Association of Vitamin D Metabolism Gene Polymorphisms with Autoimmunity: Evidence in Population Genetic Studies
Abstract
1. Introduction
2. Genetic Susceptibility to Autoimmune Diseases
3. Genetic Polymorphisms Overview
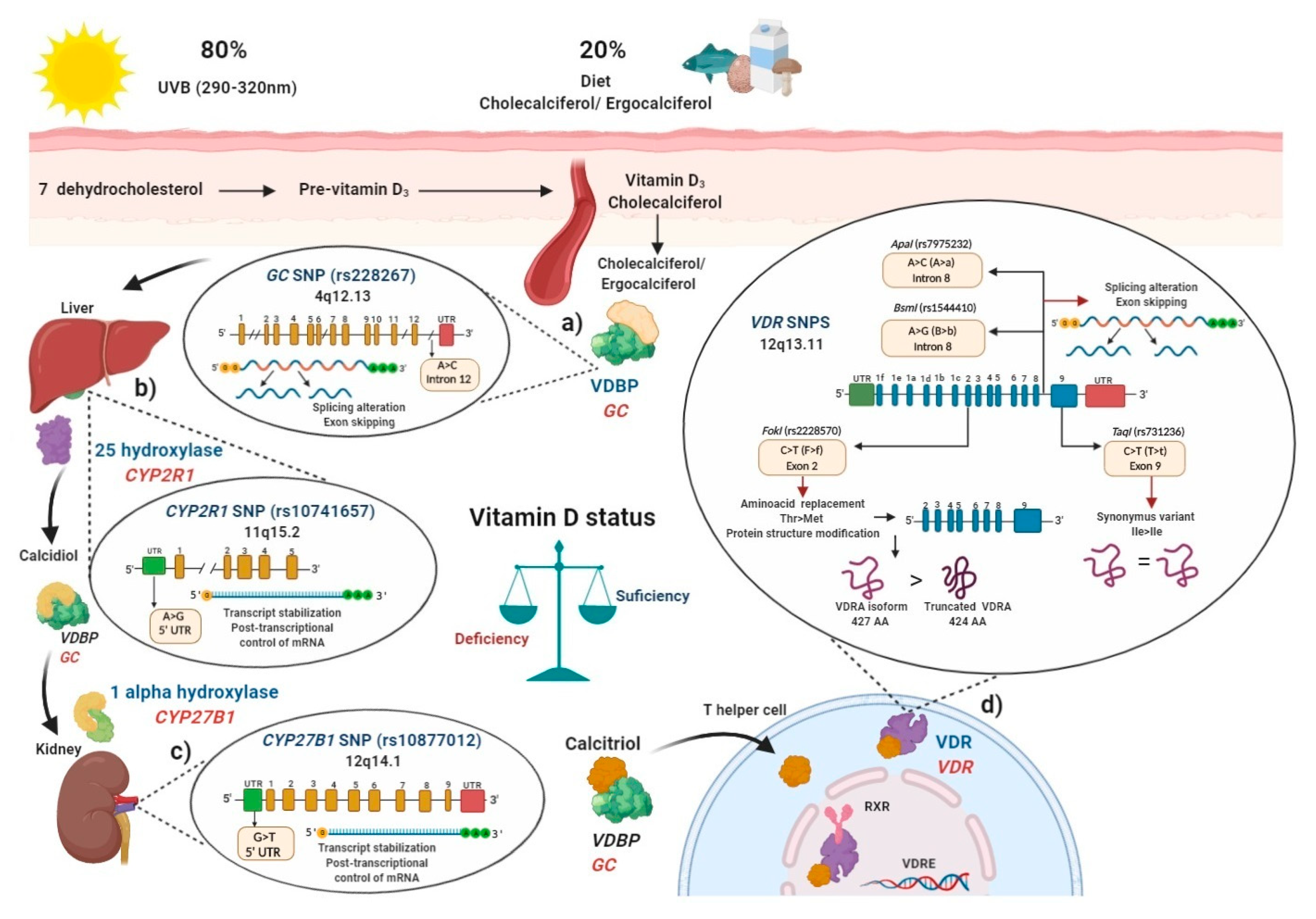
4. Functional Effects of Genetic Polymorphisms
5. Vitamin D Status and Genetic Evidence in the Populations
6. Polymorphisms in the Main Key Genes Related to Vitamin D Metabolism
6.1. Vitamin D Binding Protein (VDBP) (SNP rs2282679 GC)
6.2. Vitamin D 25-Hydroxylase (SNP rs10741657 CYP2R1)
6.3. Vitamin D 1-α Hydroxylase (SNP rs10877012 CYP27B1)
6.4. Polymorphisms in Vitamin D Receptor (VDR)
6.4.1. FokI (rs2228570) VDR SNP
6.4.2. BsmI (rs1544410) and ApaI (rs7975232) VDR SNPs
6.4.3. TaqI (rs731236) VDR SNP
7. Polymorphisms in Main Vitamin D Metabolism Genes Associated with Autoimmune Diseases
7.1. Multiple Sclerosis (MS)
7.2. Rheumatoid Arthritis (RA)
7.3. Systemic Lupus Erythematosus (SLE)
8. Methods
Literature Search Strategy
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AID | Autoimmune disease |
| CS | Control subject |
| HLA | Human leukocyte antigen |
| LD | Linkage disequilibrium |
| MS | Multiple sclerosis |
| OR | Odds ratio |
| RA | Rheumatoid arthritis |
| RNA | Ribonucleic acid |
| SLE | Systemic lupus erythematosus |
| SNP | Single nucleotide polymorphism |
| UTR | Untranslated region |
| VDBP | Vitamin D binding protein |
| VDR | Vitamin D receptor |
References
- Selmi, C.; Lu, Q.; Humble, M.C. Heritability versus the role of the environment in autoimmunity. J. Autoimmun. 2012, 39, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Floreani, A.; Leung, P.; Gershwin, M. Enviromental basis of autoimmunity. Clin. Rev. Allergy Immunol. 2016, 50, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Adorini, L.; Penna, G. Control of autoimmune diseases by the vitamin D endocrine system. Nat. Clin. Pract. Rheumatol. 2008, 4, 404–412. [Google Scholar] [CrossRef]
- Arnson, Y.; Amital, H.; Shoenfeld, Y. Vitamin D and autoimmunity: New aetiological and therapeutic considerations. Ann. Rheum. Dis. 2007, 66, 1137–1142. [Google Scholar] [CrossRef]
- Dankers, W.; Colin, E.M.; van Hamburg, J.P.; Lubberts, E. Vitamin D in Autoimmunity: Molecular Mechanisms and Therapeutic Potential. Front. Immunol. 2017, 20, 697. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Lv, Z.; Fan, X.; Wang, L.; Han, F.; Wang, H.; Bi, S. Vitamin D status and the risk of multiple sclerosis: A systematic review and meta-analysis. Neurosci. Lett. 2014, 570, 108–113. [Google Scholar] [CrossRef]
- Lin, J.; Liu, J.; Davies, M.L.; Chen, W. Serum Vitamin D Level and Rheumatoid Arthritis Disease Activity: Review and Meta-Analysis. PLoS ONE 2016, 11, e0146351. [Google Scholar] [CrossRef]
- Shoenfeld, Y.; Giacomelli, R.; Azrielant, S.; Berardicurti, O.; Reynolds, J.A.; Bruce, I.N. Vitamin D and systemic lupus erythematosus—The hype and the hope. Autoimmun. Rev. 2018, 17, 19–23. [Google Scholar] [CrossRef]
- Sakthiswary, R.; Raymond, A.A. The Clinical Significance of Vitamin D in Systemic Lupus Erythematosus: A Systematic Review. PLoS ONE 2013, 8, e55275. [Google Scholar] [CrossRef]
- Brouwer-Brolsma, E.M.; Vaes, A.M.M.; van der Zwaluw, N.L.; van Wijngaarden, J.P.; Swart, K.M.A.; Ham, A.C.; van Dijk, S.C.; Enneman, A.W.; Sohl, E.; van Schoor, N.M.; et al. Relative importance of summer sun exposure, vitamin D intake, and genes to vitamin D status in Dutch older adults: The B-PROOF study. J. Steroid Biochem. Mol. Biol. 2016, 164, 168–176. [Google Scholar] [CrossRef]
- Sepulveda-Villegas, M.; Elizondo-Montemayor, L.; Trevino, V. Identification and analysis of 35 genes associated with vitamin D deficiency: A systematic review to identify genetic variants. J. Steroid Biochem. Mol. Biol. 2020, 196, 105516. [Google Scholar] [CrossRef] [PubMed]
- Hayter, S.M.; Cook, M.C. Updated assessment of the prevalence, spectrum and case definition of autoimmune disease. Autoimmun. Rev. 2012, 11, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.; Jeremias, P.; Matthias, T. The World Incidence and Prevalence of Autoimmune Diseases is Increasing. Int. J. Celiac Dis. 2016, 3, 151–155. [Google Scholar] [CrossRef]
- Baranzini, S.E. The genetics of autoimmune diseases: A networked perspective. Curr. Opin. Immunol. 2009, 21, 596–605. [Google Scholar] [CrossRef]
- Hewagama, A.; Richardson, B. The genetics and epigenetics of autoimmune diseases. J. Autoimmun. 2009, 33, 3–11. [Google Scholar] [CrossRef]
- Kuusisto, H.; Kaprio, J.; Kinnunen, E.; Luukkaala, T.; Koskenvuo, M.; Elovaara, I. Concordance and heritability of multiple sclerosis in Finland: Study on a nationwide series of twins: Concordance and heritability of MS. Eur. J. Neurol. 2008, 15, 1106–1110. [Google Scholar] [CrossRef]
- Lill, C.M. Recent Advances and Future Challenges in the Genetics of Multiple Sclerosis. Front. Neurol. 2014, 5, 130. [Google Scholar] [CrossRef]
- MacGregor, A.J.; Snieder, H.; Rigby, A.S.; Koskenvuo, M.; Kaprio, J.; Aho, K.; Silman, A.J. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheumatol. 2000, 43, 30–37. [Google Scholar] [CrossRef]
- Deng, Y.; Tsao, B.P. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat. Rev. Rheumatol. 2010, 6, 683–692. [Google Scholar] [CrossRef]
- Kochi, Y. Genetics of autoimmune diseases: Perspectives from genome-wide association studies: Table 1. Int. Immunol. 2016, 28, 155–161. [Google Scholar] [CrossRef]
- Gregersen, P.K.; Behrens, T.W. Genetics of autoimmune diseases—Disorders of immune homeostasis. Nat. Rev. Genet. 2006, 7, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, M.; Gough, S. The HLA Region and Autoimmune Disease: Associations and Mechanisms of Action. Curr. Genom. 2007, 8, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Okuda, D.T.; Srinivasan, R.; Oksenberg, J.R.; Goodin, D.S.; Baranzini, S.E.; Beheshtian, A.; Waubant, E.; Zamvil, S.S.; Leppert, D.; Qualley, P.; et al. Genotype–Phenotype correlations in multiple sclerosis: HLA genes influence disease severity inferred by 1HMR spectroscopy and MRI measures. Brain 2009, 132, 250–259. [Google Scholar] [CrossRef]
- Iniesta, R.; Guinó, E.; Moreno, V. Análisis estadístico de polimorfismos genéticos en estudios epidemiológicos. Gac. Sanit. 2005, 19, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Scally, S.W.; Petersen, J.; Law, S.C.; Dudek, N.L.; Nel, H.J.; Loh, K.L.; Wijeyewickrema, L.C.; Eckle, S.B.; Van Heemst, J.; Pike, R.N.; et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J. Exp. Med. 2013, 210, 2569–2582. [Google Scholar] [CrossRef] [PubMed]
- Castaño-Rodríguez, N.; Diaz-Gallo, L.-M.; Pineda-Tamayo, R.; Rojas-Villarraga, A.; Anaya, J.-M. Meta-analysis of HLA-DRB1 and HLA-DQB1 polymorphisms in Latin American patients with systemic lupus erythematosus. Autoimmun. Rev. 2008, 7, 322–330. [Google Scholar] [CrossRef]
- Holoshitz, J. The rheumatoid arthritis HLA–DRB1 shared epitope. Curr. Opin. Rheumatol. 2010, 22, 293–298. [Google Scholar] [CrossRef]
- Ramagopalan, S.V.; Maugeri, N.J.; Handunnetthi, L.; Lincoln, M.R.; Orton, S.-M.; Dyment, D.A.; DeLuca, G.C.; Herrera, B.M.; Chao, M.J.; Sadovnick, A.D.; et al. Expression of the Multiple Sclerosis-Associated MHC Class II Allele HLA-DRB1*1501 Is Regulated by Vitamin D. PLoS Genet. 2009, 5, e1000369. [Google Scholar] [CrossRef] [PubMed]
- Cocco, E.; Meloni, A.; Murru, M.R.; Corongiu, D.; Tranquilli, S.; Fadda, E.; Murru, R.; Schirru, L.; Secci, M.A.; Costa, G.; et al. Vitamin D Responsive Elements within the HLA-DRB1 Promoter Region in Sardinian Multiple Sclerosis Associated Alleles. Campbell M, editor. PLoS ONE 2012, 7, e41678. [Google Scholar] [CrossRef] [PubMed]
- Serrano, N.C.; Millan, P.; Páez, M.-C. Non-HLA associations with autoimmune diseases. Autoimmun. Rev. 2006, 5, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Emerah, A.A.; El-Shal, A.S. Role of vitamin D receptor gene polymorphisms and serum 25-hydroxyvitamin D level in Egyptian female patients with systemic lupus erythematosus. Mol. Biol. Rep. 2013, 40, 6151–6162. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.G.; Choi, E.-H.; Foster, C.B.; Chanock, S.J. Using genetic variation to study human disease. Trends Mol. Med. 2001, 7, 507–512. [Google Scholar] [CrossRef]
- The International SNP Map Working Group. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 2001, 409, 928–933. [Google Scholar] [CrossRef] [PubMed]
- Vignal, A.; Milan, D.; SanCristobal, M.; Eggen, A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 2002, 34, 275. [Google Scholar] [CrossRef] [PubMed]
- Sirota, M.; Schaub, M.A.; Batzoglou, S.; Robinson, W.H.; Butte, A.J. Autoimmune Disease Classification by Inverse Association with SNP Alleles. PLoS Genet. 2009, 5, e1000792. [Google Scholar] [CrossRef] [PubMed]
- Cardon, L.R.; Bell, J.I. Association study designs for complex diseases. Nat. Rev. Genet. 2001, 2, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Hurst, L.D. Determinants of the Usage of Splice-Associated cis-Motifs Predict the Distribution of Human Pathogenic SNPs. Mol. Biol. Evol. 2016, 33, 518–529. [Google Scholar] [CrossRef]
- Guo, Y.; Jamison, D.C. The distribution of SNPs in human gene regulatory regions. BMC Genom. 2005, 6, 140. [Google Scholar] [CrossRef]
- Nishimura, D. Functional analysis of human promoter polymorphisms. Hum. Mol. Genet. 2003, 12, 2249–2254. [Google Scholar]
- Oubounyt, M.; Louadi, Z.; Tayara, H.; Chong, K.T. DeePromoter: Robust Promoter Predictor Using Deep Learning. Front. Genet. 2019, 10, 286. [Google Scholar] [CrossRef]
- Carlon, E.; Malki, M.L.; Blossey, R. Exons, Introns, and DNA Thermodynamics. Phys. Rev. Lett. 2005, 94, 178101. [Google Scholar] [CrossRef] [PubMed]
- Shaul, O. How introns enhance gene expression. Int. J. Biochem. Cell Biol. 2017, 91, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Krawczak, M.; Thomas, N.S.T.; Hundrieser, B.; Mort, M.; Wittig, M.; Hampe, J.; Cooper, D.N. Single base-pair substitutions in exon-intron junctions of human genes: Nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007, 28, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.N. Functional intronic polymorphisms: Buried treasure awaiting discovery within our genes. Hum. Genom. 2010, 4, 284. [Google Scholar] [CrossRef] [PubMed]
- Mignone, F.; Gissi, C.; Liuni, S.; Pesole, G. Untranslated regions of mRNAs. Genome Biol. 2002, 3, reviews0004.1. [Google Scholar] [CrossRef] [PubMed]
- Maodobra, M. The Role of Single Nucleotide Polymorphisms of Untranslated Regions (Utrs) in Insulin Resistance Pathogenesis in Patients with Type 2 Diabetes. In Medical Complications of Type 2 Diabetes; Croniger, C., Ed.; IntechOpen: London, UK, 2011. [Google Scholar]
- Carmody, S.R.; Wente, S.R. mRNA nuclear export at a glance. J. Cell Sci. 2009, 122, 1933–1937. [Google Scholar] [CrossRef]
- Pesole, G.; Mignone, F.; Gissi, C.; Grillo, G.; Licciulli, F.; Liuni, S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene 2001, 276, 73–81. [Google Scholar] [CrossRef]
- Attia, J.; Ioannidis, J.P.A.; Thakkinstian, A.; McEvoy, M.; Scott, R.J.; Minelli, C.; Thompson, J.; Infante-Rivard, C.; Guyatt, G. How to Use an Article About Genetic Association: A: Background Concepts. JAMA 2009, 301, 74–81. [Google Scholar] [CrossRef]
- The International HapMap Consortium. A haplotype map of the human genome. Nature 2005, 437, 1299–1320. [Google Scholar] [CrossRef]
- Zhao, H.; Pfeiffer, R.; Gail, M.H. Haplotype analysis in population genetics and association studies. Pharmacogenomics 2003, 4, 171–178. [Google Scholar] [CrossRef]
- Van den Oord, E.J.C.G.; Neale, B.M. Will haplotype maps be useful for finding genes? Mol. Psychiatry 2004, 9, 227–236. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, N.; Zhang, K.; Zhao, H. Haplotype-Association Analysis. Adv. Genet. 2008, 60, 335–405. [Google Scholar]
- Khayyatzadeh, S.S.; Mehramiz, M.; Esmaeily, H.; Mirmousavi, S.J.; Khajavi, L.; Salehkhani, F.N.; Hanachi, P.; Bahrami-Taghanaki, H.; Eslami, S.; Vatanparast, H.; et al. A variant in CYP2R1 predicts circulating vitamin D levels after supplementation with high-dose of vitamin D in healthy adolescent girls. J. Cell. Physiol. 2019, 234, 13977–13983. [Google Scholar] [CrossRef]
- Christakos, S.; Dhawan, P.; Verstuyf, A.; Verlinden, L.; Carmeliet, G. Vitamin D: Metabolism, Molecular Mechanism of Action, and Pleiotropic Effects. Physiol. Rev. 2016, 96, 365–408. [Google Scholar] [CrossRef]
- Slater, N.A.; Rager, M.L.; Havrda, D.E.; Harralson, A.F. Genetic Variation in CYP2R1 and GC Genes Associated with Vitamin D Deficiency Status. J. Pharm. Pract. Res. 2017, 30, 31–36. [Google Scholar] [CrossRef]
- Nissen, J.; Vogel, U.; Ravn-Haren, G.; Andersen, E.W.; Madsen, K.H.; Nexø, B.A.; Andersen, R.; Mejborn, H.; Bjerrum, P.J.; Rasmussen, L.B.; et al. Common variants in CYP2R1 and GC genes are both determinants of serum 25-hydroxyvitamin D concentrations after UVB irradiation and after consumption of vitamin D3–fortified bread and milk during winter in Denmark. Am. J. Clin. Nutr. 2015, 101, 218–227. [Google Scholar] [CrossRef]
- Cheung, C.-L.; Lau, K.-S.; Sham, P.-C.; Tan, K.C.; Kung, A.W. Genetic variant in vitamin D binding protein is associated with serum 25-hydroxyvitamin D and vitamin D insufficiency in southern Chinese. J. Hum. Genet. 2013, 58, 749–751. [Google Scholar] [CrossRef]
- Holick, M.F. Vitamin D Status: Measurement, Interpretation, and Clinical Application. Ann. Epidemiol. 2009, 19, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Xue, Z.; Ji, H.; Zhang, D.; Wang, Y. Effects of CYP2R1 gene variants on vitamin D levels and status: A systematic review and meta-analysis. Gene 2018, 678, 361–369. [Google Scholar] [CrossRef]
- Ramos-Lopez, E.; Kahles, H.; Weber, S.; Kukic, A.; Penna-Martinez, M.; Badenhoop, K.; Louwen, F. Gestational diabetes mellitus and vitamin D deficiency: Genetic contribution of CYP27B1 and CYP2R1 polymorphisms. Diabetes Obes. Metab. 2008, 10, 683–685. [Google Scholar]
- Haussler, M.R.; Jurutka, P.W.; Mizwicki, M.; Norman, A.W. Vitamin D receptor (VDR)-mediated actions of 1α,25(OH)2vitamin D3: Genomic and non-genomic mechanisms. Best Pract. Res. Clin. Endocrinol. Metab. 2011, 25, 543–559. [Google Scholar] [CrossRef]
- Triantos, C.; Aggeletopoulou, I.; Kalafateli, M.; Spantidea, P.I.; Vourli, G.; Diamantopoulou, G.; Tapratzi, D.; Michalaki, M.; Manolakopoulos, S.; Gogos, C.; et al. Prognostic significance of vitamin D receptor (VDR) gene polymorphisms in liver cirrhosis. Sci. Rep. 2018, 8, 14065. [Google Scholar] [CrossRef]
- Charoenngam, N.; Holick, M.F. Immunologic Effects of Vitamin D on Human Health and Disease. Nutrients 2020, 12, 2097. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, M.; Guo, Y.; Song, Z.; Liu, B. 1,25-Dihydroxyvitamin D3 Promotes High Glucose-Induced M1 Macrophage Switching to M2 via the VDR-PPARγ Signaling Pathway. Biomed. Res. Int. 2015, 2015, 157834. [Google Scholar] [PubMed]
- Powe, C.E.; Evans, M.K.; Wenger, J.; Zonderman, A.B.; Berg, A.H.; Nalls, M.; Tamez, H.; Zhang, D.; Bhan, I.; Karumanchi, S.A.; et al. Vitamin D–Binding Protein and Vitamin D Status of Black Americans and White Americans. N. Engl. J. Med. 2013, 369, 1991–2000. [Google Scholar] [CrossRef] [PubMed]
- Witke, W.F.; Gibbs, P.E.M.; Zielinski, R.; Yang, F.; Bowman, B.H.; Dugaiczyk, A. Complete Structure of the Human Gc Gene: Differences and Similarities between Members of the Albumin Gene Family. Genomics 1993, 16, 751–754. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Yu, K.; Stolzenberg-Solomon, R.; Simon, K.C.; McCullough, M.L.; Gallicchio, L.; Jacobs, E.J.; Ascherio, A.; Helzlsouer, K.; Jacobs, K.B.; et al. Genome-wide association study of circulating vitamin D levels. Hum. Mol. Genet. 2010, 19, 2739–2745. [Google Scholar] [CrossRef] [PubMed]
- Thanapirom, K.; Suksawatamnuay, S.; Sukeepaisarnjareon, W.; Tanwandee, T.; Charatcharoenwitthaya, P.; Thongsawat, S.; Leerapun, A.; Piratvisuth, T.; Boonsirichan, R.; Bunchorntavakul, C.; et al. Genetic variation in the vitamin D pathway CYP2R1 gene predicts sustained HBeAg seroconversion in chronic hepatitis B patients treated with pegylated interferon: A multicenter study. PLoS ONE 2017, 12, e0173263. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.J.; Zhang, F.; Richards, J.B.; Kestenbaum, B.; van Meurs, J.B.; Berry, D.; Kiel, D.P.; Streeten, E.A.; Ohlsson, C.; Koller, D.L.; et al. Common genetic determinants of vitamin D insufficiency: A genome-wide association study. Lancet 2010, 376, 180–188. [Google Scholar] [CrossRef]
- Laursen, J.H.; Søndergaard, H.B.; Albrechtsen, A.; Frikke-Schmidt, R.; Koch-Henriksen, N.; Sørensen, P.S.; Sellebjerg, F.; Oturai, A. Genetic and environmental determinants of 25-hydroxyvitamin D levels in multiple sclerosis. Mult. Scler. J. 2015, 21, 1414–1422. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhao, Y.; Pan, J.; Fang, K.; Wang, Y.; Li, Z.; Chang, X. Vitamin D-binding protein (group-specific component) has decreased expression in rheumatoid arthritis. Clin. Exp. Rheumatol. 2012, 30, 525–533. [Google Scholar] [PubMed]
- Yoshida, S.; Ikari, K.; Furuya, T.; Toyama, Y.; Taniguchi, A.; Yamanaka, H.; Momohara, S. A GC polymorphism associated with serum 25-hydroxyvitamin D level is a risk factor for hip fracture in Japanese patients with rheumatoid arthritis: 10-year follow-up of the Institute of Rheumatology, Rheumatoid Arthritis cohort study. Arthritis Res. Ther. 2014, 16, R75. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.-C.; Lee, Y.H. Vitamin D level and risk of systemic lupus erythematosus and rheumatoid arthritis: A Mendelian randomization. Clin. Rheumatol. 2018, 37, 2415–2421. [Google Scholar] [CrossRef]
- Zhu, J.; DeLuca, H.F. Vitamin D 25-hydroxylase—Four decades of searching, are we there yet? Arch. Biochem. Biophys. 2012, 523, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.-M.; Shin, E.-A. Exploring vitamin D metabolism and function in cancer. Exp. Mol. Med. 2018, 50, 20. [Google Scholar] [CrossRef] [PubMed]
- Falleti, E.; Cmet, S.; Fabris, C.; Fattovich, G.; Cussigh, A.; Bitetto, D.; Ceriani, E.; Lenisa, I.; Dissegna, D.; Ieluzzi, D.; et al. Genetic Polymorphisms of Vitamin D Pathway Predict Antiviral Treatment Outcome in Slow Responder Naïve Patients with Chronic Hepatitis C. PLoS ONE 2013, 8, e80764. [Google Scholar] [CrossRef]
- Hewison, M.; Zehnder, D.; Bland, R.; Stewart, P. 1alpha-Hydroxylase and the action of vitamin D. J. Mol. Endocrinol. 2000, 25, 141–148. [Google Scholar] [CrossRef]
- Lange, C.M.; Bojunga, J.; Ramos-Lopez, E.; Von Wagner, M.; Hassler, A.; Vermehren, J.; Herrmann, E.; Badenhoop, K.; Zeuzem, S.; Sarrazin, C. Vitamin D deficiency and a CYP27B1-1260 promoter polymorphism are associated with chronic hepatitis C and poor response to interferon-alfa based therapy. J. Hepatol. 2011, 54, 887–893. [Google Scholar] [CrossRef]
- Sundqvist, E.; Bäärnhielm, M.; Alfredsson, L.; Hillert, J.; Olsson, T.; Kockum, I. Confirmation of association between multiple sclerosis and CYP27B1. Eur. J. Hum. Genet. 2010, 18, 1349–1352. [Google Scholar] [CrossRef]
- Hyppönen, E.; Berry, D.J.; Wjst, M.; Power, C. Serum 25-hydroxyvitamin D and IgE—A significant but nonlinear relationship. Allergy 2009, 64, 613–620. [Google Scholar] [CrossRef]
- Gado, K.H.; Gado, T.H.; Samie, R.M.A.; Khalil, N.M.; Emam, S.L.; Fouad, H.H. Clinical significance of vitamin D deficiency and receptor gene polymorphism in systemic lupus erythematosus patients. Egypt. Rheumatol. 2017, 39, 159–164. [Google Scholar] [CrossRef]
- Zenata, O.; Vrzal, R. Fine tuning of vitamin D receptor (VDR) activity by post-transcriptional and post-translational modifications. Oncotarget 2017, 8, 35390–35402. [Google Scholar] [CrossRef] [PubMed]
- Sunn, K.L.; Cock, T.-A.; Crofts, L.A.; Eisman, J.A.; Gardiner, E.M. Novel N-Terminal Variant of Human VDR. J. Mol. Endocrinol. 2001, 15, 1599–1609. [Google Scholar] [CrossRef]
- Hasan, H.A.; AbuOdeh, R.O.; Muda, W.A.M.B.W.; Mohamed, H.J.B.J.; Samsudin, A.R. Association of Vitamin D receptor gene polymorphisms with metabolic syndrome and its components among adult Arabs from the United Arab Emirates. Diabetes Metab. Syndr. 2017, 11, S531–S537. [Google Scholar] [CrossRef]
- Arai, H.; Miyamoto, K.-I.; Taketani, Y.; Yamamoto, H.; Iemori, Y.; Morita, K.; Tonai, T.; Nishisho, T.; Mori, S.; Takeda, E. A Vitamin D Receptor Gene Polymorphism in the Translation Initiation Codon: Effect on Protein Activity and Relation to Bone Mineral Density in Japanese Women. J. Bone Miner. Res. 1997, 12, 915–921. [Google Scholar] [CrossRef]
- Al-Nahas, Z. 25-hydroxyvitamin D3 deficiency and vitamin D receptor polymorphisms in Egyptian patients with Behçet’s disease: A pilot study. Clin. Rheumatol. 2017, 12, 20–27. [Google Scholar]
- Uitterlinden, A.G.; Fang, Y.; van Meurs, J.B.J.; Pols, H.A.P.; van Leeuwen, J.P.T.M. Genetics and biology of vitamin D receptor polymorphisms. Gene 2004, 338, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Gross, C.; Krishnan, A.V.; Malloy, P.J.; Eccleshall, T.R.; Zhao, X.-Y.; Feldman, D. The Vitamin D Receptor Gene Start Codon Polymorphism: A Functional Analysis of FokI Variants. J. Bone Miner. Res. 1998, 13, 1691–1699. [Google Scholar] [CrossRef] [PubMed]
- Mahto, H.; Tripathy, R.; Das, B.K.; Panda, A.K. Association between vitamin D receptor polymorphisms and systemic lupus erythematosus in an Indian cohort. Int. J. Rheum. Dis. 2018, 21, 468–476. [Google Scholar] [CrossRef]
- Yiallourou, A.I.; Ekonomou, E.; Tsamadias, V.; Nastos, K.; Karapanos, K.; Papaconstantinou, I.; Theodosopoulos, T.; Contis, J.; Papalambros, E.; Voros, D.; et al. Association of FokI and PvuII polymorphisms with breast cancer staging and survival among Caucasian women: A prospective study. J. BU ON Off. J. Balk. Union Oncol. 2014, 19, 633–642. [Google Scholar]
- Li, L.; Wu, B.; Liu, J.-Y.; Yang, L.-B. Vitamin D Receptor Gene Polymorphisms and Type 2 Diabetes: A Meta-analysis. Arch. Med. Res. 2013, 44, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Goswami, R. Primer on the metabolic bone diseases and disorders of mineral metabolism. Indian J. Med. Res. 2016, 144, 489–490. [Google Scholar] [CrossRef]
- Garavito, G.; Egea, E.; Fang, L.; Malagón, C.; Olmos, C.; González, L.; Guarnizo, P.; Aroca, G.; López, G.; Iglesias, A. Association of polymorphic variants of PTPN22, TNF and VDR systems in children with lupus nephritis: A study in trios of Colombian families. Biomedica 2017, 37, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Salimi, S.; Eskandari, F.; Rezaei, M.; Sandoughi, M. Vitamin D Receptor rs2228570 and rs731236 Polymorphisms are Susceptible Factors for Systemic Lupus Erythematosus. Adv. Biomed. Res. 2019, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- López-Mejías, R.; Genre, F.; Remuzgo-Martínez, S.; Robledo, G.; Llorca, J.; Corrales, A.; González-Juanatey, C.; Ubilla, B.; Mijares, V.; Pina, T.; et al. Vitamin D receptor GATG haplotype association with atherosclerotic disease in patients with rheumatoid arthritis. Atherosclerosis 2016, 245, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Ghaly, M.S.; Badra, D.I.; Dessouki, O.; Elmaraghy, N.N.; Hassan, R. Vitamin D receptor Fok1 & Bsm 1 Gene Polymorphisms in Systemic Lupus Erythematosus and Osteoarthritis: Autoimmune Inflammatory versus Degenerative Model. Egypt. J. Immunol. 2017, 24, 151–164. [Google Scholar]
- Babenko, S.A.; Alifirova, V.M.; Orlova, I.; Puzyrev, V.P. The VDR gene polymorphism in patients with multiple sclerosis. Zhurnal Nevrol. Psikhiatrii Imeni SS Korsakova 2009, 109 (Suppl. 2), 23–27. [Google Scholar]
- Dendrou, C.A.; Fugger, L.; Friese, M.A. Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 2015, 15, 545–558. [Google Scholar] [CrossRef]
- Cox, M.B.; Ban, M.; Bowden, N.A.; Baker, A.; Scott, R.J.; Lechner-Scott, J. Potential association of vitamin D receptor polymorphism Taq1 with multiple sclerosis. Mult. Scler. 2012, 18, 16–22. [Google Scholar] [CrossRef]
- García-Martín, E.; Agúndez, J.A.G.; Martinez, C.; Benito-León, J.; Millán-Pascual, J.; Calleja, P.; Díaz-Sánchez, M.; Pisa, D.; Turpín-Fenoll, L.; Alonso-Navarro, H.; et al. Vitamin D3 Receptor (VDR) Gene rs2228570 (Fok1) and rs731236 (Taq1) Variants Are Not Associated with the Risk for Multiple Sclerosis: Results of a New Study and a Meta-Analysis. PLoS ONE 2013, 8, e65487. [Google Scholar] [CrossRef]
- Orton, S.-M.; Ramagopalan, S.V.; Para, A.E.; Lincoln, M.R.; Handunnetthi, L.; Chao, M.J.; Morahan, J.; Morrison, K.M.; Sadovnick, A.D.; Ebers, G.C. Vitamin D metabolic pathway genes and risk of multiple sclerosis in Canadians. J. Neurol. Sci. 2011, 305, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Simon, K.C.; Munger, K.L.; Yang, X.; Ascherio, A. Polymorphisms in vitamin D metabolism related genes and risk of multiple sclerosis. Mult. Scler. 2010, 16, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, A.; Shojapour, M.; Ghassami, K.; Mosayebi, G. The BsmI, FokI, ApaI and TaqI Polymorphisms in Vitamin D Receptor Gene in Iranian Multiple Sclerosis Patients: A Case-Control Study. J. Iran. Clin. Res. 2015, 3, 28–32. [Google Scholar]
- Křenek, P.; Benešová, Y.; Bienertová-Vašků, J.; Vašků, A. The Impact of Five VDR Polymorphisms on Multiple Sclerosis Risk and Progression: A Case-Control and Genotype-Phenotype Study. J. Mol. Neurosci. 2018, 64, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Narooie-Nejad, M.; Moossavi, M.; Torkamanzehi, A.; Moghtaderi, A.; Salimi, S. Vitamin D Receptor Gene Polymorphism and the Risk of Multiple Sclerosis in South Eastern of Iran. J. Mol. Neurosci. 2015, 56, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Imani, D.; Razi, B.; Motallebnezhad, M.; Rezaei, R. Association between vitamin D receptor (VDR) polymorphisms and the risk of multiple sclerosis (MS): An updated meta-analysis. BMC Neurol. 2019, 19, 339. [Google Scholar] [CrossRef]
- Mohammadi, A.; Azarnezhad, A.; Khanbabaei, H.; Izadpanah, E.; Abdollahzadeh, R.; Barreto, G.E.; Sahebkar, A. Vitamin D receptor genetic polymorphisms and the risk of multiple sclerosis: A systematic review and meta-analysis. Steroids 2020, 158, 108615. [Google Scholar] [CrossRef]
- Huang, J.; Xie, Z.-F. Polymorphisms in the vitamin D receptor gene and multiple sclerosis risk: A meta-analysis of case–control studies. J. Neurol. Sci. 2012, 313, 79–85. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Zhang, L.; Chen, S.-Y.; Yang, G.-J.; Huang, X.-L.; Duan, Y.; Yang, L.-J.; Ye, D.-Q.; Wang, J. Association between VDR polymorphisms and multiple sclerosis: Systematic review and updated meta-analysis of case-control studies. Neurol. Sci. 2018, 39, 225–234. [Google Scholar] [CrossRef]
- Smolders, J.; Damoiseaux, J.; Menheere, P.; Tervaert, J.W.C.; Hupperts, R. Fok-I vitamin D receptor gene polymorphism (rs10735810) and vitamin D metabolism in multiple sclerosis. J. Neuroimmunol. 2009, 207, 117–121. [Google Scholar] [CrossRef]
- Tizaoui, K.; Kaabachi, W.; Hamzaoui, A.; Hamzaoui, K. Association between vitamin D receptor polymorphisms and multiple sclerosis: Systematic review and meta-analysis of case–control studies. Cell. Mol. Immunol. 2014, 12, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Čierny, D.; Michalik, J.; Kurča, E.; Dobrota, D.; Lehotský, J. FokI vitamin D receptor gene polymorphism in association with multiple sclerosis risk and disability progression in Slovaks. Neurol. Res. 2015, 37, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt, A.; Boleixa, D.; Guimarães, A.L.; Leal, B.; Carvalho, C.; Brás, S.; Samões, R.; Santos, E.; Costa, P.P.; Silva, B.; et al. The vitamin D receptor gene FokI polymorphism and Multiple Sclerosis in a Northern Portuguese population. J. Neuroimmunol. 2017, 309, 34–37. [Google Scholar] [CrossRef]
- Chen, X.-L.; Zhang, M.-L.; Zhu, L.; Peng, M.-L.; Liu, F.-Z.; Zhang, G.-X.; Wang, L.-M.; Zhao, J. Vitamin D receptor gene polymorphisms and the risk of multiple sclerosis: An updated meta-analysis. Microb. Pathog. 2017, 110, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Čierny, D.; Michalik, J.; Škereňová, M.; Kantorová, E.; Sivák, Š.; Javor, J.; Kurča, E.; Dobrota, D.; Lehotský, J. ApaI, BsmI and TaqI VDR gene polymorphisms in association with multiple sclerosis in Slovaks. Neurol. Res. 2016, 38, 678–684. [Google Scholar] [CrossRef]
- Kamisli, O.; Acar, C.; Sozen, M.; Tecellioğlu, M.; Yücel, F.E.; Vaizoglu, D.; Özcan, C. The association between vitamin D receptor polymorphisms and multiple sclerosis in a Turkish population. Mult. Scler. Relat. Disord. 2018, 20, 78–81. [Google Scholar] [CrossRef]
- Niino, M.; Fukazawa, T.; Yabe, I.; Kikuchi, S.; Sasaki, H.; Tashiro, K. Vitamin D receptor gene polymorphism in multiple sclerosis and the association with HLA class II alleles. J. Neurol. Sci. 2000, 177, 65–71. [Google Scholar] [CrossRef]
- Irizar, H.; Muñoz-Culla, M.; Zuriarrain, O.; Goyenechea, E.; Castillo-Triviño, T.; Prada, A.; Saenz-Cuesta, M.; De Juan, D.; De Munain, A.L.; Olascoaga, J.; et al. HLA-DRB1*15:01 and multiple sclerosis: A female association? Mult. Scler. 2011, 18, 569–577. [Google Scholar] [CrossRef]
- Smolen, J.S.; Aletaha, D.; Barton, A.; Burmester, G.R.; Emery, P.; Firestein, G.S.; Kavanaugh, A.; McInnes, I.B.; Solomon, D.H.; Strand, V.; et al. Rheumatoid arthritis. Nat. Rev. Dis. Primers 2018, 4, 18001. [Google Scholar] [CrossRef]
- Rodríguez-Carrio, J.; Alperi-López, M.; Naves-Díaz, M.; Dusso, A.; López, P.; Ballina-García, F.J.; Cannata-Andía, J.B.; Suárez, A. Vitamin D Receptor Polymorphism and DHCR7 Contribute to the Abnormal Interplay Between Vitamin D and Lipid Profile in Rheumatoid Arthritis. Sci. Rep. 2019, 9, 2546. [Google Scholar] [CrossRef]
- Mosaad, Y.M.; Hammad, E.M.; Fawzy, Z.; Abdal Aal, I.A.; Youssef, H.M.; ElSaid, T.O.; Monir, R.; El-Deek, B.S. Vitamin D receptor gene polymorphism as possible risk factor in rheumatoid arthritis and rheumatoid related osteoporosis. Hum. Immunol. 2014, 75, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.N.; Mabrouk, M.S.; Eldeib, A.M.; Shaker, O.G. Genetic Case-Control Study for Eight Polymorphisms Associated with Rheumatoid Arthritis. PLoS ONE 2015, 10, e0131960. [Google Scholar] [CrossRef] [PubMed]
- Hitchon, C.A.; Sun, Y.; Robinson, D.B.; Peschken, C.A.; Bernstein, C.N.; Siminovitch, K.A.; El-Gabalawy, H. Vitamin D Receptor Polymorphism rs2228570 (Fok1) Is Associated with Rheumatoid Arthritis in North American Natives. J. Rheumatol. 2012, 39, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Karray, E.F.; Ben Dhifallah, I.; Ben Abdelghani, K.; Ben Ghorbel, I.; Khanfir, M.; Houman, H.; Hamzaoui, K.; Zakraoui, L. Associations of vitamin D receptor gene polymorphisms FokI and BsmI with susceptibility to rheumatoid arthritis and Behçet’s disease in Tunisians. Jt. Bone Spine 2012, 79, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Maalej, A.; Petit-Teixeira, E.; Michou, L.; Rebai, A.; Cornelis, F.; Ayadi, H. Association study of VDR gene with rheumatoid arthritis in the French population. Genes Immun. 2005, 6, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Song, G.G.; Bae, S.-C.; Lee, Y.H. Vitamin D receptor FokI, BsmI, and TaqI polymorphisms and susceptibility to rheumatoid arthritis: A meta-analysis. Z. Rheumatol. 2016, 75, 322–329. [Google Scholar] [CrossRef]
- Tizaoui, K.; Hamzaoui, K. Association between VDR polymorphisms and rheumatoid arthritis disease: Systematic review and updated meta-analysis of case–control studies. Immunobiology 2015, 220, 807–816. [Google Scholar] [CrossRef]
- Lee, Y.H.; Bae, S.-C.; Choi, S.J.; Ji, J.D.; Song, G.G. Associations between vitamin D receptor polymorphisms and susceptibility to rheumatoid arthritis and systemic lupus erythematosus: A meta-analysis. Mol. Biol. Rep. 2011, 38, 3643–3651. [Google Scholar] [CrossRef]
- Kaviani, N.; Yazdani, Y.; Hadi, B. The Association of Vitamin D Receptor Polymorphisms of FokI and TaqI with Rheumatoid Arthritis in North-East of Iran. Jorjani Biomed. J. 2019, 7, 20–29. [Google Scholar] [CrossRef]
- Bernardes, M.; Machado, J.C.; Gonçalves, D.; Abelha-Aleixo, J.; Fonseca, R.; Madureira, P.; Vieira, R.; Bernardo, A.; Martins, M.; Costa, L.; et al. AB0272 Bsmi and Foki VDR Gene Polymorphisms Influence Disease Activity in Established Rheumatoid Arthritis Patients. Ann. Rheum. Dis. 2014, 73 (Suppl. 2), 894. [Google Scholar] [CrossRef]
- Saad, M.N.; Mabrouk, M.S.; Eldeib, A.M.; Shaker, O.G. Vitamin D receptor gene polymorphisms in rheumatoid arthritis patients associating osteoporosis. In Proceedings of the 2014 Cairo International Biomedical Engineering Conference (CIBEC), Giza, Egypt, 11–13 December 2014; pp. 75–78. [Google Scholar]
- Bagheri-Hosseinabadi, Z.; Imani, D.; Yousefi, H.; Abbasifard, M. Vitamin D receptor (VDR) gene polymorphism and risk of rheumatoid arthritis (RA): Systematic review and meta-analysis. Clin. Rheumatol. 2020, 39, 3555–3569. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Lozano, J.R.; Gonzalez-Escribano, M.F.; Valenzuela, A.; Garcia, A.; Núñez-Roldán, A. Association BlackwellScience, Ltd of vitamin D receptor genotypes with early onset rheumatoid arthritis. Eur. J. Immunogenet. 2001, 28, 89–93. [Google Scholar]
- Tizaoui, K.; Kaabachi, W.; Ouled Salah, M.; Ben Amor, A.; Hamzaoui, A.; Hamzaoui, K. Vitamin D receptor TaqI and ApaI polymorphisms: A comparative study in patients with Behçet’s disease and Rheumatoid arthritis in Tunisian population. Cell. Immunol. 2014, 290, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Di Spigna, G.; Del Puente, A.; Covelli, B.; Abete, E.; Varriale, E.; Salzano, S.; Postiglione, L. Vitamin D receptor polymorphisms as tool for early screening of severe bone loss in women patients with rheumatoid arthritis. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4664–4669. [Google Scholar] [PubMed]
- Al Haj-Mahmoud, S.A.; Fayiz-Atoum, M.; Al-Houran, H.M.; Bateineh, S.; Abderrahman, S.; Alzoughool, F. Vitamin d deficiency and rs731236 (Taq1) vitamin D receptor gene polymorphism as possible risk factors for rheumatoid arthritis and osteoarthritis. Acta Med. Mediterr. 2018, 34, 209–213. [Google Scholar]
- Ghodke-Puranik, Y.; Niewold, T.B. Immunogenetics of systemic lupus erythematosus: A comprehensive review. J. Autoimmun. 2015, 64, 125–136. [Google Scholar] [CrossRef]
- Mostowska, A.; Lianeri, M.; Wudarski, M.; Olesińska, M.; Jagodziński, P.P. Vitamin D receptor gene BsmI, FokI, ApaI and TaqI polymorphisms and the risk of systemic lupus erythematosus. Mol. Biol. Rep. 2013, 40, 803–810. [Google Scholar] [CrossRef]
- Huang, C.-M.; Wu, M.-C.; Wu, J.-Y.; Tsai, F.-J. No Association of Vitamin D Receptor Gene Start Codon Fok I Polymorphisms in Chinese Patients with Systemic Lupus Erythematosus. J. Rheumatol. 2002, 29, 1211–1213. [Google Scholar]
- Imam, A.A.; Ibrahim, H.E.; Farghaly, M.A.A.; Alkholy, U.M.; Gawish, H.H.; Abdalmonem, N.; Sherif, A.M.; Ali, Y.F.; Hamed, M.E.; Waked, N.M.; et al. Vitamin D receptor gene FokI polymorphism in Egyptian children and adolescents with SLE: A case-control study. Lupus 2017, 26, 1426–1434. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Huang, S. Association between vitamin D receptor gene BsmI, FokI, ApaI and TaqI polymorphisms and the risk of systemic lupus erythematosus: A meta-analysis. Rheumatol. Int. 2014, 34, 381–388. [Google Scholar] [CrossRef]
- Xiong, J.; He, Z.; Zeng, X.; Zhang, Y.; Hu, Z. Association of vitamin D receptor gene polymorphisms with systemic lupus erythematosus: A meta-analysis. Clin. Exp. Rheumatol. 2014, 32, 174–181. [Google Scholar]
- Bae, S.-C.; Lee, Y.H. Vitamin D receptor FokI, TaqI, and ApaI polymorphisms and susceptibility to systemic lupus erythematosus: An updated meta-analysis. Clin. Rheumatol. 2018, 37, 1529–1537. [Google Scholar] [CrossRef] [PubMed]
- Monticielo, O.A.; Brenol, J.C.T.; Chies, J.A.B.; Longo, M.G.F.; Rucatti, G.G.; Scalco, R.; Xavier, R.M. The role of Bsm I and Fok I vitamin D receptor gene polymorphisms and serum 25-hydroxyvitamin D in Brazilian patients with systemic lupus erythematosus. Lupus 2012, 21, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.; Marinho, A.; Leal, B.; Bettencourt, A.; Boleixa, D.; Almeida, I.F.; Farinha, F.; E Costa, P.P.; Vasconcelos, C.; Martins-Silva, B. Association between vitamin D receptor ( VDR ) gene polymorphisms and systemic lupus erythematosus in Portuguese patients. Lupus 2015, 24, 846–853. [Google Scholar] [CrossRef]
- Kaleta, B.; Bogaczewicz, J.; Robak, E.; Sysa-Jędrzejowska, A.; Wrzosek, M.; Szubierajska, W.; Mróz, P.; Łukaszkiewicz, J.; Woźniacka, A. Vitamin D Receptor Gene BsmI Polymorphism in Polish Patients with Systemic Lupus Erythematosus. ISRN Endocrinol. 2013, 2013, 427818. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.-B.; Jiang, Z.-P.; Lin, Z.-J.; Su, N. Association of vitamin D receptor gene polymorphism with the risk of systemic lupus erythematosus. J. Recept. 2015, 35, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Dzhebir, G.; Kamenarska, Z.; Hristova, M.; Savov, A.; Vinkov, A.; Kaneva, R.; Mitev, V.; Dourmishev, L.A. Association of vitamin D receptor gene Bsm I B/b and Fok I F/f polymorphisms with adult dermatomyositis and systemic lupus erythematosus. Int. J. Dermatol. 2016, 55, e465–e468. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Ballesteros, A.I.; Meza-Meza, M.R.; Vizmanos-Lamotte, B.; Parra-Rojas, I.; de la Cruz-Mosso, U. Association of Vitamin D Metabolism Gene Polymorphisms with Autoimmunity: Evidence in Population Genetic Studies. Int. J. Mol. Sci. 2020, 21, 9626. https://doi.org/10.3390/ijms21249626
Ruiz-Ballesteros AI, Meza-Meza MR, Vizmanos-Lamotte B, Parra-Rojas I, de la Cruz-Mosso U. Association of Vitamin D Metabolism Gene Polymorphisms with Autoimmunity: Evidence in Population Genetic Studies. International Journal of Molecular Sciences. 2020; 21(24):9626. https://doi.org/10.3390/ijms21249626
Chicago/Turabian StyleRuiz-Ballesteros, Adolfo I., Mónica R. Meza-Meza, Barbara Vizmanos-Lamotte, Isela Parra-Rojas, and Ulises de la Cruz-Mosso. 2020. "Association of Vitamin D Metabolism Gene Polymorphisms with Autoimmunity: Evidence in Population Genetic Studies" International Journal of Molecular Sciences 21, no. 24: 9626. https://doi.org/10.3390/ijms21249626
APA StyleRuiz-Ballesteros, A. I., Meza-Meza, M. R., Vizmanos-Lamotte, B., Parra-Rojas, I., & de la Cruz-Mosso, U. (2020). Association of Vitamin D Metabolism Gene Polymorphisms with Autoimmunity: Evidence in Population Genetic Studies. International Journal of Molecular Sciences, 21(24), 9626. https://doi.org/10.3390/ijms21249626







