M.; Chen, C.-J.; et al. Serum Levels of M2BPGi as Short-Term Predictors of Hepatocellular Carcinoma in Untreated Chronic Hepatitis B Patients. Sci. Rep. 2017, 7, 14352. [Google Scholar] [CrossRef] [PubMed]
- Mak, L.Y.; To, W.P.; Fung, J.; Wong, D.K.H.; Liu, F.; Seto, W.K.; Lai, C.L.; Yuen, M.F. Serum Mac-2 binding protein glycosylation isomer level predicts hepatocellular carcinoma development in E-negative chronic hepatitis B patients. World J. Gastroenterol. 2019, 25, 1398–1408. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.C.; Jun, T.; Huang, Y.T.; Yeh, M.L.; Lee, C.L.; Ogawa, S.; Cho, S.H.; Lin, J.T.; Yu, M.L.; Nguyen, M.H.; et al. Serum M2BPGi level and risk of hepatocellular carcinoma after oral anti-viral therapy in patients with chronic hepatitis B. Aliment. Pharmacol. Ther. 2018, 48, 1128–1137. [Google Scholar] [CrossRef]
- Shinkai, N.; Nojima, M.; Iio, E.; Matsunami, K.; Toyoda, H.; Murakami, S.; Inoue, T.; Ogawa, S.; Kumada, T.; Tanaka, Y. High levels of serum Mac-2-binding protein glycosylation isomer (M2BPGi) predict the development of hepatocellular carcinoma in hepatitis B patients treated with nucleot(s)ide analogues. J. Gastroenterol. 2018, 53, 883–889. [Google Scholar] [CrossRef]
- Cheung, K.S.; Seto, W.K.; Wong, D.K.; Mak, L.Y.; Lai, C.L.; Yuen, M.F. Wisteria floribunda agglutinin-positive human Mac-2 binding protein predicts liver cancer development in chronic hepatitis B patients under antiviral treatment. Oncotarget 2017, 8, 47507–47517. [Google Scholar] [CrossRef]
- Li, Q.; Song, J.; Huang, Y.; Li, X.; Zhuo, Q.; Li, W.; Chen, C.; Lu, C.; Qi, X.; Chen, L. The Gamma-Glutamyl-Transpeptidase to Platelet Ratio Does not Show Advantages than APRI and Fib-4 in Diagnosing Significant Fibrosis and Cirrhosis in Patients With Chronic Hepatitis B: A Retrospective Cohort Study in China. Medicine (Baltimore) 2016, 95, e3372. [Google Scholar] [CrossRef]
- Nishikawa, H.; Enomoto, H.; Iwata, Y.; Kishino, K.; Shimono, Y.; Hasegawa, K.; Nakano, C.; Takata, R.; Nishimura, T.; Yoh, K.; et al. Serum Wisteria floribunda agglutinin-positive Mac-2-binding protein for patients with chronic hepatitis B and C: A comparative study. J. Viral Hepat. 2016, 23, 977–984. [Google Scholar] [CrossRef]
- Ichikawa, Y.; Joshita, S.; Umemura, T.; Shobugawa, Y.; Usami, Y.; Shibata, S.; Yamazaki, T.; Fujimori, N.; Komatsu, M.; Matsumoto, A.; et al. Serum Wisteria floribunda agglutinin-positive human Mac-2 binding protein may predict liver fibrosis and progression to hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Hepatol. Res. 2017, 47, 226–233. [Google Scholar] [CrossRef]
- Liu, N.; Zhao, F.; Zhan, P.; Liu, X. Review of small synthetic molecules targeting HBV capsid assembly. Med. Chem. 2015, 11, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Wooddell, C.I.; Yuen, M.F.; Chan, H.L.; Gish, R.G.; Locarnini, S.A.; Chavez, D.; Ferrari, C.; Given, B.D.; Hamilton, J.; Kanner, S.B.; et al. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
| Category | Findings | HBcrAg level (log U/mL) and point | References |
|---|---|---|---|
| cccDNA activity | Lower amounts of intrahepatic cccDNA and lower cccDNA activity | <3 log U/mL | [44] |
| Identification of inactive carriers with a high accuracy (any HBV genotype) | HBcrAg ≤ 3 log U/mL plus HBV DNA ≤ 2000 U/mL | [45] | |
| HCC occurrence/recurrence | Incidence of HCC for treatment-naïve patients | >2.9 log U/mL during the follow-up period | [48] |
| At high risk for HCC with intermediate viral load (HBV DNA 2000–19,999 U/mL) | ≥4.0 log U/mL | [49] | |
| Incidence of HCC for treatment-experienced patients | >4.67 log U/mL at pre-treatment, >3.89 log U/mL at post-treatment | [50] | |
| HCC development during NA treatment | Detectable HBcrAg during NA treatment | [51] | |
| Long-term effect of NA treatment on HCC development | Higher serum levels of HBcrAg and BCP mutations were associated with HCC development, independent of NA therapy | [52] | |
| Evaluation of HCC occurrence | HBcrAg > 3.0 log U/mL and HBsAg > 3.0 log U/mL (cut-off values) | [53] | |
| HCC recurrence within 2 years | >4.8 log U/mL at the time of HCC diagnosis | [54] |
| Category | Findings | M2BPGi level (C.O.I.) and point | References |
|---|---|---|---|
| Liver fibrosis | Significant fibrosis (≥F2) in CHB | ≥1.06 C.O.I. (AUC 0.753) | [79] |
| Survival rate | Lower cumulative survival rate in patients with cirrhosis | ≥5.0 C.O.I. | [78] |
| HCC development | A >15-fold-increased risk of HCC development in CHC patients after SVR | ≥2.80 C.O.I. | [73] |
| Higher risk of HCC development in CHB patients | ≥1.8 C.O.I. for patients without cirrhosis (p < 0.001) ≥1.8 C.O.I. for patients with cirrhosis (p = 0.073) | [83] | |
| Low risk of HCC in HBeAg-negative patients | ≤0.68 C.O.I. at baseline | [85] | |
| Risk for HCC development in CHB patients with cirrhosis treated with NAs | M2BPGi-based score* ≥ 652.5 at baseline | [86] | |
| Risk for HCC development in CHB patients treated with NAs | ≥1.215 C.O.I. at 48 weeks | [87] | |
| HCC development in CHB patients | ≥0.69 C.O.I. | [88] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baudi, I.; Inoue, T.; Tanaka, Y. Novel Biomarkers of Hepatitis B and Hepatocellular Carcinoma: Clinical Significance of HBcrAg and M2BPGi. Int. J. Mol. Sci. 2020, 21, 949. https://doi.org/10.3390/ijms21030949
Baudi I, Inoue T, Tanaka Y. Novel Biomarkers of Hepatitis B and Hepatocellular Carcinoma: Clinical Significance of HBcrAg and M2BPGi. International Journal of Molecular Sciences. 2020; 21(3):949. https://doi.org/10.3390/ijms21030949
Chicago/Turabian StyleBaudi, Ian, Takako Inoue, and Yasuhito Tanaka. 2020. "Novel Biomarkers of Hepatitis B and Hepatocellular Carcinoma: Clinical Significance of HBcrAg and M2BPGi" International Journal of Molecular Sciences 21, no. 3: 949. https://doi.org/10.3390/ijms21030949
APA StyleBaudi, I., Inoue, T., & Tanaka, Y. (2020). Novel Biomarkers of Hepatitis B and Hepatocellular Carcinoma: Clinical Significance of HBcrAg and M2BPGi. International Journal of Molecular Sciences, 21(3), 949. https://doi.org/10.3390/ijms21030949




