How p53 Molecules Solve the Target DNA Search Problem: A Review
Abstract
1. Introduction
2. p53 As a Model System for Target Search Studies
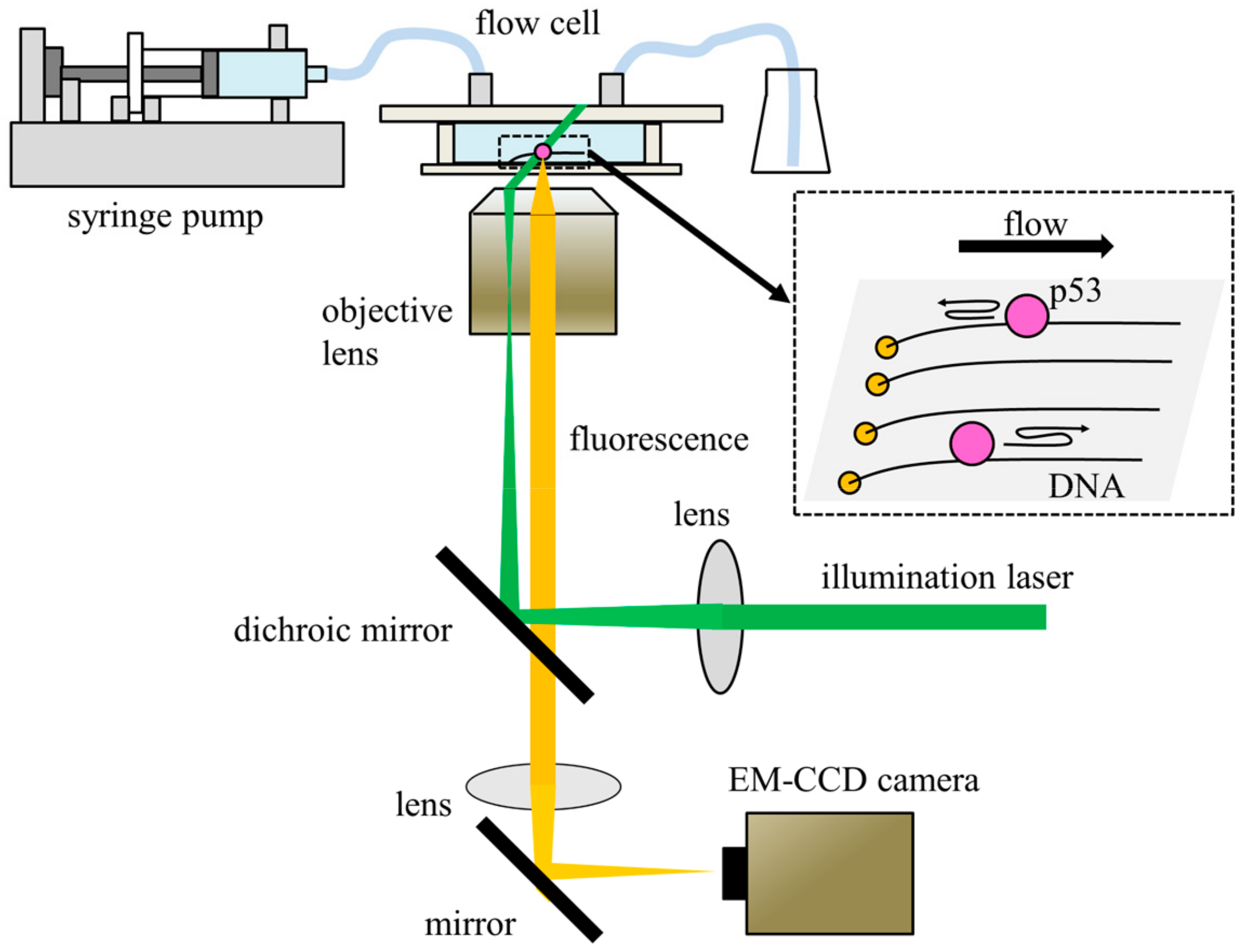
3. Single-Molecule Fluorescence Microscopy
4. 1D Diffusion of p53 along DNA
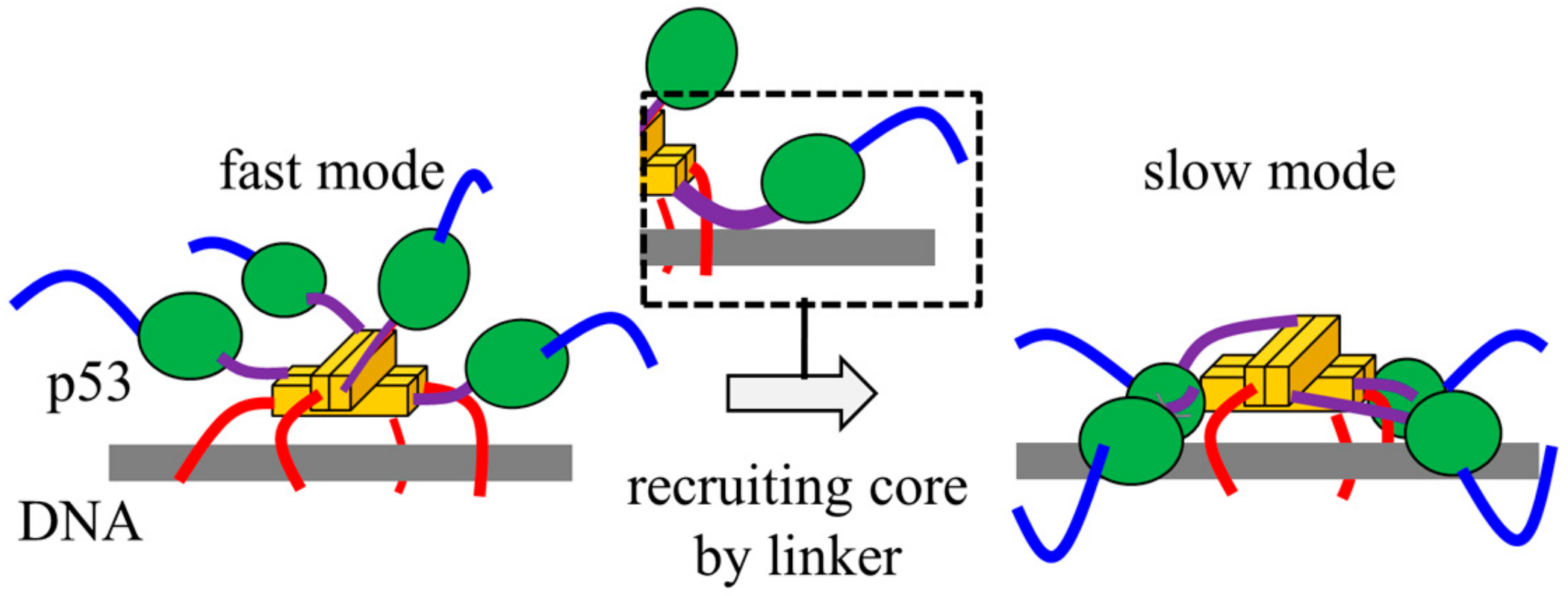
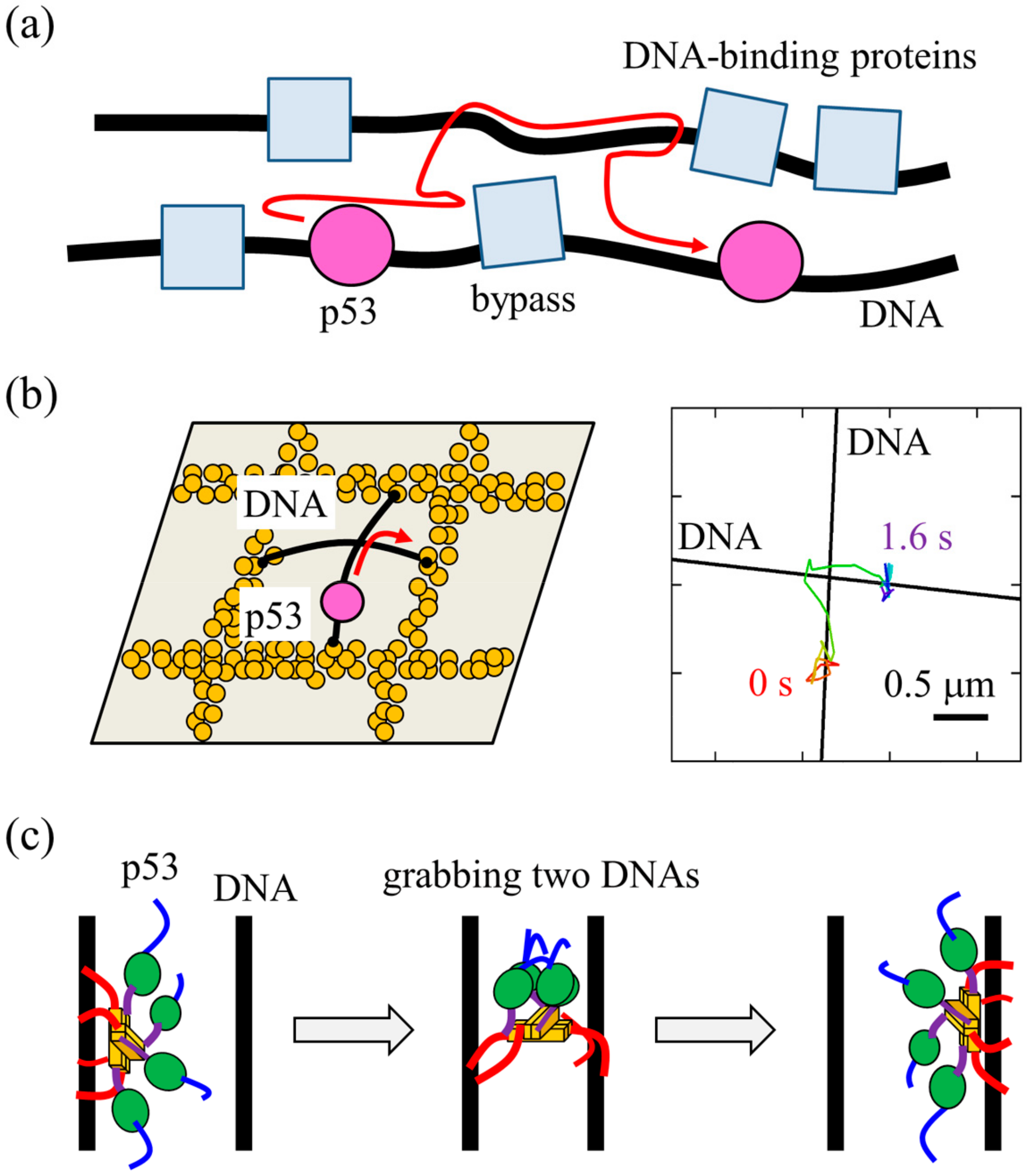
5. Ultrafast Intersegmental Transfer of p53
6. Target Search by p53 May Be Well Designed by Biological Requirements
7. Target Search of p53 in Live Cells
8. Common Target Search in Biology
9. Summary and Future Perspectives
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| NT | N-terminal |
| Tet | tetramerization |
| CT | C-terminal |
| TIRF | total internal reflection fluorescence |
| HILO | highly inclined and laminated optical sheet |
| PDMS | polydimethylsiloxane |
| MPC | 2-methacryloyloxyethyl phosphorylcholine |
| MD | molecular dynamics |
References
- Halford, S.E.; Marko, J.F. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004, 32, 3040–3052. [Google Scholar] [CrossRef] [PubMed]
- Tafvizi, A.; Mirny, L.A.; van Oijen, A.M. Dancing on DNA: Kinetic aspects of search processes on DNA. Chemphyschem 2011, 12, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Kamagata, K.; Murata, A.; Itoh, Y.; Takahashi, S. Characterization of facilitated diffusion of tumor suppressor p53 along DNA using single-molecule fluorescence imaging. J. Photochem. Photobiol. C Photochem. Reviews 2017, 30, 36–50. [Google Scholar] [CrossRef]
- Slutsky, M.; Mirny, L.A. Kinetics of protein-DNA interaction: Facilitated target location in sequence-dependent potential. Biophys. J. 2004, 87, 4021–4035. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.G.; Sewitz, S.; Andrews, S.S.; Lipkow, K. An integrated model of transcription factor diffusion shows the importance of intersegmental transfer and quaternary protein structure for target site finding. PLoS ONE 2014, 9, e108575. [Google Scholar] [CrossRef] [PubMed]
- Joerger, A.C.; Fersht, A.R. The tumor suppressor p53: From structures to drug discovery. Cold Spring Harb. Perspect. Biol. 2010, 2, a000919. [Google Scholar] [CrossRef] [PubMed]
- Laptenko, O.; Tong, D.R.; Manfredi, J.; Prives, C. The Tail That Wags the Dog: How the Disordered C-Terminal Domain Controls the Transcriptional Activities of the p53 Tumor-Suppressor Protein. Trends Biochem. Sci. 2016, 41, 1022–1034. [Google Scholar] [CrossRef]
- Kamada, R.; Toguchi, Y.; Nomura, T.; Imagawa, T.; Sakaguchi, K. Tetramer formation of tumor suppressor protein p53: Structure, function, and applications. Biopolymers 2016, 106, 598–612. [Google Scholar] [CrossRef]
- Anderson, M.E.; Woelker, B.; Reed, M.; Wang, P.; Tegtmeyer, P. Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: Implications for regulation. Mol. Cell. Biol. 1997, 17, 6255–6264. [Google Scholar] [CrossRef]
- Vuzman, D.; Levy, Y. Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Mol. Biosyst. 2012, 8, 47–57. [Google Scholar] [CrossRef]
- Hainaut, P.; Hollstein, M. p53 and human cancer: The first ten thousand mutations. Adv. Cancer Res. 2000, 77, 81–137. [Google Scholar] [PubMed]
- Kabata, H.; Kurosawa, O.; Arai, I.; Washizu, M.; Margarson, S.A.; Glass, R.E.; Shimamoto, N. Visualization of single molecules of RNA-polymerase sliding along DNA. Science 1993, 262, 1561–1563. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.M.; Austin, R.H.; Cox, E.C. Single molecule measurements of repressor protein 1D diffusion on DNA. Phys. Rev. Lett. 2006, 97, 048302. [Google Scholar] [CrossRef] [PubMed]
- Gorman, J.; Chowdhury, A.; Surtees, J.A.; Shimada, J.; Reichman, D.R.; Alani, E.; Greene, E.C. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol. Cell 2007, 28, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Komazin-Meredith, G.; Mirchev, R.; Golan, D.E.; van Oijen, A.M.; Coen, D.M. Hopping of a processivity factor on DNA revealed by single-molecule assays of diffusion. Proc. Natl. Acad. Sci. USA 2008, 105, 10721–10726. [Google Scholar] [CrossRef] [PubMed]
- Blainey, P.C.; Luo, G.; Kou, S.C.; Mangel, W.F.; Verdine, G.L.; Bagchi, B.; Xie, X.S. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009, 16, 1224–1229. [Google Scholar] [CrossRef] [PubMed]
- Biebricher, A.; Wende, W.; Escude, C.; Pingoud, A.; Desbiolles, P. Tracking of single quantum dot labeled EcoRV sliding along DNA manipulated by double optical tweezers. Biophys. J. 2009, 96, L50–L52. [Google Scholar] [CrossRef]
- Gorman, J.; Plys, A.J.; Visnapuu, M.L.; Alani, E.; Greene, E.C. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat. Struct. Mol. Biol. 2010, 17, 932–938. [Google Scholar] [CrossRef]
- Dunn, A.R.; Kad, N.M.; Nelson, S.R.; Warshaw, D.M.; Wallace, S.S. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011, 39, 7487–7498. [Google Scholar] [CrossRef]
- Forget, A.L.; Dombrowski, C.C.; Amitani, I.; Kowalczykowski, S.C. Exploring protein-DNA interactions in 3D using in situ construction, manipulation and visualization of individual DNA dumbbells with optical traps, microfluidics and fluorescence microscopy. Nat. Protoc. 2013, 8, 525–538. [Google Scholar] [CrossRef]
- Lee, J.B.; Cho, W.K.; Park, J.; Jeon, Y.; Kim, D.; Lee, S.H.; Fishel, R. Single-molecule views of MutS on mismatched DNA. DNA Repair 2014, 20, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Countryman, P.; Buncher, N.; Kaur, P.; E, L.; Zhang, Y.; Gibson, G.; You, C.; Watkins, S.C.; Piehler, J.; et al. TRF1 and TRF2 use different mechanisms to find telomeric DNA but share a novel mechanism to search for protein partners at telomeres. Nucleic Acids Res. 2014, 42, 2493–2504. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.D.; Lopez, M.A., Jr.; Hanne, J.; Peake, M.B.; Lee, J.B.; Fishel, R.; Yoder, K.E. Retroviral intasomes search for a target DNA by 1D diffusion which rarely results in integration. Nat. Commun. 2016, 7, 11409. [Google Scholar] [CrossRef] [PubMed]
- Kostiuk, G.; Dikic, J.; Schwarz, F.W.; Sasnauskas, G.; Seidel, R.; Siksnys, V. The dynamics of the monomeric restriction endonuclease BcnI during its interaction with DNA. Nucleic Acids Res. 2017, 45, 5968–5979. [Google Scholar] [CrossRef] [PubMed]
- Kamagata, K.; Itoh, Y.; Subekti, D.R.G. How does tumor suppressor protein p53 solve the target DNA search problem? (Japanese). BUTSURI 2019, 74, 472–475. [Google Scholar]
- Yamamoto, T.; Kurosawa, O.; Kabata, H.; Shimamoto, N.; Washizu, M. Molecular surgery of DNA based on electrostatic micromanipulation. Ieee Trans. Ind. Appl. 2000, 36, 1010–1017. [Google Scholar] [CrossRef]
- Fazio, T.; Visnapuu, M.L.; Wind, S.; Greene, E.C. DNA curtains and nanoscale curtain rods: High-throughput tools for single molecule imaging. Langmuir 2008, 24, 10524–10531. [Google Scholar] [CrossRef]
- Igarashi, C.; Murata, A.; Itoh, Y.; Subekti, D.R.G.; Takahashi, S.; Kamagata, K. DNA garden: A simple method for producing arrays of stretchable DNA for single-molecule fluorescence imaging of DNA binding proteins. Bull. Chem. Soc. Jpn. 2017, 90, 34–43. [Google Scholar] [CrossRef]
- Tafvizi, A.; Huang, F.; Leith, J.S.; Fersht, A.R.; Mirny, L.A.; van Oijen, A.M. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys. J. 2008, 95, L01–L03. [Google Scholar] [CrossRef]
- McKinney, K.; Mattia, M.; Gottifredi, V.; Prives, C. p53 linear diffusion along DNA requires its C terminus. Mol. Cell 2004, 16, 413–424. [Google Scholar] [CrossRef]
- Murata, A.; Ito, Y.; Kashima, R.; Kanbayashi, S.; Nanatani, K.; Igarashi, C.; Okumura, M.; Inaba, K.; Tokino, T.; Takahashi, S.; et al. One-dimensional sliding of p53 along DNA is accelerated in the presence of Ca(2+) or Mg(2+) at millimolar concentrations. J. Mol. Biol. 2015, 427, 2663–2678. [Google Scholar] [CrossRef] [PubMed]
- Khazanov, N.; Levy, Y. Sliding of p53 along DNA can be modulated by its oligomeric state and by cross-talks between its constituent domains. J. Mol. Biol. 2011, 408, 335–355. [Google Scholar] [CrossRef] [PubMed]
- Terakawa, T.; Kenzaki, H.; Takada, S. p53 searches on DNA by rotation-uncoupled sliding at C-terminal tails and restricted hopping of core domains. J. Am. Chem. Soc. 2012, 134, 14555–14562. [Google Scholar] [CrossRef] [PubMed]
- Tafvizi, A.; Huang, F.; Fersht, A.R.; Mirny, L.A.; van Oijen, A.M. A single-molecule characterization of p53 search on DNA. Proc. Natl. Acad. Sci. USA 2011, 108, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Leith, J.S.; Tafvizi, A.; Huang, F.; Uspal, W.E.; Doyle, P.S.; Fersht, A.R.; Mirny, L.A.; van Oijen, A.M. Sequence-dependent sliding kinetics of p53. Proc. Natl. Acad. Sci. USA 2012, 109, 16552–16557. [Google Scholar] [CrossRef]
- Murata, A.; Itoh, Y.; Mano, E.; Kanbayashi, S.; Igarashi, C.; Takahashi, H.; Takahashi, S.; Kamagata, K. One-dimensional search dynamics of tumor suppressor p53 regulated by a disordered C-terminal domain. Biophys. J. 2017, 112, 2301–2314. [Google Scholar] [CrossRef]
- Subekti, D.R.G.; Murata, A.; Itoh, Y.; Fukuchi, S.; Takahashi, H.; Kanbayashi, S.; Takahashi, S.; Kamagata, K. The disordered linker in p53 participates in nonspecific binding to and one-dimensional sliding along DNA revealed by single-molecule fluorescence measurements. Biochemistry 2017, 56, 4134–4144. [Google Scholar] [CrossRef]
- Benichou, O.; Loverdo, C.; Moreau, M.; Voituriez, R. Intermittent search strategies. Rev. Modern Phys. 2011, 83, 81–129. [Google Scholar] [CrossRef]
- Itoh, Y.; Murata, A.; Sakamoto, S.; Nanatani, K.; Wada, T.; Takahashi, S.; Kamagata, K. Activation of p53 facilitates the target search in DNA by enhancing the target recognition probability. J. Mol. Biol. 2016, 428, 2916–2930. [Google Scholar] [CrossRef]
- Terakawa, T.; Takada, S. p53 dynamics upon response element recognition explored by molecular simulations. Sci. Rep. 2015, 5, 17107. [Google Scholar] [CrossRef]
- Itoh, Y.; Murata, A.; Takahashi, S.; Kamagata, K. Intrinsically disordered domain of tumor suppressor p53 facilitates target search by ultrafast transfer between different DNA strands. Nucleic Acids Res. 2018, 46, 7261–7269. [Google Scholar] [CrossRef] [PubMed]
- Gorman, J.; Wang, F.; Redding, S.; Plys, A.J.; Fazio, T.; Wind, S.; Alani, E.E.; Greene, E.C. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc. Natl. Acad. Sci. USA 2012, 109, E3074–E3083. [Google Scholar] [CrossRef] [PubMed]
- Takada, S.; Kanada, R.; Tan, C.; Terakawa, T.; Li, W.; Kenzaki, H. Modeling Structural Dynamics of Biomolecular Complexes by Coarse-Grained Molecular Simulations. Acc. Chem. Res. 2015, 48, 3026–3035. [Google Scholar] [CrossRef] [PubMed]
- Neylon, C.; Brown, S.E.; Kralicek, A.V.; Miles, C.S.; Love, C.A.; Dixon, N.E. Interaction of the Escherichia coli replication terminator protein (Tus) with DNA: A model derived from DNA-binding studies of mutant proteins by surface plasmon resonance. Biochemistry 2000, 39, 11989–11999. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Takayama, Y.; Clore, G.M. Impact of protein/protein interactions on global intermolecular translocation rates of the transcription factors Sox2 and Oct1 between DNA cognate sites analyzed by z-exchange NMR spectroscopy. J. Biol. Chem. 2012, 287, 26962–26970. [Google Scholar] [CrossRef] [PubMed]
- Spinner, D.S.; Liu, S.; Wang, S.W.; Schmidt, J. Interaction of the myogenic determination factor myogenin with E12 and a DNA target: Mechanism and kinetics. J. Mol. Biol. 2002, 317, 431–445. [Google Scholar] [CrossRef]
- Kim, J.G.; Takeda, Y.; Matthews, B.W.; Anderson, W.F. Kinetic studies on Cro repressor-operator DNA interaction. J. Mol. Biol. 1987, 196, 149–158. [Google Scholar] [CrossRef]
- Luo, Y.; North, J.A.; Rose, S.D.; Poirier, M.G. Nucleosomes accelerate transcription factor dissociation. Nucleic Acids Res. 2014, 42, 3017–3027. [Google Scholar] [CrossRef]
- Esadze, A.; Iwahara, J. Stopped-flow fluorescence kinetic study of protein sliding and intersegment transfer in the target DNA search process. J. Mol. Biol. 2014, 426, 230–244. [Google Scholar] [CrossRef]
- Carlsson, B.; Haggblad, J. Quantitative determination of DNA-binding parameters for the human estrogen receptor in a solid-phase, nonseparation assay. Anal. Biochem. 1995, 232, 172–179. [Google Scholar] [CrossRef]
- Cho, S.; Wensink, P.C. DNA binding by the male and female doublesex proteins of Drosophila melanogaster. J. Biol. Chem. 1997, 272, 3185–3189. [Google Scholar] [CrossRef] [PubMed]
- Golebiowski, F.M.; Gorecki, A.; Bonarek, P.; Rapala-Kozik, M.; Kozik, A.; Dziedzicka-Wasylewska, M. An investigation of the affinities, specificity and kinetics involved in the interaction between the Yin Yang 1 transcription factor and DNA. FEBS J. 2012, 279, 3147–3158. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, F.; Kawamura, S.; Yamasaki, N.; Tanaka, I.; Kimura, M. Arginine-55 in the beta-arm is essential for the activity of DNA-binding protein HU from Bacillus stearothermophilus. Biosci. Biotechnol. Biochem. 1999, 63, 2232–2235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nakadai, T.; Shimada, M.; Shima, D.; Handa, H.; Tamura, T.A. Specific interaction with transcription factor IIA and localization of the mammalian TATA-binding protein-like protein (TLP/TRF2/TLF). J. Biol. Chem. 2004, 279, 7447–7455. [Google Scholar] [CrossRef] [PubMed]
- Wanandi, I.; Waldschmidt, R.; Seifart, K.H. Mammalian transcription factor PBP. Characterization of its binding properties to the proximal sequence element of U6 genes. J. Biol. Chem. 1993, 268, 6629–6640. [Google Scholar]
- Kwon, H.; Park, S.; Lee, S.; Lee, D.K.; Yang, C.H. Determination of binding constant of transcription factor AP-1 and DNA. Application of inhibitors. Eur. J. Biochem. 2001, 268, 565–572. [Google Scholar] [CrossRef]
- Okahata, Y.; Niikura, K.; Sugiura, Y.; Sawada, M.; Morii, T. Kinetic studies of sequence-specific binding of GCN4-bZIP peptides to DNA strands immobilized on a 27-MHz quartz-crystal microbalance. Biochemistry 1998, 37, 5666–5672. [Google Scholar] [CrossRef]
- Sugo, N.; Morimatsu, M.; Arai, Y.; Kousoku, Y.; Ohkuni, A.; Nomura, T.; Yanagida, T.; Yamamoto, N. Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence. Sci. Rep. 2015, 5, 10662. [Google Scholar] [CrossRef]
- Iwahara, J.; Zweckstetter, M.; Clore, G.M. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc. Natl. Acad. Sci. USA 2006, 103, 15062–15067. [Google Scholar] [CrossRef]
- Iwahara, J.; Clore, G.M. Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J. Am. Chem. Soc. 2006, 128, 404–405. [Google Scholar] [CrossRef]
- Takayama, Y.; Sahu, D.; Iwahara, J. NMR studies of translocation of the Zif268 protein between its target DNA Sites. Biochemistry 2010, 49, 7998–8005. [Google Scholar] [CrossRef] [PubMed]
- Takayama, Y.; Clore, G.M. Interplay between minor and major groove-binding transcription factors Sox2 and Oct1 in translocation on DNA studied by paramagnetic and diamagnetic NMR. J. Biol. Chem. 2012, 287, 14349–14363. [Google Scholar] [CrossRef] [PubMed]
- Esadze, A.; Kemme, C.A.; Kolomeisky, A.B.; Iwahara, J. Positive and negative impacts of nonspecific sites during target location by a sequence-specific DNA-binding protein: Origin of the optimal search at physiological ionic strength. Nucleic Acids Res. 2014, 42, 7039–7046. [Google Scholar] [CrossRef]
- Giuntoli, R.D.; Linzer, N.B.; Banigan, E.J.; Sing, C.E.; de la Cruz, M.O.; Graham, J.S.; Johnson, R.C.; Marko, J.F. DNA-segment-facilitated Dissociation of Fis and NHP6A from DNA detected via single-molecule mechanical response. J. Mol. Biol. 2015, 427, 3123–3136. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.V.; Wade, M.; Wong, E.; Li, Y.C.; Rodewald, L.W.; Wahl, G.M. Quantitative analyses reveal the importance of regulated Hdmx degradation for P53 activation. Proc. Natl. Acad. Sci. USA 2007, 104, 12365–12370. [Google Scholar] [CrossRef]
- Wu, M.; Ye, H.; Tang, Z.; Shao, C.; Lu, G.; Chen, B.; Yang, Y.; Wang, G.; Hao, H. p53 dynamics orchestrates with binding affinity to target genes for cell fate decision. Cell Death Dis. 2017, 8, e3130. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Kato, S.; Otsuka, K.; Watanabe, G.; Kumabe, T.; Tominaga, T.; Yoshimoto, T.; Ishioka, C. The relationship among p53 oligomer formation, structure and transcriptional activity using a comprehensive missense mutation library. Oncogene 2005, 24, 6976–6981. [Google Scholar] [CrossRef]
- Rajagopalan, S.; Huang, F.; Fersht, A.R. Single-Molecule characterization of oligomerization kinetics and equilibria of the tumor suppressor p53. Nucleic Acids Res. 2011, 39, 2294–2303. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.W.; Prodeus, A.; Malkin, D.; Gariepy, J. p53 oligomerization status modulates cell fate decisions between growth, arrest and apoptosis. Cell Cycle 2016, 15, 3210–3219. [Google Scholar] [CrossRef] [PubMed]
- Gaglia, G.; Guan, Y.; Shah, J.V.; Lahav, G. Activation and control of p53 tetramerization in individual living cells. Proc. Natl. Acad. Sci. USA 2013, 110, 15497–15501. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, R.L.; Veprintsev, D.B.; Fersht, A.R. Cooperative binding of tetrameric p53 to DNA. J. Mol. Biol. 2004, 341, 1145–1159. [Google Scholar] [CrossRef] [PubMed]
- Beckerman, R.; Prives, C. Transcriptional regulation by p53. Cold Spring Harb. Perspect. Biol. 2010, 2, a000935. [Google Scholar] [CrossRef] [PubMed]
- Bieging, K.T.; Mello, S.S.; Attardi, L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Canc. 2014, 14, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Hamard, P.J.; Lukin, D.J.; Manfredi, J.J. p53 basic C terminus regulates p53 functions through DNA binding modulation of subset of target genes. J. Biol. Chem. 2012, 287, 22397–22407. [Google Scholar] [CrossRef] [PubMed]
- Hamard, P.J.; Barthelery, N.; Hogstad, B.; Mungamuri, S.K.; Tonnessen, C.A.; Carvajal, L.A.; Senturk, E.; Gillespie, V.; Aaronson, S.A.; Merad, M.; et al. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev. 2013, 27, 1868–1885. [Google Scholar] [CrossRef] [PubMed]
- Marouco, D.; Garabadgiu, A.V.; Melino, G.; Barlev, N.A. Lysine-specific modifications of p53: A matter of life and death? Oncotarget 2013, 4, 1556–1571. [Google Scholar] [CrossRef]
- Laptenko, O.; Shiff, I.; Freed-Pastor, W.; Zupnick, A.; Mattia, M.; Freulich, E.; Shamir, I.; Kadouri, N.; Kahan, T.; Manfredi, J.; et al. The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol. Cell 2015, 57, 1034–1346. [Google Scholar] [CrossRef]
- Retzlaff, M.; Rohrberg, J.; Kupper, N.J.; Lagleder, S.; Bepperling, A.; Manzenrieder, F.; Peschek, J.; Kessler, H.; Buchner, J. The regulatory domain stabilizes the p53 tetramer by intersubunit contacts with the DNA binding domain. J. Mol. Biol. 2013, 425, 144–155. [Google Scholar] [CrossRef]
- Friedler, A.; Veprintsev, D.B.; Freund, S.M.; von Glos, K.I.; Fersht, A.R. Modulation of binding of DNA to the C-terminal domain of p53 by acetylation. Structure 2005, 13, 629–636. [Google Scholar] [CrossRef]
- Loffreda, A.; Jacchetti, E.; Antunes, S.; Rainone, P.; Daniele, T.; Morisaki, T.; Bianchi, M.E.; Tacchetti, C.; Mazza, D. Live-cell p53 single-molecule binding is modulated by C-terminal acetylation and correlates with transcriptional activity. Nat. Commun. 2017, 8, 313. [Google Scholar] [CrossRef]
- Mazza, D.; Abernathy, A.; Golob, N.; Morisaki, T.; McNally, J.G. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 2012, 40, e119. [Google Scholar] [CrossRef] [PubMed]
- Morisaki, T.; Muller, W.G.; Golob, N.; Mazza, D.; McNally, J.G. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat. Commun. 2014, 5, 4456. [Google Scholar] [CrossRef] [PubMed]
- Kanada, R.; Terakawa, T.; Kenzaki, H.; Takada, S. Nucleosome Crowding in Chromatin Slows the Diffusion but Can Promote Target Search of Proteins. Biophys. J. 2019, 116, 2285–2295. [Google Scholar] [CrossRef] [PubMed]
- Visnapuu, M.L.; Greene, E.C. Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition. Nat. Struct. Mol. Biol. 2009, 16, 1056–1062. [Google Scholar] [CrossRef]
- Jagelska, E.B.; Brazda, V.; Pecinka, P.; Palecek, E.; Fojta, M. DNA topology influences p53 sequence-specific DNA binding through structural transitions within the target sites. Biochem. J. 2008, 412, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Jagelska, E.B.; Pivonkova, H.; Fojta, M.; Brazda, V. The potential of the cruciform structure formation as an important factor influencing p53 sequence-specific binding to natural DNA targets. Biochem. Biophys. Res. Commun. 2010, 391, 1409–1414. [Google Scholar] [CrossRef] [PubMed]
- Coufal, J.; Jagelska, E.B.; Liao, J.C.; Brazda, V. Preferential binding of p53 tumor suppressor to p21 promoter sites that contain inverted repeats capable of forming cruciform structure. Biochem. Biophys. Res. Commun. 2013, 441, 83–88. [Google Scholar] [CrossRef]
- Brazda, V.; Cechova, J.; Battistin, M.; Coufal, J.; Jagelska, E.B.; Raimondi, I.; Inga, A. The structure formed by inverted repeats in p53 response elements determines the transactivation activity of p53 protein. Biochem. Biophys. Res. Commun. 2017, 483, 516–521. [Google Scholar] [CrossRef]
- Brazda, V.; Coufal, J. Recognition of Local DNA Structures by p53 Protein. Int. J. Mol. Sci. 2017, 18, 375. [Google Scholar] [CrossRef]
- Brazda, V.; Fojta, M. The Rich World of p53 DNA Binding Targets: The Role of DNA Structure. Int. J. Mol. Sci. 2019, 20, 5605. [Google Scholar] [CrossRef]
- Garcia-Alai, M.M.; Tidow, H.; Natan, E.; Townsley, F.M.; Veprintsev, D.B.; Fersht, A.R. The novel p53 isoform “delta p53” is a misfolded protein and does not bind the p21 promoter site. Protein Sci. 2008, 17, 1671–1678. [Google Scholar] [CrossRef] [PubMed]
- Sauer, M.; Bretz, A.C.; Beinoraviciute-Kellner, R.; Beitzinger, M.; Burek, C.; Rosenwald, A.; Harms, G.S.; Stiewe, T. C-terminal diversity within the p53 family accounts for differences in DNA binding and transcriptional activity. Nucleic Acids Res. 2008, 36, 1900–1912. [Google Scholar] [CrossRef] [PubMed]
- Meek, D.W.; Anderson, C.W. Posttranslational modification of p53: Cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 2009, 1, a000950. [Google Scholar] [CrossRef] [PubMed]
- Botchkarev, V.A.; Flores, E.R. p53/p63/p73 in the epidermis in health and disease. Cold Spring Harb. Perspect. Med. 2014, 4, a015248. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Menendez, D.; Resnick, M.A.; Anderson, C.W. Mutant TP53 posttranslational modifications: Challenges and opportunities. Hum. Mutat. 2014, 35, 738–755. [Google Scholar] [CrossRef]
- Bonnet, I.; Biebricher, A.; Porte, P.-L.; Loverdo, C.; Benichou, O.; Voituriez, R.; Escude, C.; Wende, W.; Pingoud, A.; Desbiolles, P. Sliding and jumping of single EcoRV restriction enzymes on non-cognate DNA. Nucleic Acids Res. 2008, 36, 4118–4127. [Google Scholar] [CrossRef]
- Aramayo, R.; Sherman, M.B.; Brownless, K.; Lurz, R.; Okorokov, A.L.; Orlova, E.V. Quaternary structure of the specific p53-DNA complex reveals the mechanism of p53 mutant dominance. Nucleic Acids Res. 2011, 39, 8960–8971. [Google Scholar] [CrossRef]
- Kearns, S.; Lurz, R.; Orlova, E.V.; Okorokov, A.L. Two p53 tetramers bind one consensus DNA response element. Nucleic Acids Res. 2016, 44, 6185–6199. [Google Scholar] [CrossRef]
- Martin, T.G.; Bharat, T.A.; Joerger, A.C.; Bai, X.C.; Praetorius, F.; Fersht, A.R.; Dietz, H.; Scheres, S.H. Design of a molecular support for cryo-EM structure determination. Proc. Natl. Acad. Sci. USA 2016, 113, E7456–E7463. [Google Scholar] [CrossRef]
- Kamagata, K.; Mano, E.; Itoh, Y.; Wakamoto, T.; Kitahara, R.; Kanbayashi, S.; Takahashi, H.; Murata, A.; Kameda, T. Rational design using sequence information only produces a peptide that binds to the intrinsically disordered region of p53. Sci. Rep. 2019, 9, 8584. [Google Scholar] [CrossRef]
- Wang, F.; Redding, S.; Finkelstein, I.J.; Gorman, J.; Reichman, D.R.; Greene, E.C. The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion. Nat. Struct. Mol. Biol. 2013, 20, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, S.H.; Redding, S.; Jinek, M.; Greene, E.C.; Doudna, J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014, 507, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.R.; Dunn, A.R.; Kathe, S.D.; Warshaw, D.M.; Wallace, S.S. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proc. Natl. Acad. Sci. USA 2014, 111, E2091–E2099. [Google Scholar] [CrossRef] [PubMed]
- Cuculis, L.; Abil, Z.; Zhao, H.; Schroeder, C.M. TALE proteins search DNA using a rotationally decoupled mechanism. Nat. Chem. Biol. 2016, 12, 831–837. [Google Scholar] [CrossRef]
- Kamagata, K.; Mano, E.; Ouchi, K.; Kanbayashi, S.; Johnson, R.C. High Free-Energy Barrier of 1D Diffusion Along DNA by Architectural DNA-Binding Proteins. J. Mol. Biol. 2018, 430, 655–667. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamagata, K.; Itoh, Y.; Subekti, D.R.G. How p53 Molecules Solve the Target DNA Search Problem: A Review. Int. J. Mol. Sci. 2020, 21, 1031. https://doi.org/10.3390/ijms21031031
Kamagata K, Itoh Y, Subekti DRG. How p53 Molecules Solve the Target DNA Search Problem: A Review. International Journal of Molecular Sciences. 2020; 21(3):1031. https://doi.org/10.3390/ijms21031031
Chicago/Turabian StyleKamagata, Kiyoto, Yuji Itoh, and Dwiky Rendra Graha Subekti. 2020. "How p53 Molecules Solve the Target DNA Search Problem: A Review" International Journal of Molecular Sciences 21, no. 3: 1031. https://doi.org/10.3390/ijms21031031
APA StyleKamagata, K., Itoh, Y., & Subekti, D. R. G. (2020). How p53 Molecules Solve the Target DNA Search Problem: A Review. International Journal of Molecular Sciences, 21(3), 1031. https://doi.org/10.3390/ijms21031031




