ADAM12 is A Potential Therapeutic Target Regulated by Hypomethylation in Triple-Negative Breast Cancer
Abstract
1. Introduction
2. Results
2.1. Genome-Wide DNA Methylation Pattern in TNBC Patients
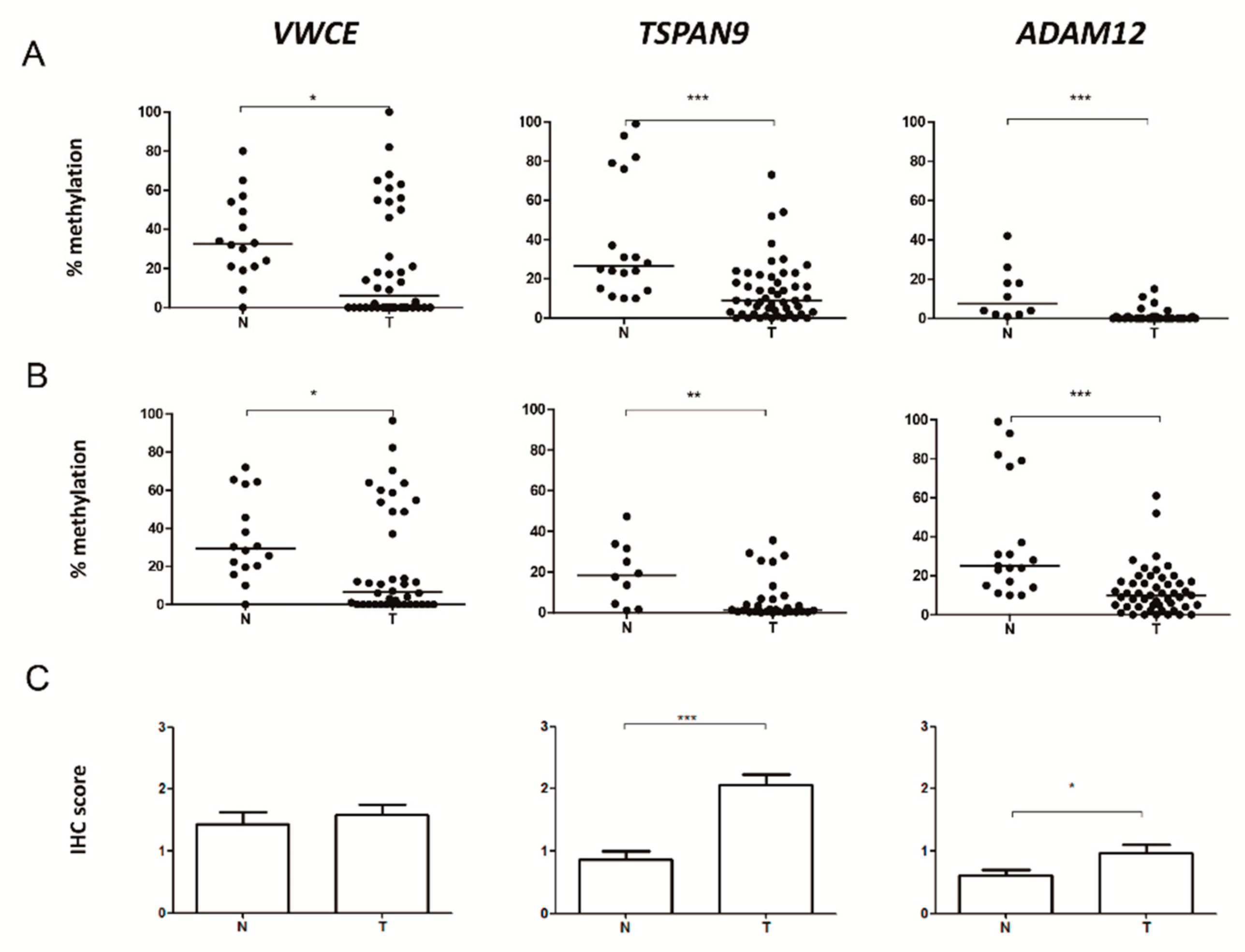
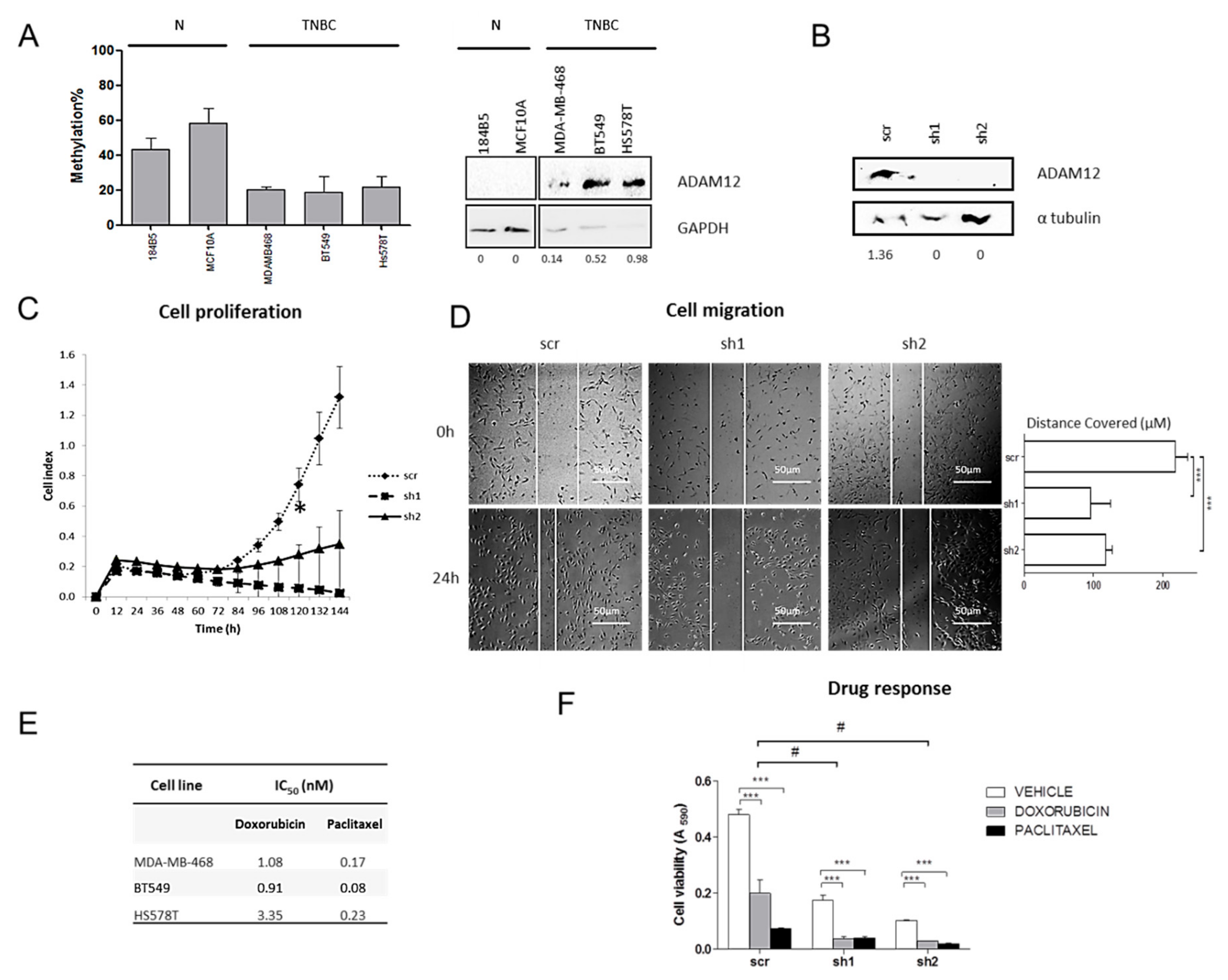
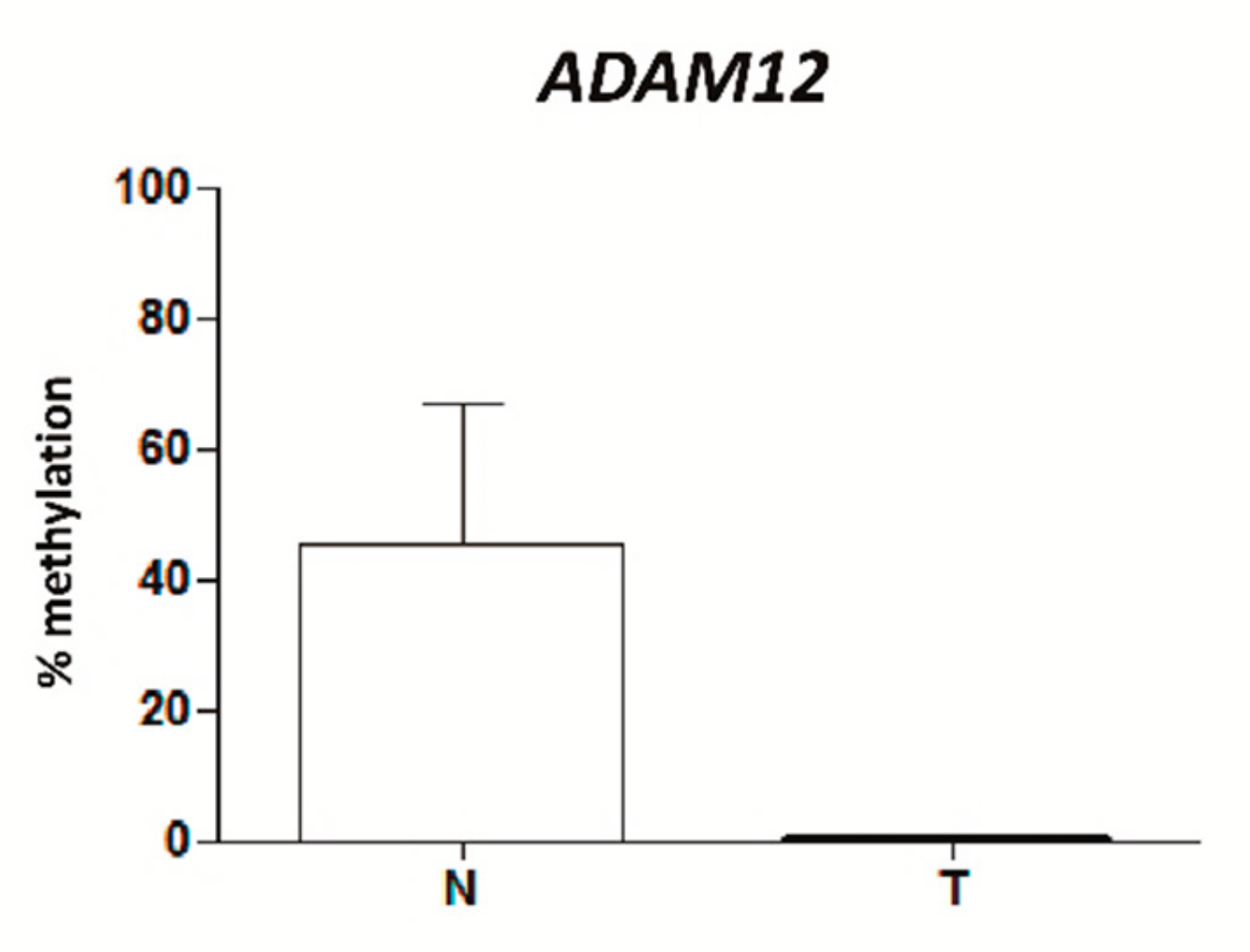
2.2. VWCE, TSPAN9 and ADAM12 Methylation Levels Are Lower in TNBCs Than in Non-Neoplastic Breast Tissues
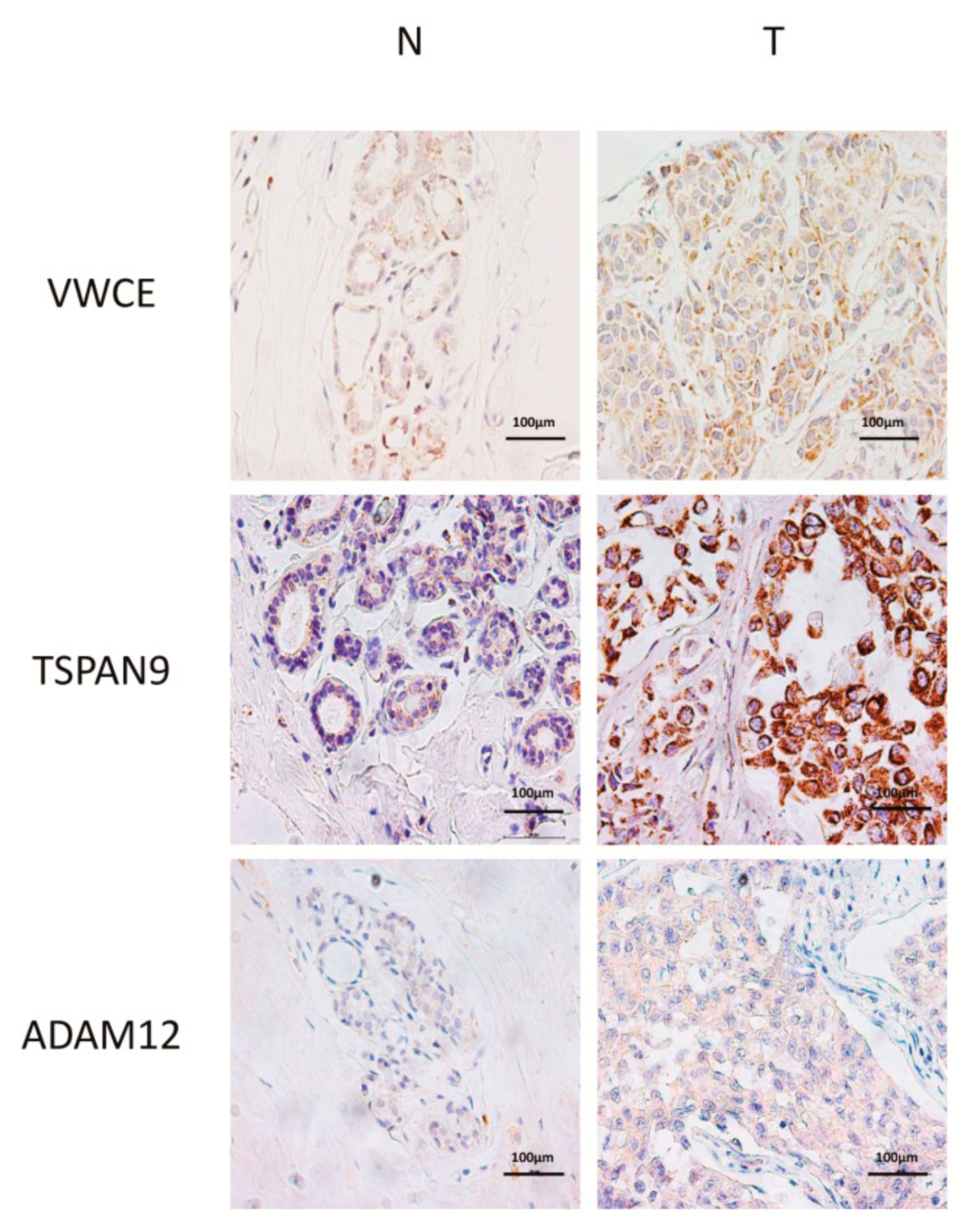
2.3. Level of Expression of TSPAN9 and ADAM12 is Higher in TNBCs Than in Non-Neoplastic Breast Tissue
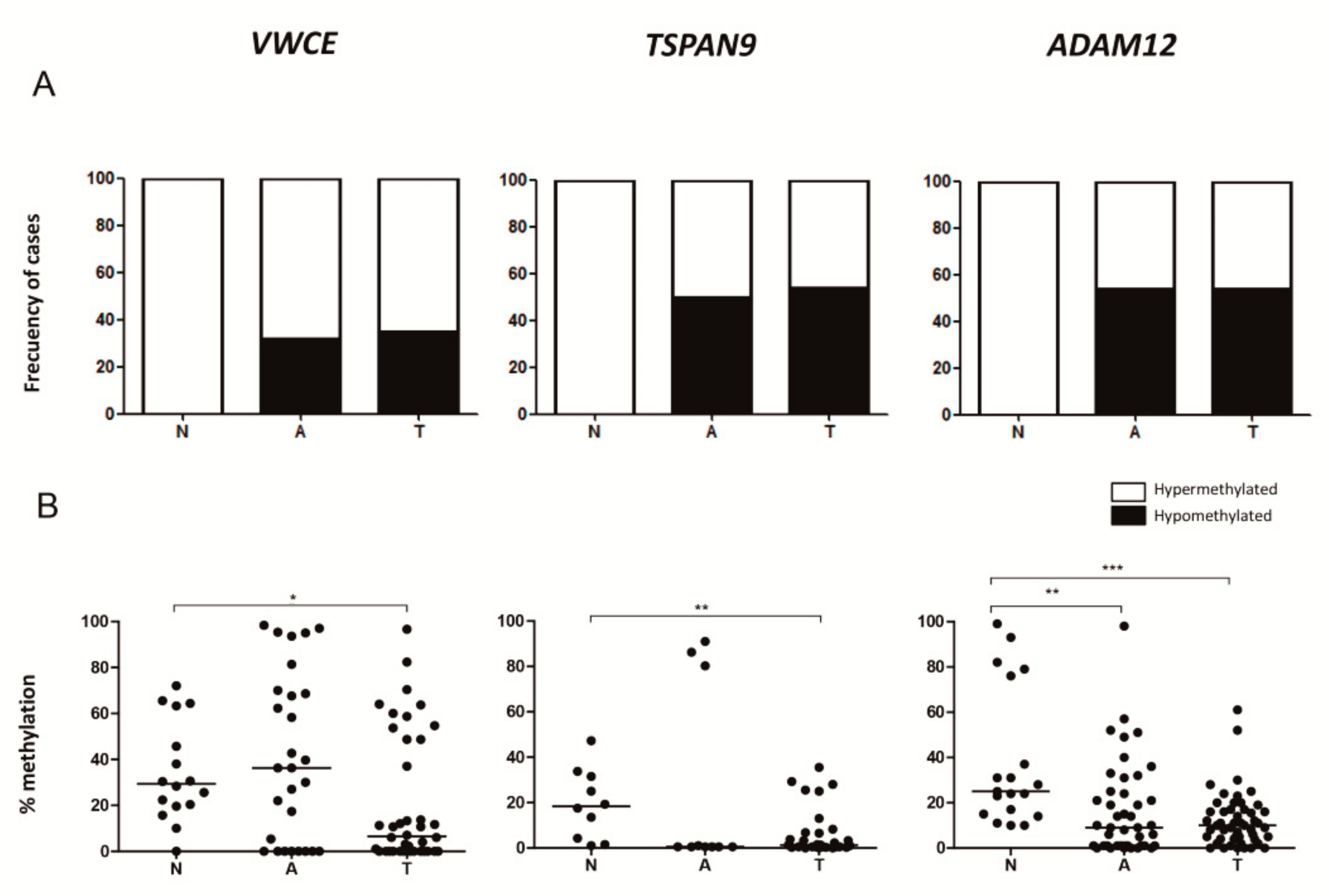
2.4. Adjacent Non-Neoplastic Tissue Has a DNA Methylation Pattern Similar to that of TNBCs but Different from that of Non-Neoplastic Mammary Tissue
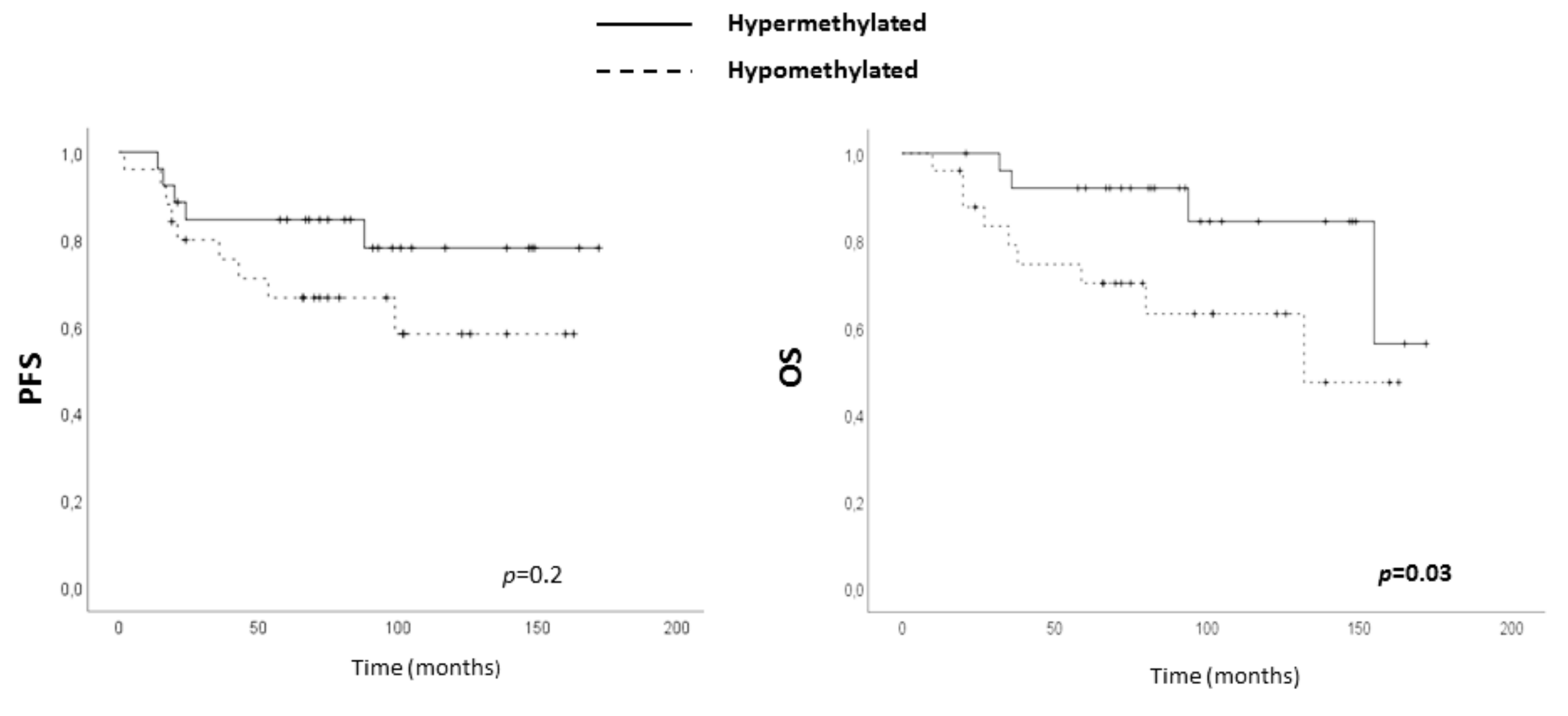
2.5. Clinical Value of ADAM12 Hypomethylation in TNBC
2.6. ADAM12 Silencing Inhibits TNBC Cell Proliferation and Migration
2.7. ADAM12 Silencing Improves Doxorubicin Sensitivity of TNBC Cells
2.8. ADAM12 Is Hypomethylated in Plasma from TNBC Patients
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. Cell Lines
4.3. DNA and Cell-Free DNA (cfDNA) Extraction and Bisulphite Conversion
4.4. DNA Methylation Array and Bioinformatics Analysis
4.5. Pyrosequencing
4.6. Immunohistochemistry
4.7. ADAM12 Silencing in TNBC Cell Lines
4.8. Western Blot
4.9. Cell Proliferation
4.10. Cell Migration
4.11. Drug Response
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Senkus, E.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rutgers, E.; Zackrisson, S.; Cardoso, F. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26 Suppl. 5, v8–v30. [Google Scholar] [CrossRef]
- Untch, M.; Gerber, B.; Harbeck, N.; Jackisch, C.; Marschner, N.; Mobus, V.; von Minckwitz, G.; Loibl, S.; Beckmann, M.W.; Blohmer, J.U.; et al. 13th st.Gallen international breast cancer conference 2013: Primary therapy of early breast cancer evidence, controversies, consensus–opinion of a german team of experts (zurich 2013). Breast Care 2013, 8, 221–229. [Google Scholar] [PubMed]
- Blows, F.M.; Driver, K.E.; Schmidt, M.K.; Broeks, A.; van Leeuwen, F.E.; Wesseling, J.; Cheang, M.C.; Gelmon, K.; Nielsen, T.O.; Blomqvist, C.; et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: A collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010, 7, e1000279. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef]
- Terry, M.B.; McDonald, J.A.; Wu, H.C.; Eng, S.; Santella, R.M. Epigenetic Biomarkers of Breast Cancer Risk: Across the Breast Cancer Prevention Continuum. Adv. Exp. Med. Biol. 2016, 882, 33–68. [Google Scholar]
- Beltran-Garcia, J.; Osca-Verdegal, R.; Mena-Molla, S.; Garcia-Gimenez, J.L. Epigenetic IVD Tests for Personalized Precision Medicine in Cancer. Front. Genet. 2019, 10, 621. [Google Scholar] [CrossRef]
- Kaminska, K.; Nalejska, E.; Kubiak, M.; Wojtysiak, J.; Zolna, L.; Kowalewski, J.; Lewandowska, M.A. Prognostic and Predictive Epigenetic Biomarkers in Oncology. Mol. Diagn. Ther. 2019, 23, 83–95. [Google Scholar] [CrossRef]
- Haynes, H.R.; Camelo-Piragua, S.; Kurian, K.M. Prognostic and predictive biomarkers in adult and pediatricgliomas: Toward personalized treatment. Front. Oncol. 2014, 4, 47. [Google Scholar] [CrossRef] [PubMed]
- Liedtke, C.; Mazouni, C.; Hess, K.R.; Andre, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M.; et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008, 26, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Turashvili, G.; Lightbody, E.D.; Tyryshkin, K.; SenGupta, S.K.; Elliott, B.E.; Madarnas, Y.; Ghaffari, A.; Day, A.; Nicol, C.J.B. Novel prognostic and predictive microRNA targets for triple-negative breast cancer. Faseb J. 2018, fj201800120R. [Google Scholar] [CrossRef]
- Gui, Y.; Xu, S.; Yang, X.; Gu, L.; Zhang, Z.; Luo, X.; Chen, L. A meta-analysis of biomarkers for the prognosis of triple-negative breast cancer patients. Biomark Med. 2016, 10, 771–790. [Google Scholar] [CrossRef] [PubMed]
- DiNome, M.L.; Orozco, J.I.J.; Matsuba, C.; Manughian-Peter, A.O.; Ensenyat-Mendez, M.; Chang, S.C.; Jalas, J.R.; Salomon, M.P.; Marzese, D.M. Clinicopathological Features of Triple-Negative Breast Cancer Epigenetic Subtypes. Ann. Surg. Oncol. 2019, 26, 3344–3353. [Google Scholar] [CrossRef] [PubMed]
- Pineda, B.; Diaz-Lagares, A.; Perez-Fidalgo, J.A.; Burgues, O.; Gonzalez-Barrallo, I.; Crujeiras, A.B.; Sandoval, J.; Esteller, M.; Lluch, A.; Eroles, P. A two-gene epigenetic signature for the prediction of response to neoadjuvant chemotherapy in triple-negative breast cancer patients. Clin. Epigenetics 2019, 11, 33. [Google Scholar] [CrossRef]
- Stirzaker, C.; Zotenko, E.; Song, J.Z.; Qu, W.; Nair, S.S.; Locke, W.J.; Stone, A.; Armstong, N.J.; Robinson, M.D.; Dobrovic, A.; et al. Methylome sequencing in triple-negative breast cancer reveals distinct methylation clusters with prognostic value. Nat. Commun. 2015, 6, 5899. [Google Scholar] [CrossRef]
- Mathe, A.; Wong-Brown, M.; Locke, W.J.; Stirzaker, C.; Braye, S.G.; Forbes, J.F.; Clark, S.J.; Avery-Kiejda, K.A.; Scott, R.J. DNA methylation profile of triple negative breast cancer-specific genes comparing lymph node positive patients to lymph node negative patients. Sci. Rep. 2016, 6, 33435. [Google Scholar] [CrossRef]
- Chai, H.; Brown, R.E. Field effect in cancer-an update. Ann. Clin. Lab. Sci. 2009, 39, 331–337. [Google Scholar]
- Baba, Y.; Ishimoto, T.; Kurashige, J.; Iwatsuki, M.; Sakamoto, Y.; Yoshida, N.; Watanabe, M.; Baba, H. Epigenetic field cancerization in gastrointestinal cancers. Cancer Lett. 2016, 375, 360–366. [Google Scholar] [CrossRef]
- Patel, A.; Tripathi, G.; Gopalakrishnan, K.; Williams, N.; Arasaradnam, R.P. Field cancerisation in colorectal cancer: A new frontier or pastures past? World J. Gastroenterol. 2015, 21, 3763–3772. [Google Scholar] [CrossRef] [PubMed]
- Dotto, G.P. Multifocal epithelial tumors and field cancerization: Stroma as a primary determinant. J. Clin. Invest. 2014, 124, 1446–1453. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.L.; Magalhaes, L.; Moreira, F.C.; Reis-das-Merces, L.; Vidal, A.F.; Ribeiro-Dos-Santos, A.M.; Demachki, S.; Anaissi, A.K.M.; Burbano, R.M.R.; Albuquerque, P.; et al. Epigenetic Field Cancerization in Gastric Cancer: microRNAs as Promising Biomarkers. J. Cancer 2019, 10, 1560–1569. [Google Scholar] [CrossRef] [PubMed]
- Martin-Sanchez, E.; Mendaza, S.; Ulazia-Garmendia, A.; Monreal-Santesteban, I.; Cordoba, A.; Vicente-Garcia, F.; Blanco-Luquin, I.; De La Cruz, S.; Aramendia, A.; Guerrero-Setas, D. CDH22 hypermethylation is an independent prognostic biomarker in breast cancer. Clin. Epigenetics 2017, 9, 7. [Google Scholar] [CrossRef]
- Spitzwieser, M.; Holzweber, E.; Pfeiler, G.; Hacker, S.; Cichna-Markl, M. Applicability of HIN-1, MGMT and RASSF1A promoter methylation as biomarkers for detecting field cancerization in breast cancer. Breast Cancer Res. 2015, 17, 125. [Google Scholar] [CrossRef]
- Spitzwieser, M.; Entfellner, E.; Werner, B.; Pulverer, W.; Pfeiler, G.; Hacker, S.; Cichna-Markl, M. Hypermethylation of CDKN2A exon 2 in tumor, tumor-adjacent and tumor-distant tissues from breast cancer patients. BMC Cancer 2017, 17, 260. [Google Scholar] [CrossRef]
- Pan, B.; Ye, Y.; Liu, H.; Zhen, J.; Zhou, H.; Li, Y.; Qu, L.; Wu, Y.; Zeng, C.; Zhong, W. URG11 Regulates Prostate Cancer Cell Proliferation, Migration, and Invasion. Biomed. Res. Int. 2018, 2018, 4060728. [Google Scholar] [CrossRef]
- Feng, T.; Sun, L.; Qi, W.; Pan, F.; Lv, J.; Guo, J.; Zhao, S.; Ding, A.; Qiu, W. Prognostic significance of Tspan9 in gastric cancer. Mol. Clin. Oncol. 2016, 5, 231–236. [Google Scholar] [CrossRef]
- Qi, Y.; Lv, J.; Liu, S.; Sun, L.; Wang, Y.; Li, H.; Qi, W.; Qiu, W. TSPAN9 and EMILIN1 synergistically inhibit the migration and invasion of gastric cancer cells by increasing TSPAN9 expression. BMC Cancer 2019, 19, 630. [Google Scholar] [CrossRef]
- Li, P.Y.; Lv, J.; Qi, W.W.; Zhao, S.F.; Sun, L.B.; Liu, N.; Sheng, J.; Qiu, W.S. Tspan9 inhibits the proliferation, migration and invasion of human gastric cancer SGC7901 cells via the ERK1/2 pathway. Oncol. Rep. 2016, 36, 448–454. [Google Scholar] [CrossRef]
- Veenstra, V.L.; Damhofer, H.; Waasdorp, C.; van Rijssen, L.B.; van de Vijver, M.J.; Dijk, F.; Wilmink, H.W.; Besselink, M.G.; Busch, O.R.; Chang, D.K.; et al. ADAM12 is a circulating marker for stromal activation in pancreatic cancer and predicts response to chemotherapy. Oncogenesis 2018, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.L.; Zhou, Z.; Sun, L.; Yu, L.; Liu, J.; Yang, Z.; Ran, Y.; Yao, Y.; Hu, H. An ADAM12 and FAK positive feedback loop amplifies the interaction signal of tumor cells with extracellular matrix to promote esophageal cancer metastasis. Cancer Lett. 2018, 422, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Dagher, A.; Butterfield, C.; Moses, M.A. ADAM12 Is a Novel Regulator of Tumor Angiogenesis via STAT3 Signaling. Mol. Cancer Res. 2017, 15, 1608–1622. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Rodig, S.; Bielenberg, D.; Zurakowski, D.; Moses, M.A. ADAM12 transmembrane and secreted isoforms promote breast tumor growth: A distinct role for ADAM12-S protein in tumor metastasis. J. Biol. Chem. 2011, 286, 20758–20768. [Google Scholar] [CrossRef] [PubMed]
- Frohlich, C.; Nehammer, C.; Albrechtsen, R.; Kronqvist, P.; Kveiborg, M.; Sehara-Fujisawa, A.; Mercurio, A.M.; Wewer, U.M. ADAM12 produced by tumor cells rather than stromal cells accelerates breast tumor progression. Mol. Cancer Res 2011, 9, 1449–1461. [Google Scholar] [CrossRef] [PubMed]
- Duhachek-Muggy, S.; Qi, Y.; Wise, R.; Alyahya, L.; Li, H.; Hodge, J.; Zolkiewska, A. Metalloprotease-disintegrin ADAM12 actively promotes the stem cell-like phenotype in claudin-low breast cancer. Mol. Cancer 2017, 16, 32. [Google Scholar] [CrossRef]
- Li, H.; Duhachek-Muggy, S.; Qi, Y.; Hong, Y.; Behbod, F.; Zolkiewska, A. An essential role of metalloprotease-disintegrin ADAM12 in triple-negative breast cancer. Breast Cancer Res. Treat. 2012, 135, 759–769. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Gu, J.; Zhou, D.; He, Z.; Ferrone, S. ADAM12-L confers acquired 5-fluorouracil resistance in breast cancer cells. Sci. Rep. 2017, 7, 9687. [Google Scholar] [CrossRef]
- Farmer, P.; Bonnefoi, H.; Anderle, P.; Cameron, D.; Wirapati, P.; Becette, V.; Andre, S.; Piccart, M.; Campone, M.; Brain, E.; et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat. Med. 2009, 15, 68–74. [Google Scholar] [CrossRef]
- Li, H.; Duhachek-Muggy, S.; Dubnicka, S.; Zolkiewska, A. Metalloproteinase-disintegrin ADAM12 is associated with a breast tumor-initiating cell phenotype. Breast Cancer Res. Treat. 2013, 139, 691–703. [Google Scholar] [CrossRef][Green Version]
- Ruff, M.; Leyme, A.; Le Cann, F.; Bonnier, D.; Le Seyec, J.; Chesnel, F.; Fattet, L.; Rimokh, R.; Baffet, G.; Theret, N. The Disintegrin and Metalloprotease ADAM12 Is Associated with TGF-beta-Induced Epithelial to Mesenchymal Transition. PLoS ONE 2015, 10, e0139179. [Google Scholar] [CrossRef] [PubMed]
- Hancock, B.A.; Chen, Y.-H.; Solzak, J.P.; Ahmad, M.N.; Wedge, D.C.; Brinza, D.; Scafe, C.; Veitch, J.; Gottimukkala, R.; Short, W.; et al. Profiling molecular regulators of recurrence in chemorefractory triple-negative breast cancers. Breast Cancer Res. 2019, 21, 87. [Google Scholar] [CrossRef] [PubMed]
- Sousa, B.; Cardoso, F. Neoadjuvant treatment for HER-2-positive and triple-negative breast cancers. Ann. Oncol. England 2012, 23 Suppl. 1, x237–x242. [Google Scholar] [CrossRef]
- Davalos, V.; Martinez-Cardus, A.; Esteller, M. The Epigenomic Revolution in Breast Cancer: From Single-Gene to Genome-Wide Next-Generation Approaches. Am. J. Pathol. 2017, 187, 2163–2174. [Google Scholar] [CrossRef] [PubMed]
- Gai, W.; Sun, K. Epigenetic Biomarkers in Cell-Free DNA and Applications in Liquid Biopsy. Genes 2019, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Eslami, S.Z.; Cortes-Hernandez, L.E.; Cayrefourcq, L.; Alix-Panabieres, C. The Different Facets of Liquid Biopsy: A Kaleidoscopic View. Cold Spring Harb. Perspect. Med. 2019. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, F.; Lunardi, F.; Pezzuto, F.; Fortarezza, F.; Vuljan, S.E.; Marquette, C.; Hofman, P. Are There New Biomarkers in Tissue and Liquid Biopsies for the Early Detection of Non-Small Cell Lung Cancer? J. Clin. Med. 2019, 8, 414. [Google Scholar] [CrossRef]
- Chimonidou, M.; Tzitzira, A.; Strati, A.; Sotiropoulou, G.; Sfikas, C.; Malamos, N.; Georgoulias, V.; Lianidou, E. CST6 promoter methylation in circulating cell-free DNA of breast cancer patients. Clin. Biochem. 2013, 46, 235–240. [Google Scholar] [CrossRef]
- Shan, M.; Yin, H.; Li, J.; Li, X.; Wang, D.; Su, Y.; Niu, M.; Zhong, Z.; Wang, J.; Zhang, X.; et al. Detection of aberrant methylation of a six-gene panel in serum DNA for diagnosis of breast cancer. Oncotarget 2016, 7, 18485–18494. [Google Scholar] [CrossRef]
- Kloten, V.; Becker, B.; Winner, K.; Schrauder, M.G.; Fasching, P.A.; Anzeneder, T.; Veeck, J.; Hartmann, A.; Knuchel, R.; Dahl, E. Promoter hypermethylation of the tumor-suppressor genes ITIH5, DKK3, and RASSF1A as novel biomarkers for blood-based breast cancer screening. Breast Cancer Res. 2013, 15, R4. [Google Scholar] [CrossRef]
- Gobel, G.; Auer, D.; Gaugg, I.; Schneitter, A.; Lesche, R.; Muller-Holzner, E.; Marth, C.; Daxenbichler, G. Prognostic significance of methylated RASSF1A and PITX2 genes in blood- and bone marrow plasma of breast cancer patients. Breast Cancer Res. Treat. 2011, 130, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Jezkova, E.; Kajo, K.; Zubor, P.; Grendar, M.; Malicherova, B.; Mendelova, A.; Dokus, K.; Lasabova, Z.; Plank, L.; Danko, J. Methylation in promoter regions of PITX2 and RASSF1A genes in association with clinicopathological features in breast cancer patients. Tumour Biol. 2016, 37, 15707–15718. [Google Scholar] [CrossRef] [PubMed]
- Widschwendter, M.; Evans, I.; Jones, A.; Ghazali, S.; Reisel, D.; Ryan, A.; Gentry-Maharaj, A.; Zikan, M.; Cibula, D.; Eichner, J.; et al. Methylation patterns in serum DNA for early identification of disseminated breast cancer. Genome Med. 2017, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Cheang, M.C.U.; Chia, S.K.; Voduc, D.; Gao, D.X.; Leung, S.; Snider, J.; Watson, M.; Davies, S.; Bernard, P.S.; Parker, J.S.; et al. Ki67 Index, HER2 Status, and Prognosis of Patients With Luminal B Breast Cancer. J. Natl. Cancer Inst. 2009, 101, 736–750. [Google Scholar] [CrossRef]
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology 2002, 41, 154–161. [Google Scholar] [CrossRef]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef]
- Martin-Sanchez, E.; Mendaza, S.; Ulazia-Garmendia, A.; Monreal-Santesteban, I.; Blanco-Luquin, I.; Cordoba, A.; Vicente-Garcia, F.; Perez-Janices, N.; Escors, D.; Megias, D.; et al. CHL1 hypermethylation as a potential biomarker of poor prognosis in breast cancer. Oncotarget 2017, 8, 15789–15801. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendaza, S.; Ulazia-Garmendia, A.; Monreal-Santesteban, I.; Córdoba, A.; Azúa, Y.R.d.; Aguiar, B.; Beloqui, R.; Armendáriz, P.; Arriola, M.; Martín-Sánchez, E.; et al. ADAM12 is A Potential Therapeutic Target Regulated by Hypomethylation in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2020, 21, 903. https://doi.org/10.3390/ijms21030903
Mendaza S, Ulazia-Garmendia A, Monreal-Santesteban I, Córdoba A, Azúa YRd, Aguiar B, Beloqui R, Armendáriz P, Arriola M, Martín-Sánchez E, et al. ADAM12 is A Potential Therapeutic Target Regulated by Hypomethylation in Triple-Negative Breast Cancer. International Journal of Molecular Sciences. 2020; 21(3):903. https://doi.org/10.3390/ijms21030903
Chicago/Turabian StyleMendaza, Saioa, Ane Ulazia-Garmendia, Iñaki Monreal-Santesteban, Alicia Córdoba, Yerani Ruiz de Azúa, Begoña Aguiar, Raquel Beloqui, Pedro Armendáriz, Marta Arriola, Esperanza Martín-Sánchez, and et al. 2020. "ADAM12 is A Potential Therapeutic Target Regulated by Hypomethylation in Triple-Negative Breast Cancer" International Journal of Molecular Sciences 21, no. 3: 903. https://doi.org/10.3390/ijms21030903
APA StyleMendaza, S., Ulazia-Garmendia, A., Monreal-Santesteban, I., Córdoba, A., Azúa, Y. R. d., Aguiar, B., Beloqui, R., Armendáriz, P., Arriola, M., Martín-Sánchez, E., & Guerrero-Setas, D. (2020). ADAM12 is A Potential Therapeutic Target Regulated by Hypomethylation in Triple-Negative Breast Cancer. International Journal of Molecular Sciences, 21(3), 903. https://doi.org/10.3390/ijms21030903




