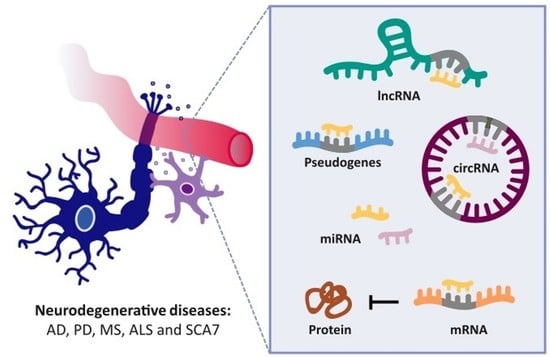
Competing Endogenous RNA Networks as Biomarkers in Neurodegenerative Diseases
Abstract
1. Introduction
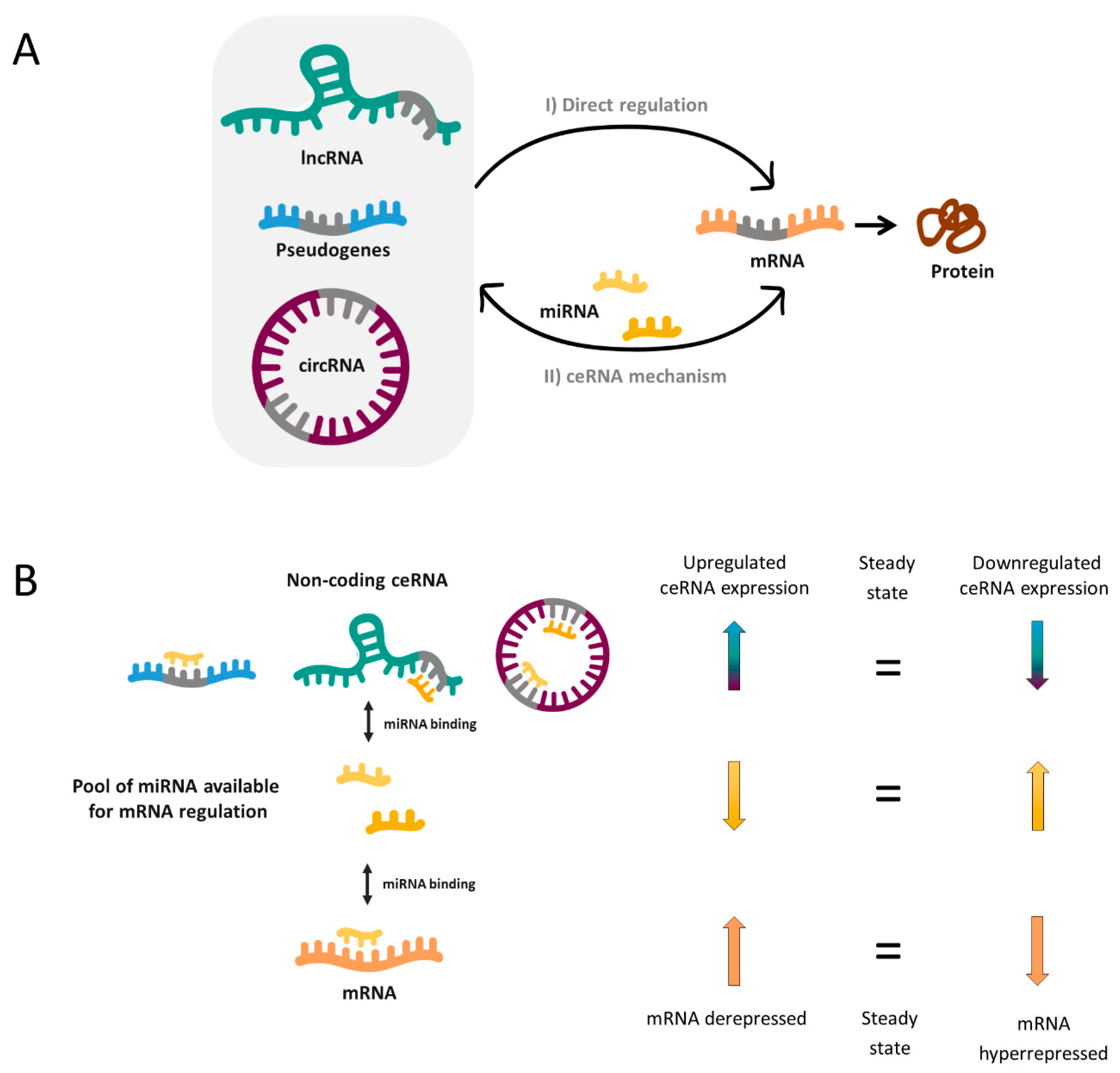
2. ceRNA Networks and Neurodegenerative Diseases
2.1. ceRNA and Alzheimer’s Disease
2.1.1. LncRNAs
2.1.2. CircRNAs
2.2. ceRNA and Parkinson’s Disease
2.2.1. Pseudogenes and lncRNAs
2.2.2. CircRNAs
2.3. ceRNA and Multiple Sclerosis
2.4. ceRNA and Amyotrophic Lateral Sclerosis
2.5. ceRNA and Spinocerebellar Ataxia Type 7
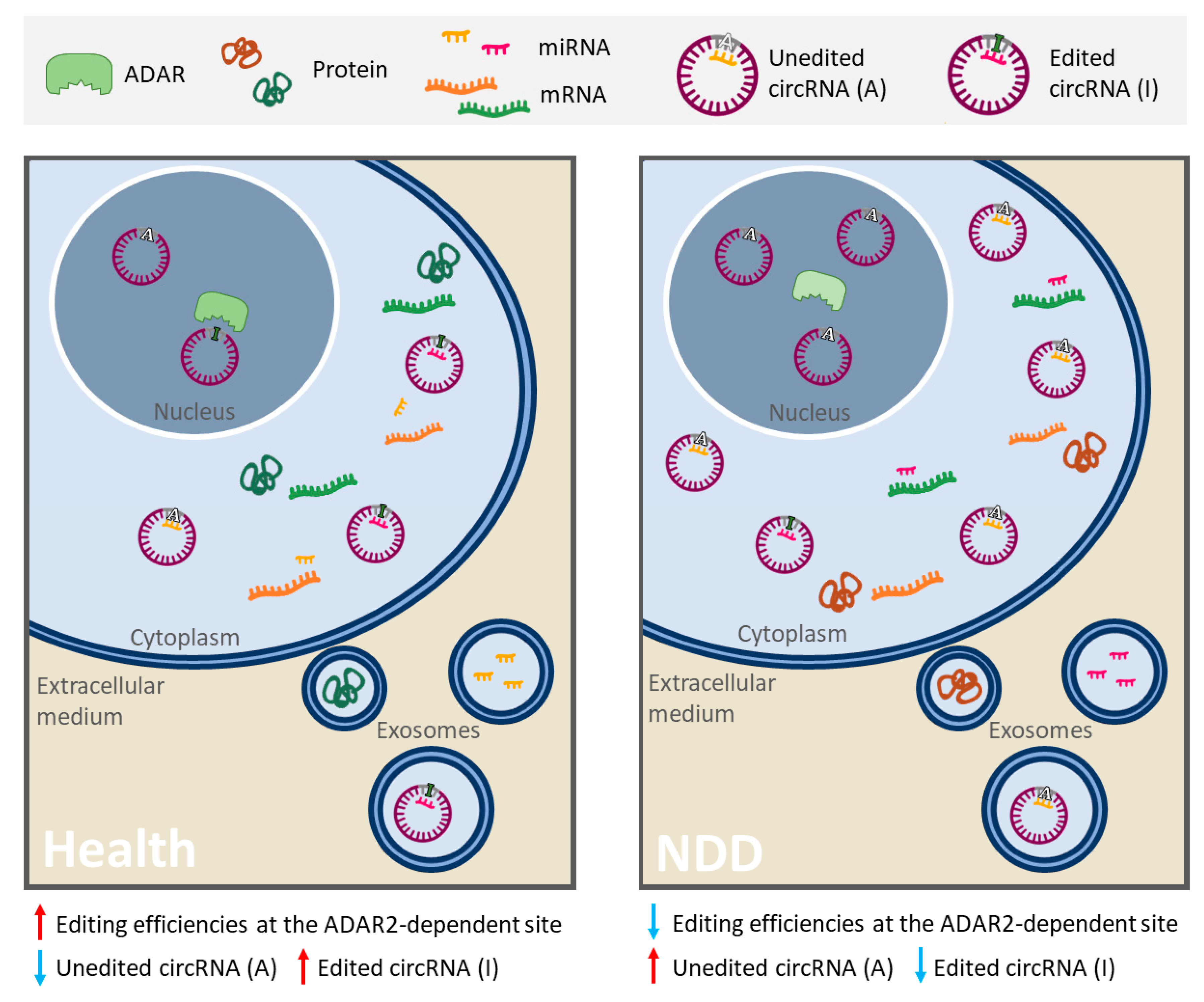
3. RNA Editing Alteration and ceRNA Networks in Neurodegenerative Diseases
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ADAR1 | Adenosine Deaminase Acting on RNA 1 |
| circRNAs | Circular Ribonucleic Acids |
| lncRNAs | Long non-coding Ribonucleic Acids |
| ceRNAs | Competing Endogenous Ribonucleic Acids |
| ceRNET | ceRNA network |
| cirCeNET | circRNA-associated ceRNA networks |
| LncACeNET | lncRNA-associated ceRNA network |
| AD | Alzheimer’s Disease |
| PD | Parkinson’s Disease |
| MS | Multiple Sclerosis |
| SCA7 | Spinocerebellar Ataxia Type 7 |
| ALS | Amyotrophic Lateral Sclerosis |
| CNS | Central Nervous System |
| MALAT1 | Metastasis-associated lung adenocarcinoma transcript 1 |
| APP/PS1 | Amyloid Precursor Protein/Presenilin 1 |
| MPTP | 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-hydrochloride |
| MPP+ | 1-methyl-4-phenylpyridinium |
| EAE | Experimental Autoimmune Encephalomyelitis |
References
- Neueder, A. RNA-Mediated Disease Mechanisms in Neurodegenerative Disorders. J. Mol. Biol. 2019, 431, 1780–1791. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.Y.; Cali, C.P.; Lee, E.B. RNA metabolism in neurodegenerative disease. Dis. Model. Mech. 2017, 10, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Volkening, K.; Strong, M.J. RNA Metabolism in Neurodegenerative Disease. Curr. Chem. Biol. 2011, 5, 90–98. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Wei, C.W.; Luo, T.; Zou, S.S.; Wu, A.S. The Role of Long Noncoding RNAs in Central Nervous System and Neurodegenerative Diseases. Front. Behav. Neurosci. 2018, 12, 175. [Google Scholar] [CrossRef]
- Salta, E.; De Strooper, B. Noncoding RNAs in neurodegeneration. Nat. Rev. Neurosci. 2017, 18, 627–640. [Google Scholar] [CrossRef]
- Amin, N.; Mcgrath, A.; Chen, Y.P.P. Evaluation of deep learning in non-coding RNA classification. Nat. Mach. Intell. 2019, 1, 246–256. [Google Scholar] [CrossRef]
- Quinlan, S.; Kenny, A.; Medina, M.; Engel, T.; Jimenez-Mateos, E.M. MicroRNAs in Neurodegenerative Diseases. Int. Rev. Cell Mol. Biol. 2017, 334, 309–343. [Google Scholar] [CrossRef]
- Cao, X.; Yeo, G.; Muotri, A.R.; Kuwabara, T.; Gage, F.H. Noncoding RNAs in the Mammalian Central Nervous System. Annu. Rev. Neurosci. 2006, 29, 77–103. [Google Scholar] [CrossRef]
- Chan, J.J.; Tay, Y. Noncoding RNA: RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell. 2015, 58, 870–885. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Lin, M.; Rockowitz, S.; Lachman, H.M.; Zheng, D. Characterization of Human Pseudogene-Derived Non-Coding RNAs for Functional Potential. PLoS ONE 2014, 9, e93972. [Google Scholar] [CrossRef] [PubMed]
- Pei, B.; Sisu, C.; Frankish, A.; Howald, C.; Habegger, L.; Mu, X.J.; Harte, R.; Balasubramanian, S.; Tanzer, A.; Diekhans, M.; et al. The GENCODE pseudogene resource. Genome Biol. 2012, 13, R51. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Ala, U.; Karreth, F.A.; Bosia, C.; Pagnani, A.; Taulli, R.; Léopold, V.; Tay, Y.; Provero, P.; Zecchina, R.; Pandolfi, P.P. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc. Natl. Acad. Sci. USA 2013, 110, 7154–7159. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Kumar, S.; Vijayan, M.; Bhatti, J.S.; Reddy, P.H. MicroRNAs as Peripheral Biomarkers in Aging and Age-Related Diseases. Prog. Mol. Biol. Transl. Sci. 2017, 146, 47–94. [Google Scholar] [CrossRef]
- Idda, M.L.; Munk, R.; Abdelmohsen, K.; Gorospe, M. Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip Rev. RNA 2018, 9. [Google Scholar] [CrossRef]
- Abdollahzadeh, R.; Daraei, A.; Mansoori, Y.; Sepahvand, M.; Amoli, M.M.; Tavakkoly-Bazzaz, J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J. Cell Physiol. 2019, 234, 10080–10100. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, J.; Ma, X.; Wang, Y. ceRNA network construction and comparison of gastric cancer with or without Helicobacter pylori infection. J. Cell Physiol. 2019, 234, 7128–7140. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, Y.; Lu, J.; Pan, T.; Ding, N.; Wang, Z.; Shao, T.; Zhang, J.; Wang, L.; Li, X. The mRNA related ceRNA–ceRNA landscape and significance across 20 major cancer types. Nucleic. Acids Res. 2015, 43, 8169–8182. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Zhang, D.-H.; Wu, N.; Xiao, J.-H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.; Ni, H.; Yu, Y.; Zhang, M.; Wu, M.; Wang, Q.; Wang, L.; Xu, S.; Xu, Z.; Xu, C.; et al. BACE1-AS prevents BACE1 mRNA degradation through the sequestration of BACE1-targeting miRNAs. J. Chem. Neuroanat. 2019, 98, 87–96. [Google Scholar] [CrossRef]
- He, W.; Chi, S.; Jin, X.; Lu, J.; Zheng, W.; Yan, J.; Zhang, D. Long non-coding RNA BACE1-AS modulates isoflurane-induced neurotoxicity to Alzheimer’s Disease through sponging miR-214-3p. Neurochem. Res. 2020, 45, 2324–2335. [Google Scholar] [CrossRef]
- Ge, Y.; Song, X.; Liu, J.; Liu, C.; Xu, C. The combined therapy of berberine treatment with lncRNA BACE1-AS depletion attenuates Aβ 25–35 induced neuronal injury through regulating the expression of miR -132-3p in neuronal cells. Neurochem. Res. 2020, 45, 741–751. [Google Scholar] [CrossRef]
- Yue, D.; Guanqun, G.; Jingxin, L.; Sen, S.; Shuang, L.; Yan, S.; Minxue, Z.; Ping, Y.; Chong, L.; Zhuobo, Z.; et al. Silencing of long noncoding RNA XIST attenuated Alzheimer’s disease-related BACE1 alteration through miR-124. Cell Biol. Int. 2020, 44, 630–636. [Google Scholar] [CrossRef]
- Wang, X.; Wang, C.; Geng, C.; Zhao, K. LncRNA XIST knockdown attenuates Aβ 25-35 -induced toxicity, oxidative stress, and apoptosis in primary cultured rat hippocampal neurons by targeting miR-132. Int. J. Clin. Exp. Pathol. 2018, 11, 3915–3924. [Google Scholar]
- Zhao, M.Y.; Wang, G.Q.; Wang, N.N.; Yu, Q.Y.; Liu, R.L.; Shi, W.Q. The long-non-coding RNA NEAT1 is a novel target for Alzheimer’s disease progression via miR-124/BACE1 axis. Neurol. Res. 2019, 41, 489–497. [Google Scholar] [CrossRef]
- Ke, S.; Yang, Z.; Yang, F.; Wang, X.; Tan, J.; Liao, B. Long noncoding RNA NEAT1 aggravates Aβ-induced neuronal damage by targeting miR-107 in Alzheimer’s disease. Yonsei Med. J. 2019, 60, 640–650. [Google Scholar] [CrossRef]
- Xu, W.; Li, K.; Fan, Q.; Zong, B.; Han, L. Knockdown of long non-coding RNA SOX21-AS1 attenuates amyloid-β-induced neuronal damage by sponging miR-107. Biosci. Rep. 2020, 40, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Spreafico, M.; Grillo, B.; Rusconi, F.; Battaglioli, E.; Venturin, M. Multiple Layers of CDK5R1 Regulation in Alzheimer’s Disease Implicate Long Non-Coding RNAs. Int. J. Mol. Sci. 2018, 19, 2022. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Li, Y.; Zhang, W.; Fang, F.; Sun, J.; Liu, M.; Li, K.; Dong, L. Long Non-coding RNA MALAT1 Inhibits Neuron Apoptosis and Neuroinflammation While Stimulates Neurite Outgrowth and Its Correlation With MiR-125b Mediates PTGS2, CDK5 and FOXQ1 in Alzheimer’s Disease. Curr. Alzheimer. Res. 2019, 16, 596–612. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xu, Y.; Zhao, M.; Gao, Z. Neuro-protective roles of long non-coding RNA MALAT1 in Alzheimer’s disease with the involvement of the microRNA-30b/CNR1 network and the following PI3K/AKT activation. Exp. Mol. Pathol. 2020, 117, 104545. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, S.W.; Li, X.L.; Yu, F.Y.; Cong, H.M. Knockdown of long non-coding RNA TUG1 depresses apoptosis of hippocampal neurons in Alzheimer’s disease by elevating microRNA-15a and repressing ROCK1 expression. Inflamm. Res. 2020, 69, 897–910. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lu, B.; Chen, J. Knockdown of lncRNA SNHG1 attenuated Ab25-35 -inudced neuronal injury via regulating KREMEN1 by acting as a ceRNA of miR-137 in neuronal cells. Biochem. Biophys. Res. Commun. 2019, 518, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, N.; Lv, C.; Li, N.; Li, X.; Li, W. LncRNA SNHG1 Knockdown Alleviates Amyloid-B-Induced Neuronal Injury by Regulating ZNF217 via Sponging miR-361-3p in Alzheimer’s Disease. J. Alzheimer Dis. 2020, 77, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, T.; Wang, T.; Wang, B. Suppression of lncRNA-ATB prevents amyloid-β-induced neurotoxicity in PC12 cells via regulating miR-200/ZNF217 axis. Biomed. Pharmacother. 2018, 108, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Lin, M.; Ma, J.; Liu, W.; Gao, L.; Wei, S.; Xue, Y.; Shang, X. The role of LINC00094/miR-224-5p (miR-497-5p)/Endophilin-1 axis in Memantine mediated protective effects on blood-brain barrier in AD microenvironment. J. Cell Mol. Med. 2019, 23, 3280–3292. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Shan, K.; Qun-Wang, X.; Zhou, R.M.; Yang, H.; Liu, C.; Li, Y.J.; Yao, J.; Li, X.M.; Shen, Y.; et al. Long non-coding RNA-MIAT promotes neurovascular remodeling in the eye and brain. Oncotarget 2016, 7, 49688–49698. [Google Scholar] [CrossRef]
- Gu, R.; Liu, R.; Wang, L.; Tang, M.; Li, S.R.; Hu, X. LncRNA RPPH1 attenuates Aβ25-35-induced endoplasmic reticulum stress and apoptosis in SH-SY5Y cells via miR-326/PKM2. Int. J. Neurosci. 2020, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ran, G.; Lu, W.; Man, T.; Shi-rong, L.; Rui, L.; Xiao, H. LncRNA Rpph1 Protects Amyloid-β Induced Neuronal Injury in SK-N-SH Cells via miR-122/Wnt1 Axis. Int. J. Neurosci. 2020, 130, 443–453. [Google Scholar] [CrossRef]
- Cai, Y.; Sun, Z.; Jia, H.; Luo, H.; Ye, X.; Wu, Q.; Xiong, Y.; Zhang, W.; Wan, J. Rpph1 Upregulates CDC42 Expression and Promotes Hippocampal Neuron Dendritic Spine Formation by Competing with miR-330-5p. Front. Mol. Neurosci. 2017, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Yan, H.; Teng, Y.; Wang, Q.; Yang, P.; Zhang, L.; Cheng, H.; Fu, S. Long non-coding RNA 00507/miRNA-181c-5p/TTBK1/MAPT axis regulates tau hyperphosphorylation in Alzheimer’s disease. J. Gene Med. 2020, 5, e3268. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Li, L.; Qiu, X.I.N.; Wu, J.; Xu, L.E.I.; Shao, W. Long non-coding RNA ANRIL knockdown suppresses apoptosis and pro-inflammatory cytokines while enhancing neurite outgrowth via binding microRNA-125a in a cellular model of Alzheimer’s disease. Mol. Med. Rep. 2020, 22, 1489–1497. [Google Scholar] [CrossRef]
- Zhao, Y.; Alexandrov, P.N.; Jaber, V.; Lukiw, W.J. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer’s Disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes 2016, 7, 116. [Google Scholar] [CrossRef]
- Shi, Z.; Chen, T.; Yao, Q.; Zheng, L.; Zhang, Z.; Wang, J.; Hu, Z.; Cui, H.; Han, Y.; Han, X.; et al. The circular RNA ciRS-7 promotes APP and BACE1 degradation in an NF-κB-dependent manner. FEBS J. 2017, 284, 1096–1109. [Google Scholar] [CrossRef]
- Ye, T.; Yang, M.; Huang, D.; Wang, X.; Xue, B.; Tian, N.; Xu, X.; Bao, L.; Hu, H.; Lv, T.; et al. MicroRNA-7 as a potential therapeutic target for aberrant NF-κB-driven distant metastasis of gastric cancer. J. Exp. Clin. Cancer Res. 2019, 38, 55. [Google Scholar] [CrossRef]
- Choi, D.C.; Chae, Y.J.; Kabaria, S.; Chaudhuri, A.D.; Jain, M.R.; Li, H.; Mouradian, M.M.; Junn, E. MicroRNA-7 protects against 1-methyl-4-phenylpyridinium-induced cell death by targeting RelA. J. Neurosci. 2014, 34, 12725–12737. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Shang, H.; Chen, X.; Yang, S.; Qu, Y.; Ding, J.; Li, X. Circular RNA circ_0000950 promotes neuron apoptosis, suppresses neurite outgrowth and elevates inflammatory cytokines levels via directly sponging miR-103 in Alzheimer’s disease. Cell Cycle 2019, 18, 2197–2214. [Google Scholar] [CrossRef]
- Lu, Y.; Tan, L.; Wang, X. Circular HDAC9/microRNA-138/Sirtuin-1 Pathway Mediates Synaptic and Amyloid Precursor Protein Processing Deficits in Alzheimer’s Disease. Neurosci. Bull. 2019, 35, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Gao, Y.; Yu, S.; Sun, X.; Shen, K. Berberine attenuates Aβ42-induced neuronal damage through regulating circHDAC9/miR-142-5p axis in human neuronal cells. Life Sci. 2020, 252, 117637. [Google Scholar] [CrossRef] [PubMed]
- Straniero, L.; Rimoldi, V.; Samarani, M.; Goldwurm, S.; Di Fonzo, A.; Krüger, R.; Deleidi, M.; Aureli, M.; Soldà, G.; Duga, S.; et al. The GBAP1 pseudogene acts as a ceRNA for the glucocerebrosidase gene GBA by sponging miR-22-3p. Sci. Rep. 2017, 7, 12702. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Geng, L.; Chen, Y.; Wu, C. SNHG1 promotes MPP+- induced cytotoxicity by regulating PTEN/AKT/mTOR signaling pathway in SH-SY5Y cells via sponging miR-153-3p. Biol. Res. 2020, 53, 1–11. [Google Scholar] [CrossRef]
- Xie, N.; Qi, J.; Li, S.; Deng, J.; Chen, Y.; Lian, Y. Upregulated lncRNA small nucleolar RNA host gene 1 promotes 1-methyl-4-phenylpyridinium ion-induced cytotoxicity and reactive oxygen species production through miR-15b-5p/GSK3β axis in human dopaminergic SH-SY5Y cells. J. Cell Biochem. 2019, 120, 5790–5801. [Google Scholar] [CrossRef]
- Chen, Y.; Lian, Y.; Ma, Y.; Wu, C.; Zheng, Y.; Xie, N. LncRNA SNHG1 promotes α-synuclein aggregation and toxicity by targeting miR-15b-5p to activate SIAH1 in human neuroblastoma SH-SY5Y cells. Neurotoxicology 2017, 68, 212–221. [Google Scholar] [CrossRef]
- Cao, B.; Wang, T.; Qu, Q.; Kang, T.; Yang, Q. Long noncoding RNA SNHG1 promotes neuroinflammation in Parkinson’s disease via regulating miR-7/NLRP3 pathway. Neuroscience 2018, 388, 118–127. [Google Scholar] [CrossRef]
- Qian, C.; Ye, Y.; Mao, H.; Yao, L.; Sun, X.; Wang, B.; Zhang, H.; Xie, L.; Zhang, H.; Zhang, Y.; et al. Downregulated lncRNA-SNHG1 enhances autophagy and prevents cell death through the miR-221/222/p27/mTOR pathway in Parkinson’s disease. Exp. Cell Res. 2019, 384, 111614. [Google Scholar] [CrossRef]
- Peng, T.; Liu, X.; Wang, J.; Liu, Y.; Fu, Z.; Ma, X.; Li, J.; Sun, G.; Ji, Y.; Lu, J.; et al. Long noncoding RNA HAGLROS regulates apoptosis and autophagy in Parkinson’s disease via regulating miR-100/ATG10 axis and PI3K/Akt/mTOR pathway activation. Artif Cells Nanomed. Biotechnol. 2019, 47, 2764–2774. [Google Scholar] [CrossRef]
- Zhao, J.; Li, H.; Chang, N. LncRNA HOTAIR promotes MPP+ -induced neuronal injury in Parkinson’s Disease by regulating the miR-874-5p/ATG10 axis. EXCLI J. 2020, 19, 1141–1153. [Google Scholar] [CrossRef]
- Lin, Q.; Hou, S.; Dai, Y.; Jiang, N.; Lin, Y. LncRNA HOTAIR targets miR-126-5p to promote the progression of Parkinson’s disease through RAB3IP. Biol. Chem. 2019, 400, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, F.; Zhao, W. Long noncoding RNA NEAT1 knockdown inhibits MPP+-induced apoptosis, inflammation and cytotoxicity in SK-N-SH cells by regulating miR-212-5p/RAB3IP axis. Neurosci. Lett. 2020, 731, 135060. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zhang, D.; Guo, J.; Chen, Z.; Chen, Y.; Zhang, J. Deficiency of NEAT1 prevented MPP + -induced inflammatory response, oxidative stress and apoptosis in dopaminergic SK-N-SH neuroblastoma cells via miR-1277-5p/ARHGAP26 axis. Brain Res. 2020, 1750, 147156. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Zhou, F.; Li, J.; Duan, S. NEAT1 regulates MPP + -induced neuronal injury by targeting miR-124 in neuroblastoma cells. Neurosci. Lett. 2019, 708, 134340. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, J.; Liu, Q.; Xiao, Z.; Dai, Q. Knockdown of long non-coding RNA AL049437 mitigates MPP+-induced neuronal injury in SH-SY5Y cells via the microRNA-205-5p/MAPK1 axis. Neurotoxicology 2020, 78, 29–35. [Google Scholar] [CrossRef]
- Chen, Q.; Huang, X.; Li, R. lncRNA MALAT1/miR-205-5p axis regulates MPP+-induced cell apoptosis in MN9D cells by directly targeting LRRK2. Am. J. Transl. Res. 2018, 10, 563–572. [Google Scholar]
- Liu, W.; Zhang, Q.; Zhang, J.; Pan, W.; Zhao, J.; Xu, Y. Long non-coding RNA MALAT1 contributes to cell apoptosis by sponging miR-124 in Parkinson disease. Cell Biosci. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Lu, Y.; Gong, Z.; Wang, Z. LncRNA MALAT1 targeting miR-124-3p regulates DAPK1 expression contributes to cell apoptosis in Parkinson’s Disease. J. Cell Biochem. 2020. Published online. [Google Scholar] [CrossRef]
- Xia, D.; Sui, R.; Zhang, Z. Administration of resveratrol improved Parkinson’s disease-like phenotype by suppressing apoptosis of neurons via modulating the MALAT1/miR-129/SNCA signaling pathway. J. Cell Biochem. 2018, 120, 4942–4951. [Google Scholar] [CrossRef]
- Zhang, L.M.; Wang, M.H.; Yang, H.C.; Tian, T.; Sun, G.F.; Ji, Y.F.; Hu, W.T.; Liu, X.; Wang, J.P.; Lu, H. Dopaminergic neuron injury in Parkinson’s disease is mitigated by interfering lncRNA SNHG14 expression to regulate the miR-133b/α-synuclein pathway. Aging 2019, 11, 9264–9279. [Google Scholar] [CrossRef]
- Xu, X.; Zhuang, C.; Wu, Z.; Qiu, H.; Feng, H.; Wu, J. LincRNA-p21 Inhibits Cell Viability and Promotes Cell Apoptosis in Parkinson’s Disease through Activating α-Synuclein Expression. Biomed. Res. Int. 2018, 2018, 8181374. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; He, X.; Lu, F.; Mao, H.; Zhu, Z.; Yao, L.; Luo, W.; Sun, X.; Wang, B.; Qian, C.; et al. A lincRNA-p21/miR-181 family feedback loop regulates microglial activation during systemic LPS- and MPTP- induced neuro inflammation. Cell Death Dis. 2018, 9, 803. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Zhao, L.; Qiao, H.; Wu, S.; Wang, X. Long non-coding RNA-p21 regulates MPP + -induced neuronal injury by targeting miR-625 and derepressing TRPM2 in SH-SY5Y cells. Chem. Biol. Interact. 2019, 307, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhang, L.; Geng, Y.; Liu, Y.; Zhang, N. Long noncoding RNA GAS5 promotes microglial inflammatory response in Parkinson’s disease by regulating NLRP3 pathway through sponging miR-223-3p. Int. Immunopharmacol. 2020, 85, 106614. [Google Scholar] [CrossRef]
- Fan, Y.; Zhao, X.; Lu, K.; Cheng, G. LncRNA BDNF-AS promotes autophagy and apoptosis in MPTP-induced Parkinson’s disease via ablating microRNA-125b-5p. Brain Res. Bull. 2020, 157, 119–127. [Google Scholar] [CrossRef]
- Han, Y.; Kang, C.; Kang, M.; Quan, W.; Gao, H.; Zhong, Z. Long non-coding RNA Mirt2 prevents TNF-α-triggered inflammation via the repression of microRNA-101. Int. Immunopharmacol. 2019, 76, 105878. [Google Scholar] [CrossRef]
- Jiang, J.; Piao, X.; Hu, S.; Gao, J.; Bao, M. LncRNA H19 diminishes dopaminergic neuron loss by mediating microRNA-301b-3p in Parkinson’s disease via the HPRT1-mediated Wnt/β-catenin signaling pathway. Aging 2020, 12, 8820–8836. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, Q.; Lin, J. LncRNA H19 Attenuates Apoptosis in MPTP-Induced Parkinson’s Disease Through Regulating miR-585-3p/PIK3R3. Neurochem. Res. 2020, 45, 1700–1710. [Google Scholar] [CrossRef]
- Junn, E.; Lee, K.; Jeong, B.S.; Chan, T.W.; Im, J.; Mouradian, M.M. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. USA 2009, 106, 13052–13057. [Google Scholar] [CrossRef]
- Hsiao, K.; Sun, H.S.; Tsai, S. Circular RNA—New member of noncoding RNA with novel functions. Exp. Biol. Med. 2017, 242, 1136–1141. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Shamsuzzama Haque, R.; Baghel, T.; Nazir, A. Circular RNAs: The Emerging Class of Non-coding RNAs and Their Potential Role in Human Neurodegenerative Diseases. Mol. Neurobiol. 2017, 54, 7224–7234. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Chen, Y. Roles of Circular RNAs in Neurologic Disease. Front. Mol. Neurosci. 2016, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- Sang, Q.; Liu, X.; Wang, L.; Qi, L.; Sun, W.; Wang, W.; Sun, Y.; Zhang, H. CircSNCA downregulation by pramipexole treatment mediates cell apoptosis and autophagy in Parkinson’s disease by targeting miR-7. Aging 2018, 10, 1281–1293. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Shamsuzzama; Jadiya, P.; Haque, R.; Shukla, S.; Nazir, A. Functional Characterization of Novel Circular RNA Molecule, circzip-2 and Its Synthesizing Gene zip-2 in C. elegans Model of Parkinson’s Disease. Mol. Neurobiol. 2018, 55, 6914–6926. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhang, L.; Wang, S.; Hong, Q. Circular RNA circDLGAP4 exerts neuroprotective effects via modulating miR-134-5p/CREB pathway in Parkinson’s disease. Biochem. Biophys. Res. Commun. 2020, 522, 388–394. [Google Scholar] [CrossRef]
- Bian, Z.; Lei, W.; Li, Q.; Xue, W.; Gao, Y.; Zeng, Y.; Wang, Y.; Tang, L.; Tang, T.; Chen, C.; et al. Gm15575 functions as a ceRNA to up-regulate CCL7 expression through sponging miR-686 in Th17 cells. Mol. Immunol. 2020, 125, 32–42. [Google Scholar] [CrossRef]
- Wu, L.; Xia, J.; Li, D.; Kang, Y.; Fang, W.; Huang, P. Mechanisms of M2 Macrophage-Derived Exosomal Long Non-coding RNA PVT1 in Regulating Th17 Cell Response in Experimental Autoimmune Encephalomyelitisa. Front. Immunol. 2020, 11, 1934. [Google Scholar] [CrossRef]
- Yue, P.; Jing, L.; Zhao, X.; Zhu, H.; Teng, J. Down-regulation of taurine-up-regulated gene 1 attenuates in fl ammation by sponging miR-9-5p via targeting NF- κ B1/p50 in multiple sclerosis. Life Sci. 2019, 233, 116731. [Google Scholar] [CrossRef]
- Duan, C.; Liu, Y.; Li, Y.; Chen, H.; Liu, X.; Chen, X.; Yue, J.; Zhou, X.; Yang, J. Sulfasalazine alters microglia phenotype by competing endogenous RNA effect of miR-136-5p and long non-coding RNA HOTAIR in cuprizone-induced demyelination. Biochem. Pharmacol. 2018, 155, 110–123. [Google Scholar] [CrossRef]
- Senousy, M.A.; Shaker, O.G.; Sayed, N.H.; Fathy, N.; Kortam, M.A. LncRNA GAS5 and miR-137 Polymorphisms and Expression are Associated with Multiple Sclerosis Risk: Mechanistic Insights and Potential Clinical Impact. ACS Chem. Neurosci. 2020, 11, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Cardamone, G.; Paraboschi, E.M.; Rimoldi, V.; Duga, S.; Sold, G.; Asselta, R. The Characterization of GSDMB Splicing and Backsplicing Profiles Identifies Novel Isoforms and a Circular RNA That Are Dysregulated in Multiple Sclerosis. Int. J. Mol. Sci. 2017, 18, 576. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Tang, X.; Wang, S. Roles of CircRNAs in Autoimmune Diseases. Front. Inmunol. 2019, 10, 639. [Google Scholar] [CrossRef] [PubMed]
- Iparraguirre, L.; Muñoz-Culla, M.; Prada-Luengo, I.; Castillo-Triviño, T.; Olascoaga, J.; Otaegui, D. Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis. Hum. Mol. Genet. 2017, 26, 3564–3572. [Google Scholar] [CrossRef]
- Tan, J.Y.; Vance, K.W.; Varela, M.A.; Sirey, T.; Watson, L.M.; Curtis, H.J.; Marinello, M.; Alves, S.; Steinkraus, B.; Cooper, S.; et al. Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7. Nat. Struct. Mol. Biol. 2014, 21, 955–961. [Google Scholar] [CrossRef]
- Ma, N.; Tie, C.; Yu, B.; Zhang, W.; Wan, J. Identifying lncRNA–miRNA–mRNA networks to investigate Alzheimer’s disease pathogenesis and therapy strategy. Aging 2020, 12, 2897–2920. [Google Scholar] [CrossRef]
- Wang, L.; Chen, X.; He, D.; Li, Y.; Fu, J. Dissection of functional lncRNAs in Alzheimer’s disease by construction and analysis of lncRNA-mRNA networks based on competitive endogenous RNAs. Biochem. Biophys. Res. Commun. 2017, 485, 569–576. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, Z.; Yu, Y.; Cao, J.; Qiao, Y. Comprehensive analysis of the lncRNA-associated ceRNA network identifies neuroinflammation biomarkers for Alzheimer’s disease. Mol. Omics 2019, 15, 459–469. [Google Scholar] [CrossRef]
- Zhang, S.; Zhu, D.; Li, H.; Li, H.; Feng, C.; Zhang, W. Characterization of circRNA-Associated-ceRNA Networks in a Senescence-Accelerated Mouse Prone 8 Brain. Mol. Ther. 2017, 25, 2053–2061. [Google Scholar] [CrossRef]
- Lee, W.J.; Moon, J.; Jeon, D.; Shin, Y.W.; Yoo, J.S.; Park, D.K.; Lee, S.T.; Jung, K.H.; Park, K.I.; Jung, K.Y.; et al. Possible epigenetic regulatory effect of dysregulated circular RNAs in Alzheimer’s disease model. Sci. Rep. 2019, 9, 11956. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, P.; Chen, B.; Zhang, Z.; Zhang, C.; Zhan, Q.; Huang, S.; Xia, Z.A.; Peng, W. Identifying circRNA-associated-ceRNA networks in the hippocampus of Aβ1-42-induced Alzheimer’s disease-like rats using microarray analysis. Aging 2018, 10, 775–788. [Google Scholar] [CrossRef]
- Huang, J.L.; Qin, M.C.; Zhou, Y.; Xu, Z.H.; Yang, S.M.; Zhang, F.; Zhong, J.; Liang, M.K.; Chen, B.; Zhang, W.Y.; et al. Comprehensive analysis of differentially expressed profiles of Alzheimer’s disease associated circular RNAs in an Alzheimer’s disease mouse model. Aging 2018, 10, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.L.; Xu, Z.H.; Yang, S.M.; Yu, C.; Zhang, F.; Qin, M.C.; Zhou, Y.; Zhong, Z.G.; Wu, D.P. Identification of Differentially Expressed Profiles of Alzheimer’s Disease Associated Circular RNAs in a Panax Notoginseng Saponins-Treated Alzheimer’s Disease Mouse Model. Comput. Struct. Biotechnol. J. 2018, 16, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Pan, J.; Ye, X.; Yu, B.; Zhang, W.; Wan, J. Whole-Transcriptome Analysis of APP/PS1 Mouse Brain and Identification of circRNA-miRNA-mRNA Networks to Investigate AD Pathogenesis. Mol. Ther. Nucleic. Acids 2019, 18, 1049–1062. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.I.; Yoon, G.; Kim, Y.K.; Song, J. Transcriptome Analysis of Pineal Glands in the Mouse Model of Alzheimer’s Disease. Front. Mol. Neurosci. 2020, 12, 318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, F.; Bao, S.; Sun, J. Systematic Characterization of Circular RNA-Associated CeRNA Network Identified Novel circRNA Biomarkers in Alzheimer’s Disease. Front. Bioeng. Biotechnol. 2019, 7, 222. [Google Scholar] [CrossRef] [PubMed]
- Dube, U.; Del-Aguila, J.L.; Li, Z.; Budde, J.P.; Jiang, S.; Hsu, S.; Ibanez, L.; Fernandez, M.V.; Farias, F.; Norton, J.; et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat. Neurosci. 2019, 22, 1903–1912. [Google Scholar] [CrossRef] [PubMed]
- Lo, I.; Hill, J.; Vilhjálmsson, B.J.; Kjems, J. Linking the association between circRNAs and Alzheimer’s disease progression by multi-tissue circular RNA characterization. RNA Biol. 2020, 3, 1–9. [Google Scholar] [CrossRef]
- Li, Y.; Fan, H.; Sun, J.; Ni, M.; Zhang, L.; Chen, C.; Hong, X.; Fang, F.; Zhang, W.; Ma, P. Circular RNA expression profile of Alzheimer’s disease and its clinical significance as biomarkers for the disease risk and progression. Int. J. Biochem. Cell Biol. 2020, 123, 105747. [Google Scholar] [CrossRef]
- Li, Y.; Lv, Z.; Zhang, J.; Ma, Q.; Li, Q.; Song, L.; Gong, L.; Zhu, Y.; Li, X.; Hao, Y.; et al. Profiling of differentially expressed circular RNAs in peripheral blood mononuclear cells from Alzheimer’s disease patients. Metab. Brain Dis. 2020, 35, 201–213. [Google Scholar] [CrossRef]
- Lin, D.; Liang, Y.; Jing, X.; Chen, Y.; Lei, M.; Zeng, Z.; Zhou, T.; Wu, X.; Peng, S.; Zheng, D.; et al. Microarray analysis of an synthetic a-synuclein induced cellular model reveals the expression profile of long non-coding RNA in Parkinson’s disease. Brain Res. 2018, 1678, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Chi, L.; Wang, L.; Jiao, D. Identification of Differentially Expressed Genes and Long Noncoding RNAs Associated with Parkinson’s Disease. Parkinsons Dis. 2019, 2019, 6078251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Feng, S.; Fan, Y.; Luo, Y.; Jin, L.; Li, S. Identifying a Comprehensive ceRNA Network to Reveal Novel Targets for the Pathogenesis of Parkinson’s Disease. Front. Neurol. 2020, 11, 810. [Google Scholar] [CrossRef] [PubMed]
- Jia, E.; Zhou, Y.; Liu, Z.; Wang, L.; Ouyang, T.; Pan, M.; Bai, Y.; Ge, Q. Transcriptomic Profiling of Circular RNA in Different Brain Regions of Parkinson’s Disease in a Mouse Model. Int. J. Mol. Sci. 2020, 21, 3006. [Google Scholar] [CrossRef]
- Hanan, M.; Simchovitz, A.; Yayon, N.; Vaknine, S.; Cohen-Fultheim, R.; Karmon, M.; Madrer, N.; Rohrlich, T.M.; Maman, M.; Bennett, E.R.; et al. A Parkinson’s disease CircRNAs Resource reveals a link between circSLC8A1 and oxidative stress. EMBO Mol. Med. 2020, 12, e11942. [Google Scholar] [CrossRef]
- Dolinar, A.; Koritnik, B.; Glavač, D.; Ravnik-Glavač, M. Circular RNAs as Potential Blood Biomarkers in Amyotrophic Lateral Sclerosis. Mol. Neurobiol. 2019, 56, 8052–8062. [Google Scholar] [CrossRef]
- Ravnik-Glavač, M.; Glavač, D. Circulating RNAs as Potential Biomarkers in Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2020, 21, 1714. [Google Scholar] [CrossRef]
- Lane, C.A.; Hardy, J.; Schott, J.M. Alzheimer’s disease. Eur. J. Neurol. 2018, 25, 59–70. [Google Scholar] [CrossRef]
- Brookmeyer, R.; Johnson, E.; Ziegler-Graham, K.; Arrighi, H.M. Forecasting the Global Burden of Alzheimer’s Disease. Alzheimer Dement. 2007, 3, 186–191. [Google Scholar] [CrossRef]
- Reitz, C.; Brayne, C.; Mayeux, R. Epidemiology of Alzheimer disease. Nat. Rev. Neurol. 2011, 7, 137–152. [Google Scholar] [CrossRef]
- Li, R.; Lindholm, K.; Yang, L.B.; Yue, X.; Citron, M.; Yan, R.; Beach, T.; Sue, L.; Sabbagh, M.; Cai, H.; et al. Amyloid β peptide load is correlated with increased β-secretase activity in sporadic Alzheimer’s disease patients. Proc. Natl. Acad. Sci. USA 2004, 101, 3632–3637. [Google Scholar] [CrossRef]
- Zhao, J.; Fu, Y.; Yasvoina, M.; Shao, P.; Hitt, B.; O’Connor, T.; Logan, S.; Maus, E.; Citron, M.; Berry, R.; et al. β-site amyloid precursor protein cleaving enzyme 1 levels become elevated in neurons around amyloid plaques: Implications for Alzheimer’s disease pathogenesis. J. Neurosci. 2007, 27, 3639–3649. [Google Scholar] [CrossRef] [PubMed]
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; St Laurent, G., 3rd; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of b-secretase. Nat. Med. 2008, 14, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Q.; Liu, C.; Gao, S.; Ping, H.; Wang, J.; Wang, P. MiR-214-3p attenuates cognition defects via the inhibition of autophagy in SAMP8 mouse model of sporadic Alzheimer’s disease. Neurotoxicology 2016, 56, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Cha, D.J.; Mengel, D.; Mustapic, M.; Liu, W.; Selkoe, D.J.; Kapogiannis, D.; Galasko, D.; Rissman, R.A.; Bennett, D.A.; Walsh, D.M. MiR-212 and miR-132 are downregulated in neurally derived plasma exosomes of Alzheimer’s patients. Front. Neurosci. 2019, 13, 1208. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, R.; Wu, J.; Wang, Q.; Pang, K.; Shi, Q.; Gao, Q.; Hu, Y.; Dong, X.; Zhang, J.; et al. Melatonin protects against Aβ -induced neurotoxicity in primary neurons via miR-132/PTEN/AKT/FOXO3a pathway. Biofactors 2018, 44, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Veremeyko, T.; Wong, A.H.; El Fatimy, R.; Wei, Z.; Cai, W.; Krichevsky, A.M. Downregulation of miR-132/212 impairs S-nitrosylation balance and induces tau phosphorylation in Alzheimer’s disease. Neurobiol. Aging 2017, 51, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Salta, E.; Sierksma, A.; Eynden, E.; De Vanden Strooper, B. miR- 132 loss de-represses ITPKB and aggravates amyloid and TAU pathology in Alzheimer’s brain. EMBO Mol. Med. 2016, 8, 1005–1018. [Google Scholar] [CrossRef]
- Xu, N.; Li, A.; Ji, L.; Ye, Y.; Wang, Z.; Tong, L. miR-132 regulates the expression of synaptic proteins in APP/PS1 transgenic mice through C1q. Eur. J. Histochem. 2019, 63, 3008. [Google Scholar] [CrossRef]
- El Fatimy, R.; Li, S.; Chen, Z.; Mushannen, T.; Gongala, S.; Wei, Z.; Balu, D.T.; Rabinovsky, R.; Cantlon, A.; Elkhal, A.; et al. MicroRNA-132 provides neuroprotection for tauopathies via multiple signaling pathways. Acta Neuropathol. 2018, 136, 537–555. [Google Scholar] [CrossRef]
- Moncini, S.; Lunghi, M.; Valmadre, A.; Grasso, M.; Del Vescovo, V.; Riva, P.; Denti, M.A.; Venturin, M. The miR-15/107 Family of microRNA Genes Regulates CDK5R1/p35 with Implications for Alzheimer’s Disease Pathogenesis. Mol. Neurobiol. 2017, 54, 4329–4342. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Rapp, J.; Rainone, S.; Goupil, C.; Dorval, V.; Smith, P.Y.; Saint-Pierre, M.; Vallée, M.; Planel, E.; Droit, A.; Calon, F.; et al. microRNA-132/212 deficiency enhances Aβ production and senile plaque deposition in Alzheimer’s disease triple transgenic mice. Sci. Rep. 2016, 6, 30953. [Google Scholar] [CrossRef] [PubMed]
- Hadar, A.; Milanes, E.; Walczak, M.; Kuźnicki, J.; Squassina, A. SIRT1, miR-132 and miR-212 link human longevity to Alzheimer’s Disease. Sci. Rep. 2018, 8, 8465. [Google Scholar] [CrossRef] [PubMed]
- Rizzi, L.; Roriz-Cruz, M. Sirtuin 1 and Alzheimer’s disease: An up-to-date review. Neuropeptides 2018, 71, 54–60. [Google Scholar] [CrossRef]
- Lee, H.R.; Shin, H.K.; Park, S.Y.; Kim, H.Y.; Lee, W.S.; Rhim, B.Y.; Hong, K.W.; Kim, C.D. Cilostazol suppresses β-amyloid production by activating a disintegrin and metalloproteinase 10 via the upregulation of SIRT1-coupled retinoic acid receptor-β. J. Neurosci. Res. 2014, 92, 1581–1590. [Google Scholar] [CrossRef]
- Qin, W.; Yang, T.; Ho, L.; Zhao, Z.; Wang, J.; Chen, L.; Zhao, W.; Thiyagarajan, M.; MacGrogan, D.; Rodgers, J.T.; et al. Neuronal SIRT1 Activation as a Novel Mechanism Underlying the Prevention of Alzheimer Disease Amyloid Neuropathology by Calorie Restriction. J. Biol. Chem. 2006, 281, 21745–21754. [Google Scholar] [CrossRef]
- Feng, X.; Liang, N.; Zhu, D.; Gao, Q.; Peng, L.; Dong, H.; Yue, Q.; Liu, H.; Bao, L.; Zhang, J.; et al. Resveratrol inhibits β-amyloid-induced neuronal apoptosis through regulation of SIRT1-ROCK1 signaling pathway. PLoS ONE 2013, 8, e59888. [Google Scholar] [CrossRef]
- Henderson, B.W.; Gentry, E.G.; Rush, T.; Troncoso, J.C.; Thambisetty, M.; Montine, T.J.; Herskowitz, J.H. Rho-associated protein kinase 1 (ROCK1) is increased in Alzheimer’s disease and ROCK1 depletion reduces amyloid-β levels in brain. J. Neurochem. 2016, 138, 525–531. [Google Scholar] [CrossRef]
- Liu, L.; Shi, Y.; Shi, J.; Wang, H.; Sheng, Y.; Jiang, Q.; Chen, H.; Li, X.; Dong, J. The long non-coding RNA SNHG1 promotes glioma progression by competitively binding to miR-194 to regulate PHLDA1 expression. Cell Death Dis. 2019, 10, 463. [Google Scholar] [CrossRef]
- Xu, M.; Chen, X.; Lin, K.; Zeng, K.; Liu, X.; Pan, B.; Xu, X.; Xu, T.; Hu, X.; Sun, L.; et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol. Cancer 2018, 17, 141. [Google Scholar] [CrossRef]
- Wang, H.; Wang, G.; Gao, Y.; Zhao, C.; Li, X.; Zhang, F.; Jiang, C.; Wu, B. Lnc-SNHG1 Activates the TGFBR2/SMAD3 and RAB11A/Wnt/β-Catenin Pathway by Sponging MiR-302/372/373/520 in Invasive Pituitary Tumors. Cell Physiol. Biochem. 2018, 48, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Gong, W.; Guo, J. Long non-coding RNA SNHG1 indicates poor prognosis and facilitates disease progression in acute myeloid leukemia. Biol. Open 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, Q.; Zhou, P.; Deng, D.; Xue, L.; Shao, N.; Peng, Y.; Zhi, F. Upregulation of the long non-coding RNA SNHG1 predicts poor prognosis, promotes cell proliferation and invasion, and reduces apoptosis in glioma. Biomed. Pharmacother. 2017, 91, 906–911. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Fu, Y.; Zhuang, Y.; Zhang, K.; Lu, D. LncRNA SNHG1 alleviates IL-1β -induced osteoarthritis by inhibiting miR-16-5p-mediated p38 MAPK and NF-κB signaling pathways. Biosci. Rep. 2019, 39, BSR20191523. [Google Scholar] [CrossRef]
- Mulvaney, J.F.; Thompkins, C.; Noda, T.; Nishimura, K.; Sun, W.W.; Lin, S.Y.; Coffin, A.; Dabdoub, A. Kremen1 regulates mechanosensory hair cell development in the mammalian cochlea and the zebrafish lateral line. Sci. Rep. 2016, 6, 4–13. [Google Scholar] [CrossRef]
- Causeret, F.; Sumia, I.; Pierani, A. Kremen1 and Dickkopf1 control cell survival in a Wnt-independent manner. Cell Death Differ. 2016, 23, 323–332. [Google Scholar] [CrossRef]
- Quinlan, K.G.R.; Verger, A.; Yaswen, P.; Crossley, M. Amplification of zinc finger gene 217 (ZNF217) and cancer: When good fingers go bad. Biochim. Biophys. Acta 2007, 1775, 333–340. [Google Scholar] [CrossRef]
- Yin, Y.; Cha, C.; Wu, F.; Li, J.; Li, S. Endophilin 1 knockdown prevents synaptic dysfunction induced by oligomeric amyloid β. Mol. Med. Rep. 2019, 19, 4897–4905. [Google Scholar] [CrossRef]
- Liu, W.; Cai, H.; Lin, M.; Zhu, L.; Gao, L.; Zhong, R.; Bi, S.; Xue, Y.; Shang, X. MicroRNA-107 prevents amyloid-beta induced blood-brain barrier disruption and endothelial cell dysfunction by targeting Endophilin-1. Exp. Cell Res. 2016, 343, 248–257. [Google Scholar] [CrossRef]
- Tanzi, R.E.; Moir, R.D.; Wagner, S.L. Clearance of Alzheimer’s Aβ peptide: The many roads to perdition. Neuron 2004, 43, 605–608. [Google Scholar] [CrossRef]
- Rodrigues, M.; Xin, X.; Jee, K.; Babapoor-Farrokhran, S.; Kashiwabuchi, F.; Ma, T.; Bhutto, I.; Hassan, S.J.; Daoud, Y.; Baranano, D.; et al. VEGF secreted by hypoxic Müller cells induces MMP-2 expression and activity in endothelial cells to promote retinal neovascularization in proliferative diabetic retinopathy. Diabetes 2013, 62, 3863–3873. [Google Scholar] [CrossRef] [PubMed]
- Reichenbanch, A.; Bringmann, A. New functions of Müller cells. Glia 2013, 61, 651–678. [Google Scholar] [CrossRef] [PubMed]
- Hagmann, H.; Taniguchi, Y.; Pippin, J.W.; Kauerz, H.M.; Benzing, T.; Shankland, S.J.; Brinkkoetter, P.T. Cyclin I and p35 determine the subcellular distribution of Cdk5. Am. J. Physiol. Cell Physiol. 2015, 308, C339–C347. [Google Scholar] [CrossRef] [PubMed]
- Patrick, G.N.; Zukerberg, L.; Nikolic, M.; De La Monte, S.; Dikkes, P.; Tsai, L.H. Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature 1999, 402, 615–622. [Google Scholar] [CrossRef]
- Guo, Z.; Cao, Q.; Zhao, Z.; Song, C. Biogenesis, Features, Functions, and Disease Relationships of a Specific Circular RNA: CDR1as. Aging Dis. 2020, 11, 1009–1020. [Google Scholar] [CrossRef]
- Hansen, T.B.; Wiklund, E.D.; Bramsen, J.B.; Villadsen, S.B.; Statham, A.L.; Clark, S.J.; Kjems, J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011, 30, 4414–4422. [Google Scholar] [CrossRef]
- Song, J.; Kim, Y.K. Identification of the Role of miR-142-5p in Alzheimer’s Disease by Comparative Bioinformatics and Cellular Analysis. Front. Mol. Neurosci. 2017, 10, 227. [Google Scholar] [CrossRef]
- Tian, J.; Guo, L.; Sui, S.; Driskill, C.; Phensy, A.; Wang, Q.; Gauba, E.; Zigman, J.M.; Swerdlow, R.H.; Kroener, S.; et al. Disrupted hippocampal growth hormone secretagogue receptor 1α interaction with dopamine receptor D1 plays a role in Alzheimer’s disease. Sci. Transl. Med. 2019, 11, eaav6278. [Google Scholar] [CrossRef]
- Yamanaka, Y.; Faghihi, M.A.; Magistri, M.; Alvarez-Garcia, O.; Lotz, M.; Wahlestedt, C. Antisense RNA controls LRP1 sense transcript expression through interaction with a chromatin-associated protein, HMGB2. Cell Rep. 2015, 11, 967–976. [Google Scholar] [CrossRef]
- Martiskainen, H.; Haapasalo, A.; Kurkinen, K.M.; Pihlajamäki, J.; Soininen, H.; Hiltunen, M. Targeting ApoE4/ApoE receptor LRP1 in Alzheimer’s disease. Expert Opin. Ther. Targets 2013, 17, 781–794. [Google Scholar] [CrossRef]
- Humphries, C.E.; Kohli, M.A.; Nathanson, L.; Whitehead, P.; Beecham, G.; Martin, E.; Mash, D.C.; Pericak-Vance, M.A.; Gilbert, J. Integrated whole transcriptome and DNA methylation analysis identifies gene networks specific to late-onset Alzheimer’s disease. J. Alzheimers Dis. 2015, 44, 977–987. [Google Scholar] [CrossRef] [PubMed]
- Murata, N.; Murakami, K.; Ozawa, Y.; Kinoshita, N.; Irie, K.; Shirasawa, T.; Shimizu, T. Silymarin attenuated the amyloid β plaque burden and improved behavioral abnormalities in an Alzheimer’s disease mouse model. Biosci. Biotechnol. Biochem. 2010, 74, 2299–2306. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Cao, H.; Cui, X.; Zheng, W.; Wang, S.; Yu, J.; Chen, Z. Silymarin’s Inhibition and Treatment Effects for Alzheimer’s Disease. Molecules 2019, 24, 1748. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kumar, A.; Verma, R.K.; Shukla, R. Silymarin encapsulated nanoliquid crystals for improved activity against beta amyloid induced cytotoxicity. Int. J. Biol. Macromol. 2020, 149, 1198–1206. [Google Scholar] [CrossRef]
- Xu, M.; Xiao, M.; Li, S.; Yang, B. Aquaporins in Nervous System. Adv. Exp. Med. Biol. 2017, 969, 81–103. [Google Scholar] [CrossRef]
- Han, C.; Yang, Y.; Guan, Q.; Zhang, X.; Shen, H.; Sheng, Y.; Wang, J.; Zhou, X.; Li, W.; Guo, L.; et al. New mechanism of nerve injury in Alzheimer’s disease: β-amyloid-induced neuronal pyroptosis. J. Cell Mol. Med. 2020, 24, 8078–8090. [Google Scholar] [CrossRef]
- Xu, Y.; Tan, H.Y.; Li, S.; Wang, N.; Feng, Y. Panax notoginseng for Inflammation-Related Chronic Diseases: A Review on the Modulations of Multiple Pathways. Am. J. Chin. Med. 2018, 46, 971–996. [Google Scholar] [CrossRef]
- Grimmond, S.; Larder, R.; Van Hateren, N.; Siggers, P.; Morse, S.; Hacker, T.; Arkell, R.; Greenfield, A. Expression of a novel mammalian epidermal growth factor-related gene during mouse neural development. Mech. Dev. 2001, 102, 209–211. [Google Scholar] [CrossRef]
- Zhang, Q.; Gao, X.; Li, C.; Feliciano, C.; Wang, D.; Zhou, D.; Mei, Y.; Monteiro, P.; Anand, M.; Itohara, S.; et al. Impaired Dendritic Development and Memory in Sorbs2 Knock-Out Mice. J. Neurosci. 2016, 36, 2247–2260. [Google Scholar] [CrossRef]
- Lee, J.H.; Cheng, R.; Vardarajan, B.; Lantigua, R.; Reyes-Dumeyer, D.; Ortmann, W.; Graham, R.R.; Bhangale, T.; Behrens, T.W.; Medrano, M.; et al. Genetic Modifiers of Age at Onset in Carriers of the G206A Mutation in PSEN1 With Familial Alzheimer Disease Among Caribbean Hispanics. JAMA Neurol. 2015, 72, 1043–1051. [Google Scholar] [CrossRef]
- Canali, G.; Garcia, M.; Hivert, B.; Pinatel, D.; Goullancourt, A.; Oguievetskaia, K.; Saint-Martin, M.; Girault, J.A.; Faivre-Sarrailh, C.; Goutebroze, L. Genetic variants in autism-related CNTNAP2 impair axonal growth of cortical neurons. Hum. Mol. Genet. 2018, 27, 1941–1954. [Google Scholar] [CrossRef] [PubMed]
- Amidfar, M.; de Oliveira, J.; Kucharska, E.; Budni, J.; Kim, Y.K. The role of CREB and BDNF in neurobiology and treatment of Alzheimer’s disease. Life Sci. 2020, 257, 118020. [Google Scholar] [CrossRef] [PubMed]
- Clokie, S.J.; Lau, P.; Kim, H.H.; Coon, S.L.; Klein, D.C. MicroRNAs in the pineal gland: miR-483 regulates melatonin synthesis by targeting arylalkylamine N-acetyltransferase. J. Biol. Chem. 2012, 287, 25312–25324. [Google Scholar] [CrossRef] [PubMed]
- Hébert, S.S.; Horré, K.; Nicolaï, L.; Papadopoulou, A.S.; Mandemakers, W.; Silahtaroglu, A.N.; Kauppinen, S.; Delacourte, A.; De Strooper, B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. USA 2008, 105, 6415–6420. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.A.; Tomás, J.F.; Queiroz, J.A.; Figueiras, A.R.; Sousa, F. Recombinant pre-miR-29b for Alzheimer´s disease therapeutics. Sci. Rep. 2016, 6, 19946. [Google Scholar] [CrossRef]
- Long, J.M.; Lahiri, D.K. MicroRNA-101 downregulates Alzheimer’s amyloid-β precursor protein levels in human cell cultures and is differentially expressed. Biochem. Biophys. Res. Commun. 2011, 404, 889–895. [Google Scholar] [CrossRef]
- Luo, P.; Li, X.; Fei, Z.; Poon, W. Scaffold protein Homer 1: Implications for neurological diseases. Neurochem. Int. 2012, 61, 731–738. [Google Scholar] [CrossRef]
- Dickey, C.A.; Loring, J.F.; Montgomery, J.; Gordon, M.N.; Eastman, P.S.; Morgan, D. Selectively reduced expression of synaptic plasticity-related genes in amyloid precursor protein + presenilin-1 transgenic mice. J. Neurosci. 2003, 23, 5219–5226. [Google Scholar] [CrossRef]
- Fjell, A.M.; Sederevicius, D.; Sneve, M.H.; de Lange, A.G.; Bråthen, A.C.; Idland, A.V.; Watne, L.O.; Wang, Y.; Reinbold, C.; Dobricic, V.; et al. Self-reported Sleep Problems Related to Amyloid Deposition in Cortical Regions with High HOMER1 Gene Expression. Cereb. Cortex. 2020, 30, 2144–2156. [Google Scholar] [CrossRef]
- Pannaccione, A.; Piccialli, I.; Secondo, A.; Ciccone, R.; Molinaro, P.; Boscia, F.; Annunziato, L. The Na+/Ca2+exchanger in Alzheimer’s disease. Cell Calcium. 2020, 87, 102190. [Google Scholar] [CrossRef]
- Vargas, L.M.; Cerpa, W.; Muñoz, F.J.; Zanlungo, S.; Alvarez, A.R. Amyloid-β oligomers synaptotoxicity: The emerging role of EphA4/c-Abl signaling in Alzheimer’s disease. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, S.; Liu, A.; Wan, J.; Tang, L.; Zheng, N.; Xiong, Y. Inhibition of BDNF production by MPP+ through up-regulation of miR-210-3p contributes to dopaminergic neuron damage in MPTP model. Neurosci. Lett. 2018, 675, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tan, L.; Lu, Y.; Peng, J.; Zhu, Y.; Zhang, Y.; Sun, Z. MicroRNA-138 promotes tau phosphorylation by targeting retinoic acid receptor alpha. FEBS Lett. 2015, 589, 726–729. [Google Scholar] [CrossRef]
- Kumar, S.; Reddy, A.P.; Yin, X.; Reddy, P.H. Novel MicroRNA-455-3p and its protective effects against abnormal APP processing and amyloid beta toxicity in Alzheimer’s Disease. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1–28. [Google Scholar] [CrossRef] [PubMed]
- De Lau, L.M.L.; Breteler, M.M.B. Epidemiology of Parkinson’s disease. Lancet Neurol. 2006, 5, 525–535. [Google Scholar] [CrossRef]
- Jankovic, J. Parkinson’s disease: Clinical features and diagnosis. J. Neurosurg. Psychiatry 2008, 79, 368–376. [Google Scholar] [CrossRef]
- Beitz, J.M. Parkinson’s disease: A review. Front. Biosci. 2014, 6, 65–74. [Google Scholar] [CrossRef]
- Dauer, W.; Przedborski, S. Parkinson’s disease: Mechanisms and models. Neuron 2003, 39, 889–909. [Google Scholar] [CrossRef]
- Nichols, W.C.; Pankratz, N.; Marek, D.K.; Pauciulo, M.W.; Elsaesser, V.E.; Halter, C.A.; Rudolph, A.; Wojcieszek, J.; Pfeiffer, R.F.; Foroud, T.; et al. Mutations in GBA are associated with familial Parkinson disease susceptibility and age at onset. Neurology 2009, 72, 210–316. [Google Scholar] [CrossRef]
- Balestrino, R.; Schapira, A.H.V. Glucocerebrosidase and Parkinson Disease: Molecular, Clinical, and Therapeutic Implications. Neuroscientist 2018, 24, 540–559. [Google Scholar] [CrossRef]
- Li, W.; Jiang, Y.; Wang, Y.; Yang, S.; Bi, X.; Pan, X.; Ma, A.; Li, W. MiR-181b regulates autophagy in a model of Parkinson’s disease by targeting the PTEN/Akt/mTOR signaling pathway. Neurosci. Lett. 2018, 675, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Leikas, J.V.; Kohtala, S.; Theilmann, W.; Jalkanen, A.J.; Forsberg, M.M.; Rantamäki, T. Brief isoflurane anesthesia regulates striatal AKT-GSK3b signaling and ameliorates motor deficits in a rat model of early-stage Parkinson’s disease. J. Neurochem. 2017, 142, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.L.; Xu, J.; Xue, S.R.; Liu, Y.Y.; Zhang, Y.J.; Zhang, X.Z.; Wang, X.; Wu, F.P.; Li, X.M. The E3 ubiquitin ligase seven in absentia homolog 1 may be a potential new therapeutic target for Parkinson’s disease. Neural. Regen. Res. 2015, 10, 1286–1291. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Wheeler, T.C.; Li, L.; Chin, L.-S. Ubiquitination of alpha-synuclein by Siah-1 promotes alpha-synuclein aggregation and apoptotic cell death. Hum. Mol. Genet. 2008, 17, 906–917. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, X.; Guo, Y.; Rong, H.; Liu, T. The long noncoding RNA HOTAIR promotes Parkinson’s disease by upregulating LRRK2 expression. Oncotarget 2017, 8, 24449–24456. [Google Scholar] [CrossRef]
- Kluss, J.H.; Mamais, A.; Cookson, M.R.; Biology, C.; Section, G.E. LRRK2 links genetic and sporadic Parkinson’s disease. Biochem. Soc. Trans. 2019, 47, 651–661. [Google Scholar] [CrossRef]
- Ultanir, S.K.; Hertz, N.T.; Li, G.; Ge, W.P.; Burlingame, A.L.; Pleasure, S.J.; Shokat, K.M.; Jan, L.Y.; Jan, Y.N. Chemical genetic identification of NDR1/2 kinase substrates AAK1 and Rabin8 uncovers their roles in controlling dendrite arborization and spine development. Neuron 2012, 73, 1127–1142. [Google Scholar] [CrossRef]
- Homma, Y.; Fukuda, M. Rabin8 regulates neurite outgrowth in both GEF activity–dependent and –independent manners. Mol. Biol. Cell 2016, 27, 2107–2118. [Google Scholar] [CrossRef]
- Boros, F.A.; Maszlag-Török, R.; Vécsei, L.; Klivényi, P. Increased level of NEAT1 long non-coding RNA is detectable in peripheral blood cells of patients with Parkinson’s disease. Brain Res. 2020, 1730, 146672. [Google Scholar] [CrossRef]
- Hirota, Y.; Yamashita, S.; Kurihara, Y.; Jin, X.; Aihara, M.; Saigusa, T.; Kan, D.; Kanki, T. Mitophagy is primarily due to alternative autophagy and requires the MAPK1 and MAPK14 signaling pathways. Autophagy 2015, 11, 332–343. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, H.; Yu, J.; Zhao, Z.; Xiao, F.; Xia, T.; Wang, C.; Li, K.; Deng, J.; Guo, Y.; et al. MAPK1/3 regulate hepatic lipid metabolism via ATG7-dependent autophagy. Autophagy 2016, 12, 592–593. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Ko, A.; Hyun, H.; Min, S.; Kang, T. P2RX7-MAPK1/2-SP1 axis inhibits MTOR independent HSPB1-mediated astroglial autophagy. Cell Death Dis. 2018, 9, 546. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Ravanan, P.; Talwar, P. Death Associated Protein Kinase 1 (DAPK1): A Regulator of Apoptosis and Autophagy. Front. Mol. Neurosci. 2016, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Kim, B.M.; Chen, C.; Begley, M.J.; Cantley, L.C.; Lee, T.H. Death-associated protein kinase 1 phosphorylates NDRG2 and induces neuronal cell death. Cell Death Differ. 2017, 24, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Ye, Y.; Mao, H.; Lu, F.; He, X.; Lu, G.; Zhang, S. MicroRNA-124 regulates the expression of MEKK3 in the inflammatory pathogenesis of Parkinson’s disease. J. Neuroinflammation 2018, 15, 13. [Google Scholar] [CrossRef]
- Wang, H.; Ye, Y.; Zhu, Z.; Mo, L.; Lin, C.; Wang, Q.; Wang, H.; Gong, X.; He, X.; Lu, G.; et al. MiR-124 Regulates Apoptosis and Autophagy Process in MPTP Model of Parkinson’s Disease by Targeting to Bim. Brain Pathol. 2016, 26, 167–176. [Google Scholar] [CrossRef]
- Yao, L.; Zhu, Z.; Wu, J.; Zhang, Y.; Zhang, H.; Sun, X.; Qian, C.; Wang, B.; Xie, L.; Zhang, S.; et al. MicroRNA-124 regulates the expression of p62/p38 and promotes autophagy in the inflammatory pathogenesis of Parkinson’s disease. FASEB J. 2019, 33, 8648–8665. [Google Scholar] [CrossRef]
- Geng, L.; Liu, W.; Chen, Y. miR-124-3p attenuates MPP þ-induced neuronal injury by targeting STAT3 in SH-SY5Y cells. Exp. Biol. Med. 2017, 242, 1757–1764. [Google Scholar] [CrossRef]
- Dong, R.; Zhang, B.; Tai, L.; Liu, H.; Shi, F.-K.; Liu, N.-N. The Neuroprotective Role of miR-124-3p in a 6-Hydroxydopamine- Induced Cell Model of Parkinson’s Disease via the Regulation of ANAX5. J. Cell Biochem. 2018, 119, 269–277. [Google Scholar] [CrossRef]
- Tay, S.S.W. Downregulation of miR-124 in MPTP-treated mouse model of Parkinson’s Disease and MPP iodide-treated MN9D cells modulates the expression of the calpain/CDK5 pathway proteins. Neuroscience 2014, 272, 167–179. [Google Scholar] [CrossRef]
- Gong, X.; Wang, H.; Ye, Y.; Shu, Y.; Deng, Y.; He, X.; Lu, G.; Zhang, S. miR-124 regulates cell apoptosis and autophagy in dopaminergic neurons and protects them by regulating AMPK/mTOR pathway in Parkinson’s disease. Am. J. Transl. Res. 2016, 8, 2127–2137. [Google Scholar] [PubMed]
- Xing, R.; Li, L. Down regulation of miR-218, miR-124, and miR-144 relates to Parkinson’s disease via activating NF-κB signaling. Kaohsiung J. Med. Sci. 2020, 36, 786–792. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Zhai, H. MicroRNA-124 Enhances Dopamine Receptor Expression and Neuronal Proliferation in Mouse Models of Parkinson’s Disease via the Hedgehog Signaling Pathway by Targeting EDN2. Neuroimmunomodulation 2019, 26, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Frahm, C.; Srivastava, A.; Schmidt, S.; Mueller, J.; Groth, M.; Guenther, M.; Ji, Y.; Priebe, S.; Platzer, M.; Witte, O.W. Transcriptional profiling reveals protective mechanisms in brains of long-lived mice. Neurobiol. Aging 2017, 52, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.A.; Potashkin, J.A. Network-based metaanalysis identifies HNF4A and PTBP1 as longitudinally dynamic biomarkers for Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2015, 112, 2257–2262. [Google Scholar] [CrossRef]
- Santiago, J.A.; Potashkin, J.A. Blood biomarkers associated with cognitive decline in early stage and drug-naïve Parkinson’s disease patients. PLoS ONE 2015, 10, e0142582. [Google Scholar] [CrossRef]
- Valente, T.; Dentesano, G.; Ezquerra, M.; Fernandez-Santiago, R.; Martinez-Martin, J.; Gallastegui, E.; Domuro, C.; Compta, Y.; Martí, M.J.; Bachs, O.; et al. CCAAT/enhancer binding protein δ is a transcriptional repressor of α-synuclein. Cell Death Differ. 2020, 27, 509–524. [Google Scholar] [CrossRef]
- Bawari, S.; Tewari, D.; Argüelles, S.; Sah, A.N.; Nabavi, S.F.; Xu, S.; Vacca, R.A.; Nabavi, S.M.; Shirooie, S. Targeting BDNF signaling by natural products: Novel synaptic repair therapeutics for neurodegeneration and behavior disorders. Pharmacol. Res. 2019, 148, 104458. [Google Scholar] [CrossRef]
- D’Orsi, B.; Mateyka, J.; Prehn, J.H.M. Neurochemistry International Control of mitochondrial physiology and cell death by the Bcl-2 family proteins Bax and Bok. Neurochem. Int. 2017, 109, 162–170. [Google Scholar] [CrossRef]
- Lv, J.; Jiang, S.; Yang, Z.; Hu, W.; Wang, Z.; Li, T.; Yang, Y. PGC-1α sparks the fire of neuroprotection against neurodegenerative disorders. Ageing Res. Rev. 2018, 44, 8–21. [Google Scholar] [CrossRef]
- Jakaria, M.; Haque, M.E.; Cho, D.Y.; Azam, S.; Kim, I.S.; Choi, D.K. Molecular Insights into NR4A2(Nurr1): An Emerging Target for Neuroprotective Therapy Against Neuroinflammation and Neuronal Cell Death. Mol. Neurobiol. 2019, 56, 5799–5814. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, C.; Paiva, J.; Santos, T.; Ferreira, L.; Bernardino, L. MicroRNA-124 loaded nanoparticles enhance brain repair in Parkinson’s disease. J. Control Release 2016, 235, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Sirabella, R.; Sisalli, M.J.; Costa, G.; Omura, K.; Ianniello, G.; Pinna, A.; Morelli, M.; Di Renzo, G.M.; Annunziato, L.; Scorziello, A. NCX1 and NCX3 as potential factors contributing to neurodegeneration and neuroinflammation in the A53T transgenic mouse model of Parkinson’s Disease. Cell Death Dis. 2018. [Google Scholar] [CrossRef] [PubMed]
- Cotsapas, C.; Mitrovic, M.; Hafler, D. Multiple Sclerosis. In Handbook of Clinical Neurology, 3rd ed.; Geschwind, D.H., Paulsori, H.L., Klein, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 148, pp. 723–730. [Google Scholar]
- Oh, J.; Vidal-jordana, A.; Montalban, X. Multiple sclerosis: Clinical aspects. Curr. Opin. Neurol. 2018, 31, 752–759. [Google Scholar] [CrossRef]
- Bjelobaba, I.; Savic, D.; Lavrnja, I. Multiple Sclerosis and neuroinflammation: The overview of current and prospective therapies. Curr. Pharm. Des. 2017, 23, 693–730. [Google Scholar] [CrossRef]
- Santoro, M.; Nociti, V.; Lucchini, M.; De Fino, C.; Losavio, F.A.; Mirabella, M. Expression Profile of Long Non-Coding RNAs in Serum of Patients with Multiple Sclerosis. J. Mol. Neurosci. 2016, 59, 18–23. [Google Scholar] [CrossRef]
- Sigdel, K.R.; Cheng, A.; Wang, Y.; Duan, L.; Zhang, Y. The Emerging Functions of Long Noncoding RNA in Immune Cells: Autoimmune Diseases. J. Immunol. Res. 2015, 2015, 848790. [Google Scholar] [CrossRef]
- Dos Passos, G.R.; Sato, D.K.; Becker, J.; Fujihara, K. Th17 Cells Pathways in Multiple Sclerosis and Neuromyelitis Optica Spectrum Disorders: Pathophysiological and Therapeutic Implications. Mediat. Inflamm. 2016. [Google Scholar] [CrossRef]
- Lindner, M.; Thümmler, K.; Arthur, A.; Brunner, S.; Elliott, C.; McElroy, D.; Mohan, H.; Williams, A.; Edgar, J.M.; Schuh, C.; et al. Fibroblast growth factor signalling in multiple sclerosis: Inhibition of myelination and induction of pro-inflammatory environment by FGF9. Brain 2015, 138, 1875–1893. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Mckenzie, B.S.; Zhou, L.; Tadokoro, C.E.; Lepelley, A.; Lafaille, J.J.; Cua, D.J.; Littman, D.R. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T Helper Cells. Cell 2006, 126, 1121–1133. [Google Scholar] [CrossRef]
- Eftekharian, M.M.; Ghafouri-Fard, S.; Soudyab, M.; Omrani, M.D.; Rahimi, M.; Sayad, A.; Komaki, A.; Mazdeh, M.; Taheri, M. Expression Analysis of Long Non-coding RNAs in the Blood of Multiple Sclerosis Patients. J. Mol. Neurosci. 2017, 63, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Toghi, M.; Taheri, M.; Arsang-jang, S.; Ohadi, M.; Mirfakhraie, R. SOCS gene family expression profile in the blood of multiple sclerosis patients. J. Neurol. Sci. 2017, 375, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, J.S.; Toth, B.; Jeschke, U.; Schleussner, E.; Markert, U.R. Knocking off the suppressors of cytokine signaling (SOCS): Their roles in mammalian pregnancy. J. Reprod. Immunol. 2009, 83, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, Y.I.; Chang, K.Y.; Lee, D.W.; Cho, S.C.; Ha, Y.W.; Na, J.E.; Rhyu, I.J.; Park, S.C.; Park, H.C. Promotion of remyelination by sulfasalazine in a transgenic zebrafish model of demyelination. Mol. Cells 2015, 38, 1013–1021. [Google Scholar] [CrossRef]
- Prosiegel, M.; Neu, I.; Vogl, S.; Hoffmann, G.; Wildfeuer, A.; Ruhenstroth-Bauer, G. Suppression of experimental autoimmune encephalomyelitis by sulfasalazine. Acta Neurol. Scand. 1990, 81, 237–238. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, L.; Wang, E.; Zhang, C.; Li, X. LncRNA GAS5 regulates ischemic stroke as a competing endogenous RNA for miR-137 to regulate the Notch1 signaling pathway. Biochem. Biophys. Res. Commun. 2018, 496, 184–190. [Google Scholar] [CrossRef]
- Sun, D.; Yu, Z.; Fang, X.; Liu, M.; Pu, Y.; Shao, Q.; Wang, D.; Zhao, X.; Huang, A.; Xiang, Z.; et al. LncRNA GAS 5 inhibits microglial M2 polarization and exacerbates demyelination. EMBO Rep. 2017, 18, 1801–1816. [Google Scholar] [CrossRef]
- Cardamone, G.; Paraboschi, E.M.; Soldà, G.; Cantoni, C.; Supino, D.; Piccio, L.; Duga, S.; Asselta, R. Not only cancer: The long non-coding RNA MALAT1 affects the repertoire of alternatively spliced transcripts and circular RNAs in multiple sclerosis. Hum. Mol. Genet. 2019, 28, 1414–1428. [Google Scholar] [CrossRef]
- Shaker, O.G.; Mahmoud, R.H.; Abdelaleem, O.O.; Ibrahem, E.G.; Mohamed, A.A.; Zaki, O.M.; Abdelghaffar, N.K.; Ahmed, T.I.; Hemeda, N.F.; Ahmed, N.A.; et al. LncRNAs, MALAT1 and lnc-DC as potential biomarkers for multiple sclerosis diagnosis. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Das, S.; Miller, M.; Broide, D.H. Chromosome 17q21 Genes ORMDL3 and GSDMB in Asthma and Immune Diseases. Adv. Immunol. 2017, 135, 1–52. [Google Scholar] [CrossRef]
- Keller, A.; Leidinger, P.; Lange, J.; Borries, A.; Schroers, H.; Scheffler, M.; Lenhof, H.P.; Ruprecht, K.; Meese, E. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS ONE 2009, 4, e7440. [Google Scholar] [CrossRef]
- Hecker, M.; Thamilarasan, M.; Koczan, D.; Schröder, I.; Flechtner, K.; Freiesleben, S.; Füllen, G.; Thiesen, H.J.; Zettl, U.K. MicroRNA expression changes during interferon-beta treatment in the peripheral blood of multiple sclerosis patients. Int. J. Mol. Sci. 2013, 14, 16087–16110. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zha, X. miR-149 reduces while let-7 elevates ASIC1a expression in vitro. Int. J. Physiol. Pathophysiol. Pharmacol. 2017, 9, 147–152. [Google Scholar]
- Friese, M.A.; Craner, M.J.; Etzensperger, R.; Vergo, S.; Wemmie, J.A.; Welsh, M.J.; Vincent, A.; Fugger, L. Acid-sensing ion channel-1 contributes to axonal degeneration in autoimmune inflammation of the central nervous system. Nat. Med. 2007, 13, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Arun, T.; Tomassini, V.; Sbardella, E.; de Ruiter, M.B.; Matthews, L.; Leite, M.I.; Gelineau-Morel, R.; Cavey, A.; Vergo, S.; Craner, M.; et al. Targeting ASIC1 in primary progressive multiple sclerosis: Evidence of neuroprotection with amiloride. Brain 2015, 136, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Boiko, N.; Kucher, V.; Eaton, B.A.; Stockand, J.D. Inhibition of neuronal degenerin/epithelial Na+ channels by the multiple sclerosis drug 4-aminopyridine. J. Biol. Chem. 2013, 288, 9418–9427. [Google Scholar] [CrossRef]
- Vergo, S.; Craner, M.J.; Etzensperger, R.; Attfield, K.; Friese, M.A.; Newcombe, J.; Esiri, M.; Fugger, L. Acid-sensing ion channel 1 is involved in both axonal injury and demyelination in multiple sclerosis and its animal model. Brain 2011, 134, 571–584. [Google Scholar] [CrossRef]
- Pianta, A.; Drouin, E.E.; Crowley, J.T.; Arvikar, S.; Strle, K.; Costello, C.E.; Steere, A.C. Annexin A2 is a target of autoimmune T and B cell responses associated with synovial fi broblast proliferation in patients with antibiotic-refractory Lyme arthritis. Clin. Immunol. 2015, 160, 336–341. [Google Scholar] [CrossRef]
- Cañas, F.; Simonin, L.; Couturaud, F.; Renaudineau, Y. Annexin A2 autoantibodies in thrombosis and autoimmune diseases. Thromb. Res. 2015, 135, 226–230. [Google Scholar] [CrossRef]
- Talbott, E.O.; Malek, A.M.; Lacomis, D. The Epidemiology of Amyotrophic Lateral Sclerosis, 1st ed.; Elsevier, B.V.: Amsterdam, The Netherlands, 2016; Volume 138. [Google Scholar] [CrossRef]
- Zarei, S.; Carr, K.; Reiley, L.; Diaz, K.; Guerra, O.; Fernandez Altamirano, P.; Pagani, W.; Lodin, D.; Orozco, G.; Chinea, A. A comprehensive review of amyotrophic lateral sclerosis. Surg. Neurol. Int. 2015, 6, 171. [Google Scholar] [CrossRef]
- Strong, M.J. The evidence for altered RNA metabolism in amyotrophic lateral sclerosis (ALS). J. Neurol. Sci. 2010, 288, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Biscarini, S.; Capauto, D.; Peruzzi, G.; Lu, L.; Colantoni, A.; Santini, T.; Shneider, N.A.; Caffarelli, E.; Laneve, P.; Bozzoni, I. Characterization of the lncRNA transcriptome in mESC-derived motor neurons: Implications for FUS-ALS. Stem. Cell Res. 2018, 27, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Errichelli, L.; Dini Modigliani, S.; Laneve, P.; Colantoni, A.; Legnini, I.; Capauto, D.; Rosa, A.; De Santis, R.; Scarfò, R.; Peruzzi, G.; et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Wu, L.; Cheng, W.; Chen, C.; Wu, M.; Wang, Y.; Tseng, Y.; Chuang, T.; Shen, C.J. Transcriptomopathies of pre- and post- symptomatic frontotemporal dementia-like mice with TDP-43 depletion in forebrain neurons. Acta Neuropathol. Commun. 2019, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, Y.; Nakagawa, S.; Hirose, T.; Okano, H.J.; Takao, M.; Shibata, S.; Suyama, S.; Kuwako, K.; Imai, T.; Murayama, S.; et al. The long non-coding RNA nuclear-enriched abundant transcript 1-2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain 2013, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Tollervey, J.R.; Curk, T.; Rogelj, B.; Briese, M.; Cereda, M.; Kayikci, M.; König, J.; Hortobágyi, T.; Nishimura, A.L.; Zupunski, V.; et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011, 14, 452–458. [Google Scholar] [CrossRef]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 39. [Google Scholar] [CrossRef]
- Gagliardi, S.; Pandini, C.; Garofalo, M.; Bordoni, M.; Pansarasa, O.; Cereda, C. Long non coding RNAs and ALS: Still much to do. Non Coding RNA Res. 2018, 3, 226–231. [Google Scholar] [CrossRef]
- Gagliardi, S.; Zucca, S.; Pandini, C.; Diamanti, L.; Bordoni, M.; Sproviero, D.; Arigoni, M.; Olivero, M.; Pansarasa, O.; Ceroni, M.; et al. Long non-coding and coding RNAs characterization in Peripheral Blood Mononuclear Cells and Spinal Cord from Amyotrophic Lateral Sclerosis patients. Sci. Rep. 2018, 8, 2378. [Google Scholar] [CrossRef]
- Zucca, S.; Gagliardi, S.; Pandini, C.; Diamanti, L.; Bordoni, M.; Sproviero, D.; Arigoni, M.; Olivero, M.; Pansarasa, O.; Ceroni, M.; et al. RNA-Seq profiling in peripheral blood mononuclear cells of amyotrophic lateral sclerosis patients and controls. Sci. Data 2019, 6, 190006. [Google Scholar] [CrossRef]
- Zhou, F.; Guan, Y.; Chen, Y.; Zhang, C.; Yu, L.; Gao, H.; Du, H.; Liu, B.; Wang, X. miRNA-9 expression is upregulated in the spinal cord of G93A-SOD1 transgenic mice. Int. J. Clin. Exp. Pathol. 2013, 6, 1826–1838. [Google Scholar] [PubMed]
- Vrabec, K.; Boštjančič, E.; Koritnik, B.; Leonardis, L.; Grošelj, L.D.; Zidar, J.; Rogelj, B.; Glavač, D.; Ravnik-Glavač, M. Differential Expression of Several miRNAs and the Host Genes AATK and DNM2 in Leukocytes of Sporadic ALS Patients. Front. Mol. Neurosci. 2018, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Hawley, Z.C.E.; Campos-Melo, D.; Strong, M.J. MiR-105 and miR-9 regulate the mRNA stability of neuronal intermediate filaments. Implications for the pathogenesis of amyotrophic lateral sclerosis (ALS). Brain Res. 2019, 1706, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Campos-Melo, D.; Droppelmann, C.A.; He, Z.; Volkening, K.; Strong, M.J. Altered microRNA expression profile in amyotrophic lateral sclerosis: A role in the regulation of NFL mRNA levels. Mol. Brain 2013, 6, 26. [Google Scholar] [CrossRef]
- Xu, C.; Xu, L.; Peng, F.; Cai, Y.; Wang, Y. MiR-647 promotes proliferation and migration of ox-LDL-treated vascular smooth muscle cells through regulating PTEN/PI3K/AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7110–7119. [Google Scholar] [CrossRef]
- Yang, D.; Wang, X.; Ismail, A.; Ashman, C.J.; Valori, C.F.; Wang, G.; Gao, S.; Higginbottom, A.; Ince, P.G.; Azzouz, M.; et al. PTEN regulates AMPA receptor-mediated cell viability in iPS-derived motor neurons. Cell Death Dis. 2014, 5, e1096. [Google Scholar] [CrossRef]
- Kirby, J.; Ning, K.; Ferraiuolo, L.; Heath, P.R.; Ismail, A.; Kuo, S.; Valori, C.F.; Cox, L.; Sharrack, B.; Wharton, S.B.; et al. Phosphatase and tensin homologue/protein kinase B pathway linked to motor neuron survival in human superoxide dismutase 1-related amyotrophic lateral sclerosis. Brain 2011, 134, 506–517. [Google Scholar] [CrossRef]
- Ning, K.; Drepper, C.; Valori, C.F.; Ahsan, M.; Wyles, M.; Higginbottom, A.; Herrmann, T.; Shaw, P.; Azzouz, M.; Sendtner, M. PTEN depletion rescues axonal growth defect and improves survival in SMN-deficient motor neurons. Hum. Mol. Genet. 2010, 19, 3159–3168. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X.; Sun, H.; Wang, H. Long non-coding RNAs in the regulation of skeletal myogenesis and muscle diseases. Cancer Lett. 2018, 417, 58–64. [Google Scholar] [CrossRef]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A Long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef]
- Kallen, A.N.; Zhou, X.; Xu, J.; Qiao, C.; Mai, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.; Zhang, H.; et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell. 2013, 52, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Yang, F.; Cao, H.; Liang, Z. Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 2015, 29, 3054–3064. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Liu, J.; Xiao, J.; Yang, L.; Cai, M.; Shen, H.; Chen, X.; Ma, Y.; Hu, S.; Wang, Z.; et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat. Commun. 2017, 8, 14718. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Li, M.; Sun, Y.; Cai, H.; Lan, X.; Huang, Y.; Bai, Y.; Qi, X.; Chen, H. The developmental transcriptome sequencing of bovine skeletal muscle reveals a long noncoding RNA, lncMD, promotes muscle differentiation by sponging miR-125b. BBA Mol. Cell Res. 2016, 1863, 2835–2845. [Google Scholar] [CrossRef]
- Lu, L.; Sun, K.; Chen, X.; Zhao, Y.; Wang, L.; Zhou, L.; Sun, H.; Wang, H. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J. 2013, 32, 2575–2588. [Google Scholar] [CrossRef]
- Pardo, P.S.; Boriek, A.M. The physiological roles of Sirt1 in skeletal muscle. Aging 2011, 3, 430–437. [Google Scholar] [CrossRef]
- Holmberg, M.; Duyckaerts, C.; Dürr, A.; Cancel, G.; Gourfinkel-An, I.; Damier, P.; Faucheux, B.; Trottier, Y.; Hirsch, E.C.; Agid, Y.; et al. Spinocerebellar ataxia type 7 (SCA7): A neurodegenerative disorder with neuronal intranuclear inclusions. Hum. Mol. Genet. 1998, 7, 913–918. [Google Scholar] [CrossRef]
- Matsuura, T.; Ashizawa, T. Spinocerebellar Ataxia Type 10. GeneReviews® 2019, 1–18. [Google Scholar]
- Yang, Y.; Zhou, X.; Jin, Y. ADAR-mediated RNA editing in non-coding RNA sequences. Sci. China Life Sci. 2013, 56, 944–952. [Google Scholar] [CrossRef]
- Zipeto, M.A.; Jiang, Q.; Melese, E.; Jamieson, C.H.M. RNA rewriting, recoding, and rewiring in human disease. Trends Mol. Med. 2015, 21, 549–559. [Google Scholar] [CrossRef]
- Lorenzini, I.; Moore, S.; Sattler, R. RNA Editing Deficiency in Neurodegeneration. Adv. Neurobiol. 2018, 20, 63–83. [Google Scholar] [CrossRef] [PubMed]
- Breen, M.S.; Dobbyn, A.; Li, Q.; Roussos, P.; Hoffman, G.E.; Stahl, E.; Chess, A.; Sklar, P.; Li, J.B.; Devlin, B.; et al. Global landscape and genetic regulation of RNA editing in cortical samples from individuals with schizophrenia. Nat. Neurosci. 2019, 22, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, E.; Levanon, E.Y. A-to-I RNA editing—immune protector and transcriptome diversifier. Nat. Rev. Genet. 2018, 19, 473–490. [Google Scholar] [CrossRef] [PubMed]
- Nigita, G.; Veneziano, D.; Ferro, A. A-to-I RNA Editing: Current Knowledge Sources and Computational Approaches with Special Emphasis on Non-Coding RNA Molecules. Front. Bioeng. Biotechnol. 2015, 3, 37. [Google Scholar] [CrossRef]
- Daniel, C.; Lagergren, J.; Öhman, M. RNA editing of non-coding RNA and its role in gene regulation. Biochimie 2015, 117, 22–27. [Google Scholar] [CrossRef]
- Shevchenko, G.; Morris, K.V. All I’s on the RADAR: Role of ADAR in gene regulation. FEBS Lett. 2018, 592, 2860–2873. [Google Scholar] [CrossRef]
- Nigita, G.; Distefano, R.; Veneziano, D.; Romano, G.; Rahman, M.; Wang, K.; Pass, H.; Croce, C.M.; Acunzo, M.; Nana-Sinkam, P. Tissue and exosomal miRNA editing in Non-Small Cell Lung Cancer. Sci. Rep. 2018, 8, 10222. [Google Scholar] [CrossRef]
- Hideyama, T.; Yamashita, T.; Aizawa, H.; Tsuji, S.; Kakita, A.; Takahashi, H.; Kwak, S. Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol. Dis. 2012, 45, 1121–1128. [Google Scholar] [CrossRef]
- Aizawa, H.; Hideyama, T.; Yamashita, T.; Kimura, T.; Suzuki, N.; Aoki, M.; Kwak, S. Deficient RNA-editing enzyme ADAR2 in an amyotrophic lateral sclerosis patient with a FUS(P525L) mutation. J. Clin. Neurosci. 2016, 32, 128–129. [Google Scholar] [CrossRef]
- Hosaka, T.; Yamashita, T.; Teramoto, S.; Hirose, N.; Tamaoka, A.; Kwak, S. ADAR2-dependent A-to-I RNA editing in the extracellular linear and circular RNAs. Neurosci. Res. 2019, 147, 48–57. [Google Scholar] [CrossRef]
- Khermesh, K.; D’Erchia, A.M.; Barak, M.; Annese, A.; Wachtel, C.; Levanon, E.Y.; Picardi, E.; Eisenberg, E. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer’s disease. RNA 2016, 22, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Singh, M. Dysregulated A to I RNA editing and non-coding RNAs in neurodegeneration. Front. Genet. 2013, 3, 326. [Google Scholar] [CrossRef] [PubMed]
- Gardner, O.K.; Wang, L.; Van Booven, D.; Whitehead, P.L.; Hamilton-Nelson, K.L.; Adams, L.D.; Starks, T.D.; Hofmann, N.K.; Vance, J.M.; Cuccaro, M.L.; et al. RNA editing alterations in a multi-ethnic Alzheimer disease cohort converge on immune and endocytic molecular pathways. Hum. Mol. Genet. 2019, 28, 3053–3061. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, T.; Yamashita, T.; Tamaoka, A.; Kwak, S. Extracellular RNAs as Biomarkers of Sporadic Amyotrophic Lateral Sclerosis and Other Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 3148. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef]
- Xu, J.; Feng, L.; Han, Z.; Li, Y.; Wu, A.; Shao, T.; Ding, N.; Li, L.; Deng, W.; Di, X.; et al. Extensive ceRNA—ceRNA interaction networks mediated by miRNAs regulate development in multiple rhesus tissues. Nucleic Acids Res. 2016, 44, 9438–9451. [Google Scholar] [CrossRef][Green Version]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef]
- Patel, N.A.; Moss, L.D.; Lee, J.; Tajiri, N.; Acosta, S.; Hudson, C.; Parag, S.; Cooper, D.R.; Borlongan, C.V.; Bickford, P.C. Long noncoding RNA MALAT1 in exosomes drives regenerative function and modulates inflammation-linked networks following traumatic brain injury. J. Neuroinflamm. 2018, 15, 204. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Chen, B.; Zhao, J.; Yu, S.; Tang, Y.; Zheng, Q.; Li, Y.; Wang, P.; He, X.; et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018, 46, D106–D112. [Google Scholar] [CrossRef]
- Wang, D.; Wang, P.; Bian, X.; Xu, S.; Zhou, Q.; Zhang, Y.; Ding, M.; Han, M.; Huang, L.; Bi, J.; et al. Elevated plasma levels of exosomal BACE1-AS combined with the volume and thickness of the right entorhinal cortex may serve as a biomarker for the detection of Alzheimer’s disease. Mol. Med. Rep. 2020, 22, 227–238. [Google Scholar] [CrossRef]
- Fotuhi, S.N.; Khalaj-Kondori, M.; Hoseinpour Feizi, M.A.; Talebi, M. Long Non-coding RNA BACE1-AS May Serve as an Alzheimer’s Disease Blood-Based Biomarker. J. Mol. Neurosci. 2019, 69, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.D.; Parsons, C.; Walker, L.; Zhang, W.C.; Slack, F.J. Targeting noncoding RNAs in disease. J. Clin. Investig. 2017, 127, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Yu, B. Role of Long Noncoding RNAs and Circular RNAs in Nerve Regeneration. Front. Mol. Neurosci. 2019, 12, 165. [Google Scholar] [CrossRef]
- Carroll, A.P.; Tooney, P.A.; Cairns, M.J. Context-specific microRNA function in developmental complexity. J. Mol. Cell Biol. 2013, 5, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Noncoding RNA 2019, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Vasudevan, S.; Tong, Y.; Steitz, J. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef]
- Xiao, M.; Li, J.; Li, W.; Wang, Y.; Wu, F.; Xi, Y.; Zhang, L.; Ding, C.; Luo, H.; Li, Y.; et al. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017, 14, 1326–1334. [Google Scholar] [CrossRef]
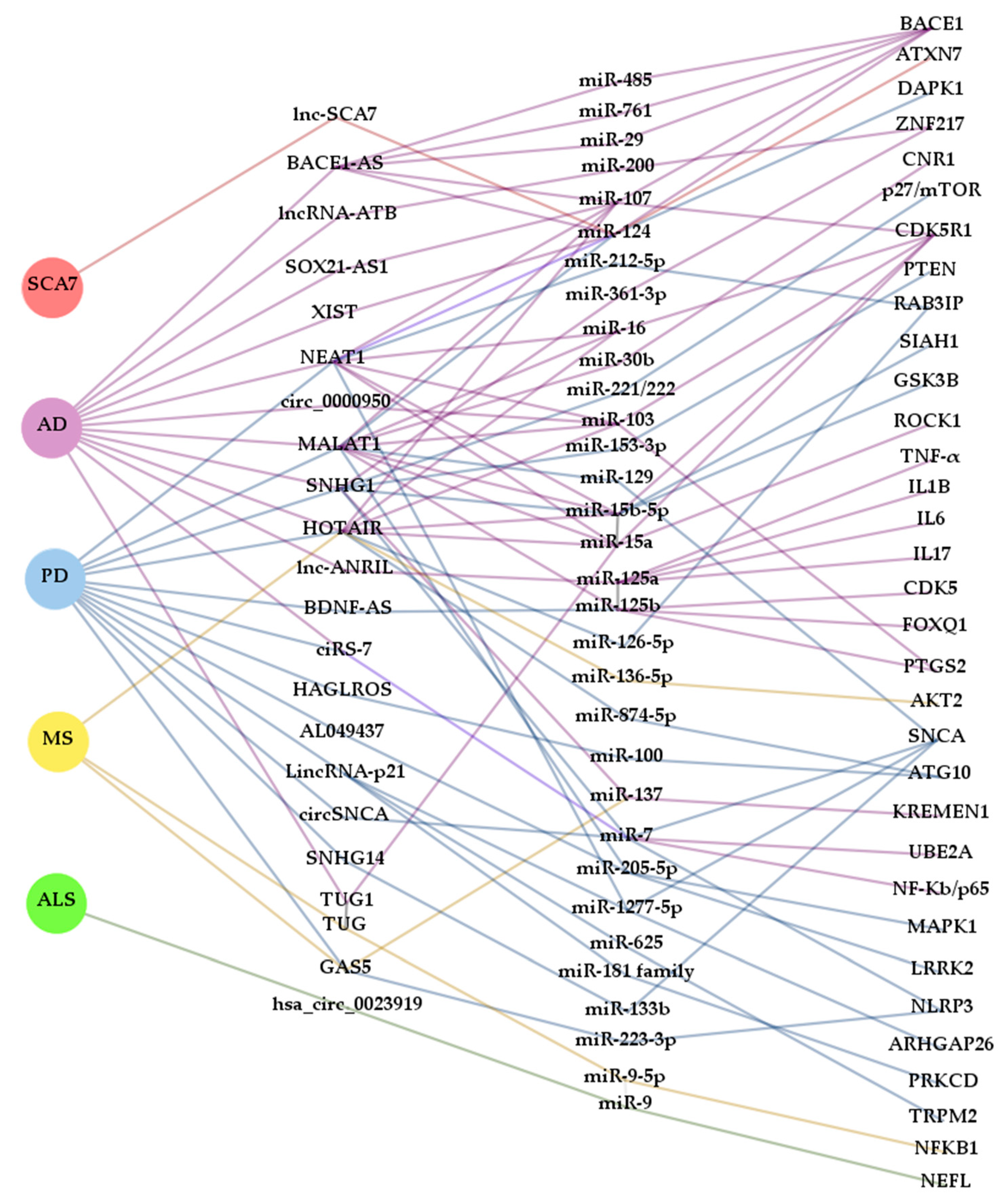
| Disease | ncRNA | miRNA | mRNA | Sample | Ref. | |
|---|---|---|---|---|---|---|
| AD | lncRNA | BACE1-AS | miR-29, miR-485, miR-761, miR-124 and miR-107 | BACE1 | Computational analysis from human data and cellular and mouse models | [24] |
| miR-214-3p | - | [25] | ||||
| miR-132-3p | - | [26] | ||||
| XIST | miR-124 | BACE1 | Cellular and mouse models | [27] | ||
| miR-132 | - | [28] | ||||
| NEAT1 | miR-124 | BACE1 | Cellular and mouse models | [29] | ||
| miR-107 | - | [30] | ||||
| SOX21-AS1 | miR-107 | - | Cellular model | [31] | ||
| NEAT1 HOTAIR MALAT1 | miR-107, miR-103, miR-16, miR-195, miR-15a and miR-15b | CDK5R1 | Cellular model | [32] | ||
| MALAT1 | miR-125b | CDK5, FOXQ1 and PTGS2 | Cellular and rat models | [33] | ||
| miR-30b | CNR1 | [34] | ||||
| TUG1 | miR-15a | ROCK1 | Cellular and mouse models | [35] | ||
| SNHG1 | miR-137 | KREMEN1 | Cellular model and human primary cell culture | [36] | ||
| miR-361-3p | ZNF217 | [37] | ||||
| lncRNA-ATB | miR-200 | ZNF217 | Cellular model | [38] | ||
| LINC00094 | miR-224-4p miR-497-5p | SH3GL2 | Cellular model | [39] | ||
| MIAT | miR-150-5p | VEGF | Cellular and mouse models | [40] | ||
| Rpph1 | miR-326 | PKM2 | Cellular and mouse models | [41] | ||
| miR-122 | Wnt1 | [42] | ||||
| miR-330-5p | CDC42 | [43] | ||||
| linc00507 | miR-181c-5p | MAPT TTBK1 | Cellular and mouse models | [44] | ||
| lnc-ANRIL | mir-125a | TNF-α, IL1B IL6 and IL17 | Cellular model | [45] | ||
| circRNA | ciRS-7 | miR-7 | UBE2A | Human brain | [46] | |
| * miR-7 | * NF-Κb/p65 | Cellular models | [47,48,49] | |||
| circ_0000950 | miR-103 | PTGS2 | Cellular models | [50] | ||
| circHDAC9 | miR-138 | Sirt1 | Cellular and mouse models | [51] | ||
| miR-142-5p | - | [52] | ||||
| PD | pseudogene | GBAP1 | miR-22-3p | GBA | Cellular models | [53] |
| lncRNA | SNHG1 | miR-153-3p miR-15b-5p miR-7 miR-221/222 | PTEN SIAH1, GSK3β NLRP3 CDKN1B (p27) | Cellular and mouse models | [54] [55,56] [57] [58] | |
| HAGLROs | miR-100 | ATG10 | Cellular and mouse models | [59] | ||
| HOTAIR | miR-874-5p | ATG10 | Cellular and mouse models | [60] | ||
| miR-126-5p | RAB3IP | [61] | ||||
| NEAT1 | miR-212-5p | RAB3IP | Cellular models | [62] | ||
| miR-1277-5p | ARHGAP26 | [63] | ||||
| miR-124 | - | [64] | ||||
| AL049437 | miR-205-5p | MAPK1 | Cellular and mouse models | [65] | ||
| MALAT1 | miR-205-5p | LRRK2 | Cellular and mouse models | [66] | ||
| miR-124 | DAPK1 | [67,68] | ||||
| miR-129 | SNCA (α-syn) | [69] | ||||
| SNHG14 | miR-133b | SNCA | Cellular and mouse models | [70] | ||
| LincRNA-p21 | miR-1277-5p | SNCA | Cellular and mouse models | [71] | ||
| miR-181 family | PRKCD (PKC-δ) | [72] | ||||
| miR-625 | TRPM2 | [73] | ||||
| GAS5 | miR-223-3p | NLRP3 | Cellular and mouse models | [74] | ||
| BDNF-AS | miR-125b-5p | - | Cellular and mouse models | [75] | ||
| Mirt2 | miR-101 | - | Cellular model | [76] | ||
| lncRNA H19 | miR-301b-3p | HPRT1 | Computational analysis from human data and cellular and mouse models | [77] | ||
| miR-585-3p | PIK3R3 | [78] | ||||
| circRNA | * ciRS-7 | miR-7 | SNCA | Cellular and mouse models | [79,80,81,82,83] | |
| circSNCA | miR-7 | SNCA | Cellular model | [84] | ||
| circzip-2 | * miR-60 | M60.4ZK470.2, igeg-2 and idhg-1 | Worm model | [85] | ||
| circDLGAP4 | miR-134-5p | CREB | Cellular and mouse models | [86] | ||
| MS | lncRNA | Gm15575 | miR-686 | CCL7 | Cellular and mouse models | [87] |
| PVT1 | miR-21-5p | SOCS5 | Cellular and mouse models | [88] | ||
| TUG | miR-9-5p | NFKB1 (p50) | Cellular and mouse models | [89] | ||
| HOTAIR | miR-136-5p | AKT2 | Cellular and mouse models | [90] | ||
| GAS5 | miR-137 | - | Human blood | [91] | ||
| circRNA | hsa_circ_0106803 | * miR-149 | * ASIC1a | Human blood (PMBCs) | [92,93] | |
| hsa_circ_0005402 hsa_circ_0035560 | * 14 miRNAs (miR-1248, miR-766) | - | Human blood (PMBCs) | [94] | ||
| SCA7 | lncRNA | lnc-SCA7 | miR-124 | ATXN7 | Human samples, and cellular and animal models | [95] |
| Disease | ceRNETs | Sample | Outcomes | Representative Networks | Ref. |
|---|---|---|---|---|---|
| AD | lncRNA-miRNA-mRNA | Mouse model (APP/PS1) brain (cortical samples) 12 months | One ceRNA network that included 4 lncRNAs, 5 miRNAs and 1082 mRNAs mainly related to AD-associated genes. |
| [43] |
| Mouse model (APP/PS1) brain (cortical samples) 6 and 9 months | 3 ceRNA networks built with lncRNAs, miRNAs and mRNAs differentially expressed according to the age at which they are found deregulated (6, 9 or 6 and 9 months). |
| [96] | ||
| Human brain (neurons from entorhinal cortex of mid-stage AD cases) | A neurofibrillary tangles-associated ceRNA network was built with 41 lncRNAs, 630 mRNAs and 2530 edges. |
| [97] | ||
| Human brain (prefrontal cortex) | An AD-associated ceRNET containing 6 lncRNAs, 3 miRNAs and 91 mRNAs. |
| [98] | ||
| circRNA-miRNA-mRNA | Mouse model (SAMP8) brain 7 months | Two ceRNA networks built with circRNAs, miRNAs and mRNAs found differentially expressed. |
| [99] | |
| Mouse model (Tg2576) brain 7 and 12 months | Four ceRNA networks built with circRNAs, miRNAs and mRNAs found differentially expressed. |
| [100] | ||
| Rat model (Aβ1-42) brain (hippocampal samples) | A ceRNA network built with 140 circRNAs, 140 miRNAs and 20 mRNAs with 503 relationships. |
| [101] | ||
| AD | circRNA-miRNA-mRNA | Mouse model (SAMP8) brain (hippocampal samples) 5 and 10 months | A ceRNA network predicted. |
| [102] |
| Mouse model (SAMP8) brain (hippocampal) 5 months | Two ceRNETs built with 2 circRNAs found differentially expressed in PNS-treated mice: mmu_circ_013636, 5 miRNAs and 442 mRNAs, mmu_circ_012180, 5 miRNAs and 631 mRNAs. |
| [103] | ||
| Mouse model (APP/PS1) brain (cortex) 6 and 9 months | Five ceRNA networks were constructed based on differentially expressed circRNAs, miRNAs and mRNAs. |
| [104] | ||
| Mouse model (5xFAD) pineal gland 5 months | A circRNA-miRNA network was constructed with 10 circRNAs. From it, a complete ceRNA net was predicted. |
| [105] | ||
| Human brain | An AD-associated ceRNET was constructed with 276 circRNAs, 14 miRNAs and 1117 mRNAs. AD risk ceRNET of KIAA1586 was stablished with 3 miRNAs (hsa-miR-29b, hsa-miR-101 and hsa-miR-15a) and 159 mRNAs. |
| [106] | ||
| Human brain | A circRNA-mRNA co-expression network was predicted. |
| [107] | ||
| AD | circRNA-miRNA-mRNA | Human brain | A circRNA-mRNA co-expression network was predicted |
| [108] |
| Human cerebrospinal fluid | A circRNA-miRNA network was built with the top 5 up- and down-regulated circRNAs (circ-LPAR1, circ-AXL, circ-GPHN, circ-ITPR3, circ-GPI, circ-HAUS4, circ-KIF18B, circ-ATP9A, circ-PCCA and circ-TTC39C). |
| [109] | ||
| Human blood (PBMCs) | Four nets constructed: (I) circRNA-miRNA network with the top 10 up- and down-regulated circRNAs (II) ceRNA network of hsa_circ_082547 (III) ceRNA network with 3 circRNAs (hsa_circ_101618, hsa_circ_405619, and hsa_circ_000843), 15 miRNAs and 223 mRNAs. (IV) ceRNA network of 4 circRNAs (hsa_circ_402265, hsa_circ_061346, hsa_circ_405836 and hsa_circ_061343), 20 miRNAs and 576 mRNAs. |
| [110] | ||
| PD | lncRNA-miRNA-mRNA | PD cell model (SY-SH5Y cells treated with α-synuclein oligomer) | PD-associated ceRNET that included the lncRNAs AC009365.4, RPS14P3 and G046036 together with the mRNAs IRF1, RIMKA, NAV1, SACS and SDC2. | [111] | |
| Human blood | A ceRNA regulatory network, including 7 lncRNAs (XIST1, PART1, MCF2L2, NOP14-AS1, LINC00328, LINC00302 and FAM215A), 3 miRNAs (miR-7, miR-433 and miR-133b), and 55 mRNAs. |
| [112] | ||
| Human brain (substantia nigra) | A ceRNA network associated with PD, that included 9 lncRNAs, 18 miRNAs, and 185 mRNA. |
| [113] | ||
| circRNA-miRNA-mRNA | Mouse model (MPTP) brain | A ceRNET built with 6 circRNAs (circ_0003292, circ_0001320, circ_0005976, circ_0005388, circ_0012384, and circ_003328), 13 miRNAs and 112 mRNAs. |
| [114] | |
| Human brain | A circRNA-associated ceRNA network predicted and validated. |
| [115] | ||
| ALS | circRNA-miRNA-mRNA | Human blood (leukocytes) | Two networks predicted. |
| [116,117] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno-García, L.; López-Royo, T.; Calvo, A.C.; Toivonen, J.M.; de la Torre, M.; Moreno-Martínez, L.; Molina, N.; Aparicio, P.; Zaragoza, P.; Manzano, R.; et al. Competing Endogenous RNA Networks as Biomarkers in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 9582. https://doi.org/10.3390/ijms21249582
Moreno-García L, López-Royo T, Calvo AC, Toivonen JM, de la Torre M, Moreno-Martínez L, Molina N, Aparicio P, Zaragoza P, Manzano R, et al. Competing Endogenous RNA Networks as Biomarkers in Neurodegenerative Diseases. International Journal of Molecular Sciences. 2020; 21(24):9582. https://doi.org/10.3390/ijms21249582
Chicago/Turabian StyleMoreno-García, Leticia, Tresa López-Royo, Ana Cristina Calvo, Janne Markus Toivonen, Miriam de la Torre, Laura Moreno-Martínez, Nora Molina, Paula Aparicio, Pilar Zaragoza, Raquel Manzano, and et al. 2020. "Competing Endogenous RNA Networks as Biomarkers in Neurodegenerative Diseases" International Journal of Molecular Sciences 21, no. 24: 9582. https://doi.org/10.3390/ijms21249582
APA StyleMoreno-García, L., López-Royo, T., Calvo, A. C., Toivonen, J. M., de la Torre, M., Moreno-Martínez, L., Molina, N., Aparicio, P., Zaragoza, P., Manzano, R., & Osta, R. (2020). Competing Endogenous RNA Networks as Biomarkers in Neurodegenerative Diseases. International Journal of Molecular Sciences, 21(24), 9582. https://doi.org/10.3390/ijms21249582