The Cardioprotective PKA-Mediated Hsp20 Phosphorylation Modulates Protein Associations Regulating Cytoskeletal Dynamics
Abstract
1. Introduction
2. Results
2.1. Identification of Hsp20/14-3-3 and 14-3-3/CFL2 Protein Complexes in Cardiac Muscle
2.2. Mapping of the Minimal Regions of Hsp20 and CFL2 Responsible for Binding to 14-3-3
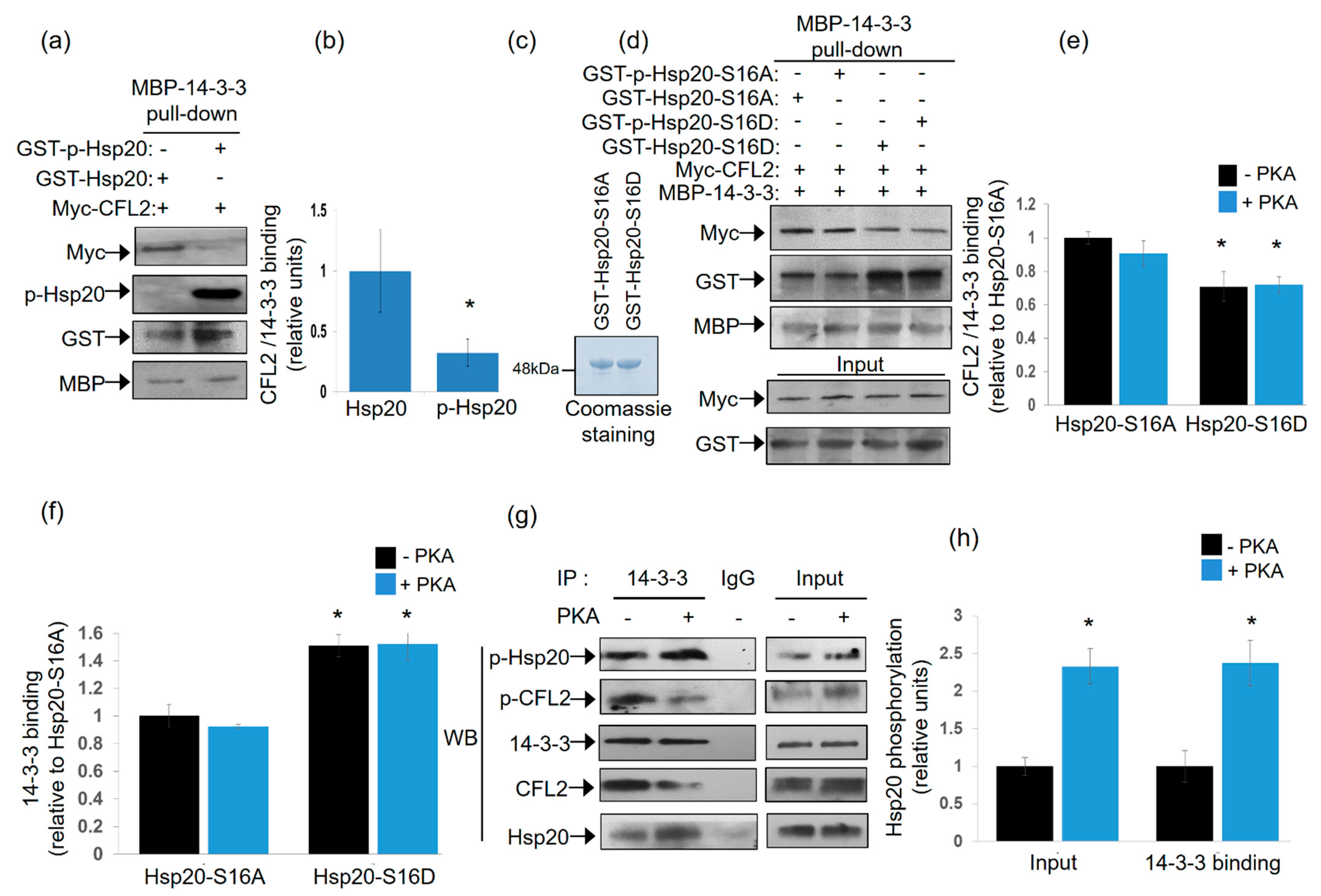
2.3. Hsp20 Competes with CFL2 for Binding to 14-3-3
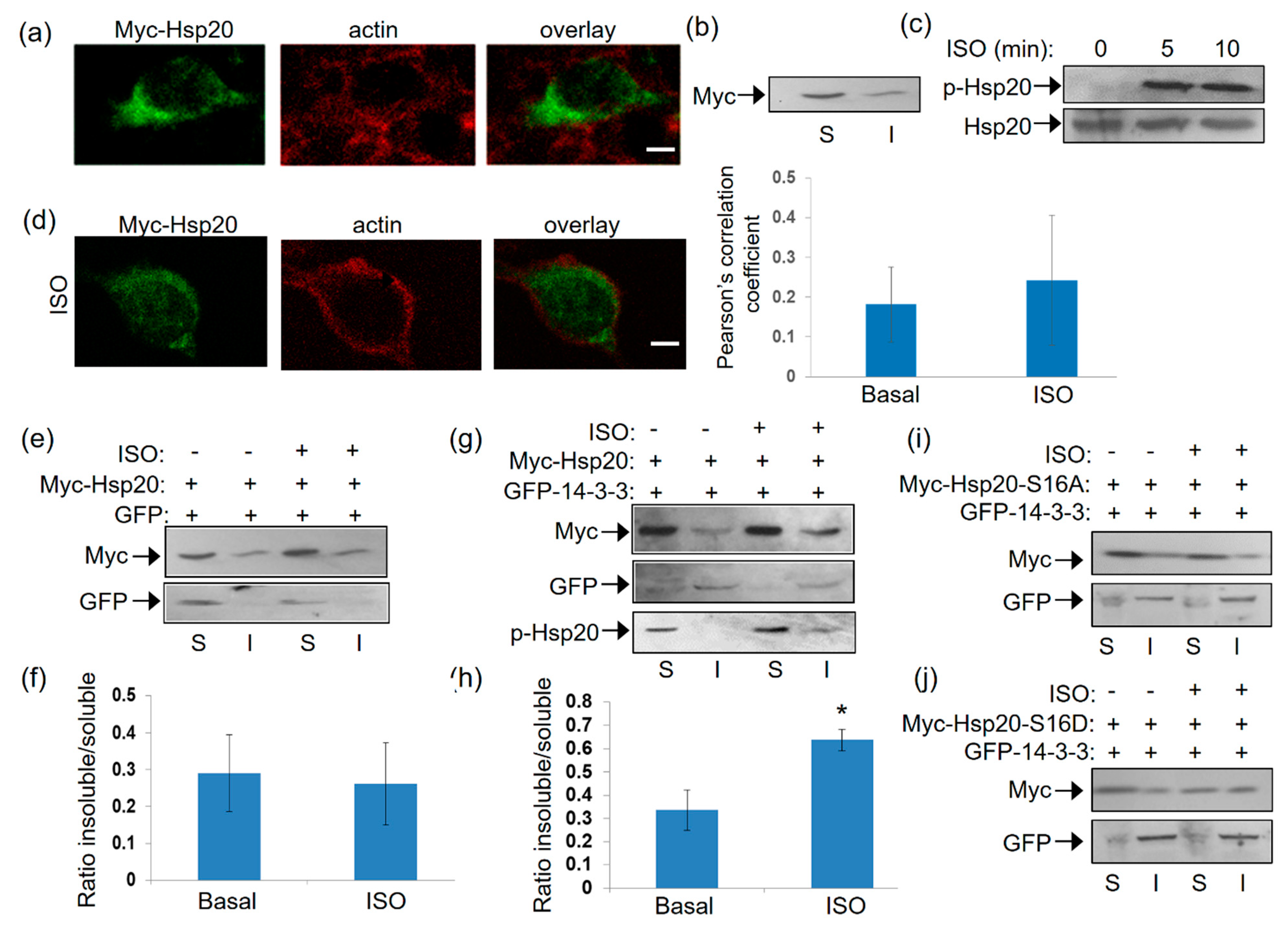
2.4. Isoproterenol-Induced Phosphorylation of Hsp20 Changes Its Subcellular Distribution by Promoting Its Interaction with 14-3-3
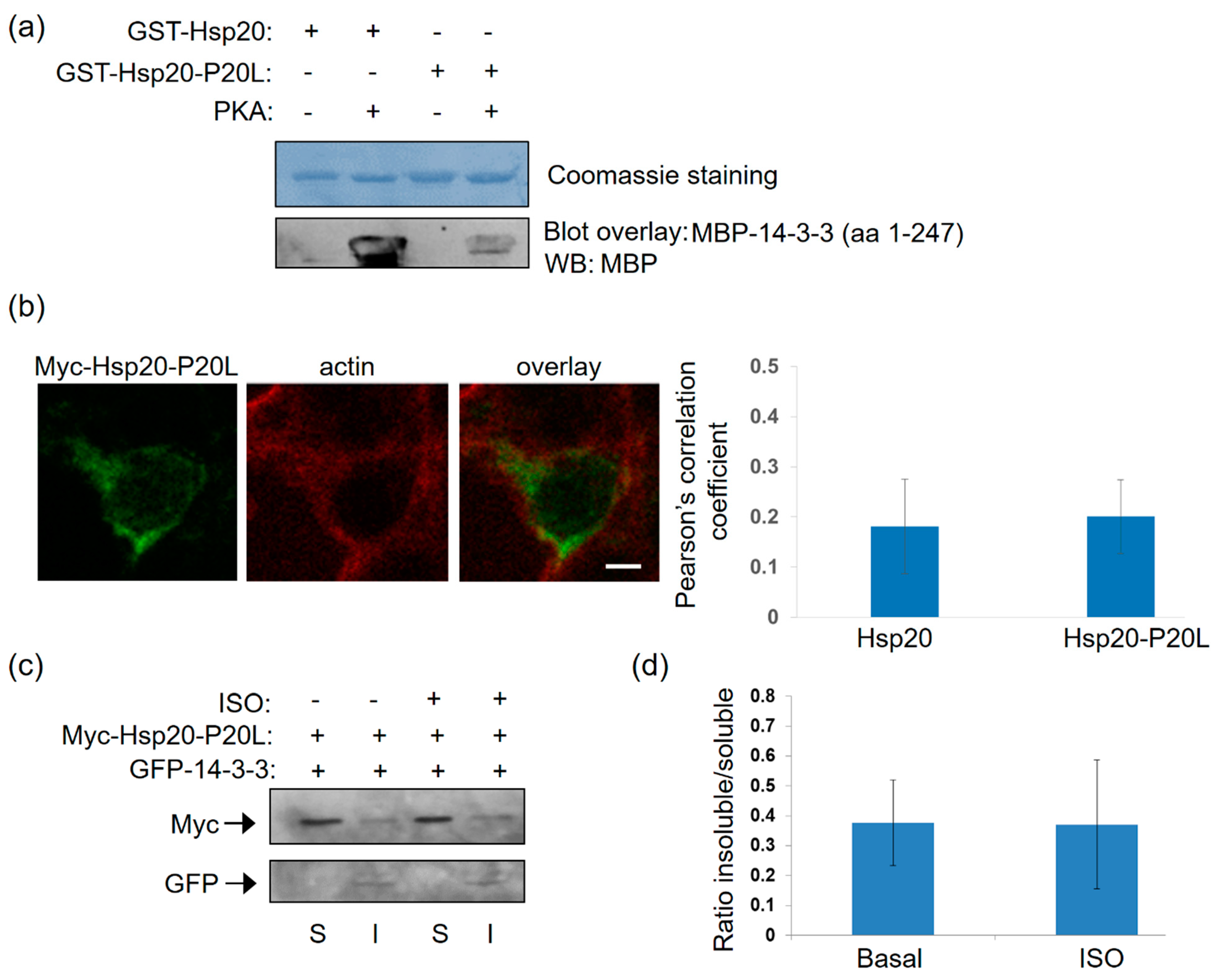
2.5. The Human Phosphorylation-Impairing Hsp20-P20L Mutation Displays Reduced 14-3-3 Interaction
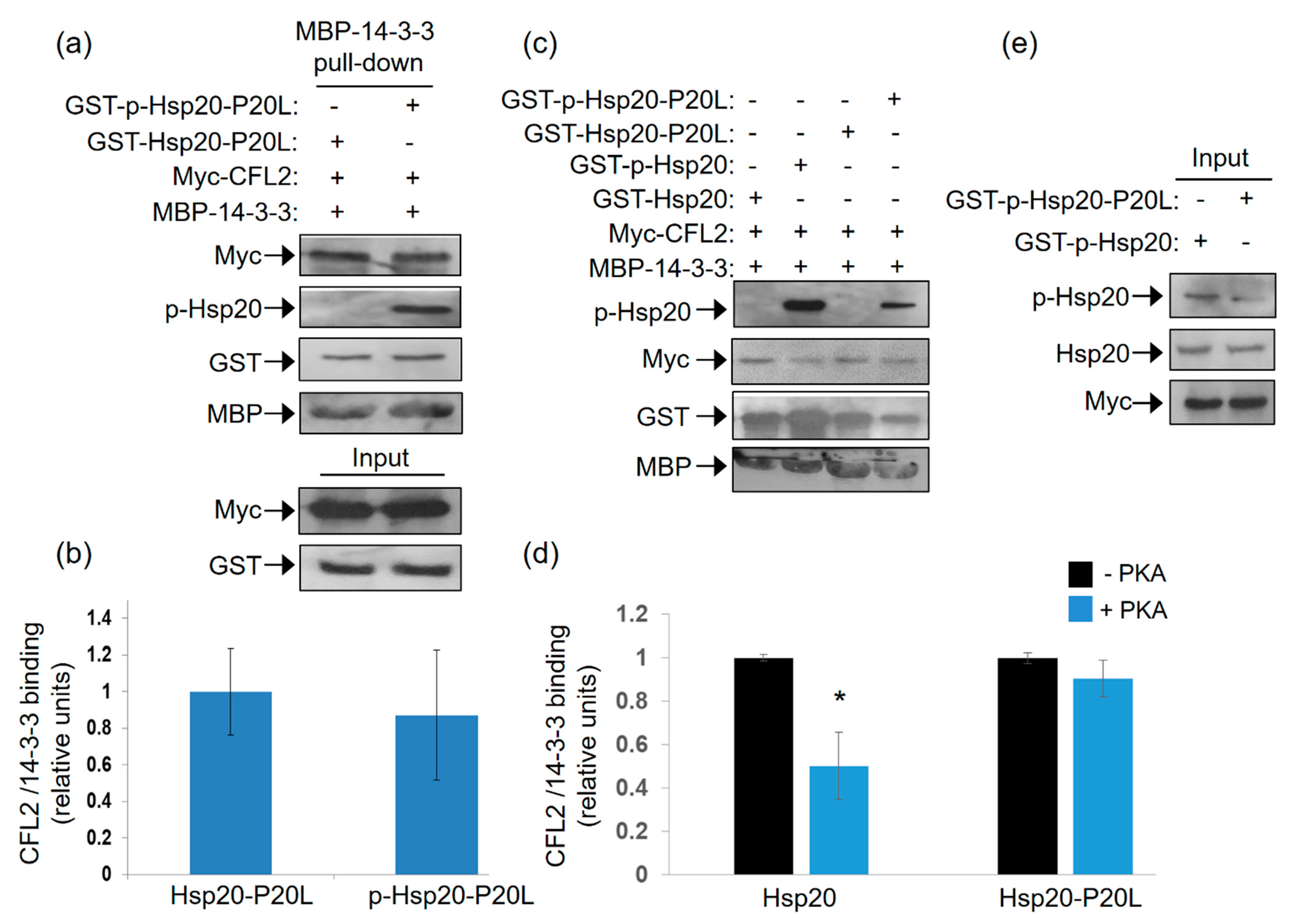
2.6. Hsp20-P20L Fails to Compete with CFL2 for Binding to 14-3-3
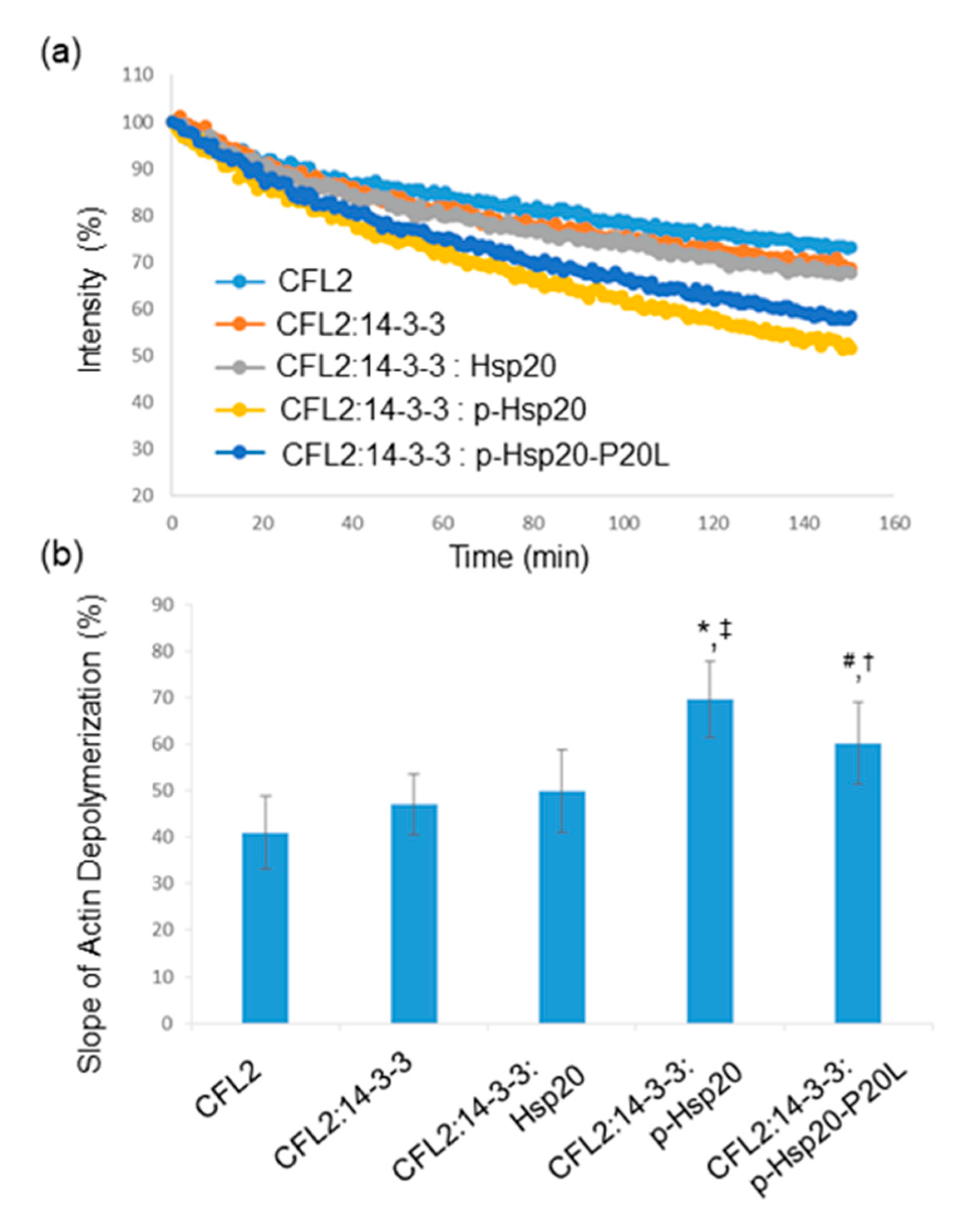
2.7. Hsp20 Phosphorylation Regulates CFL2-Mediated Actin Depolymerization
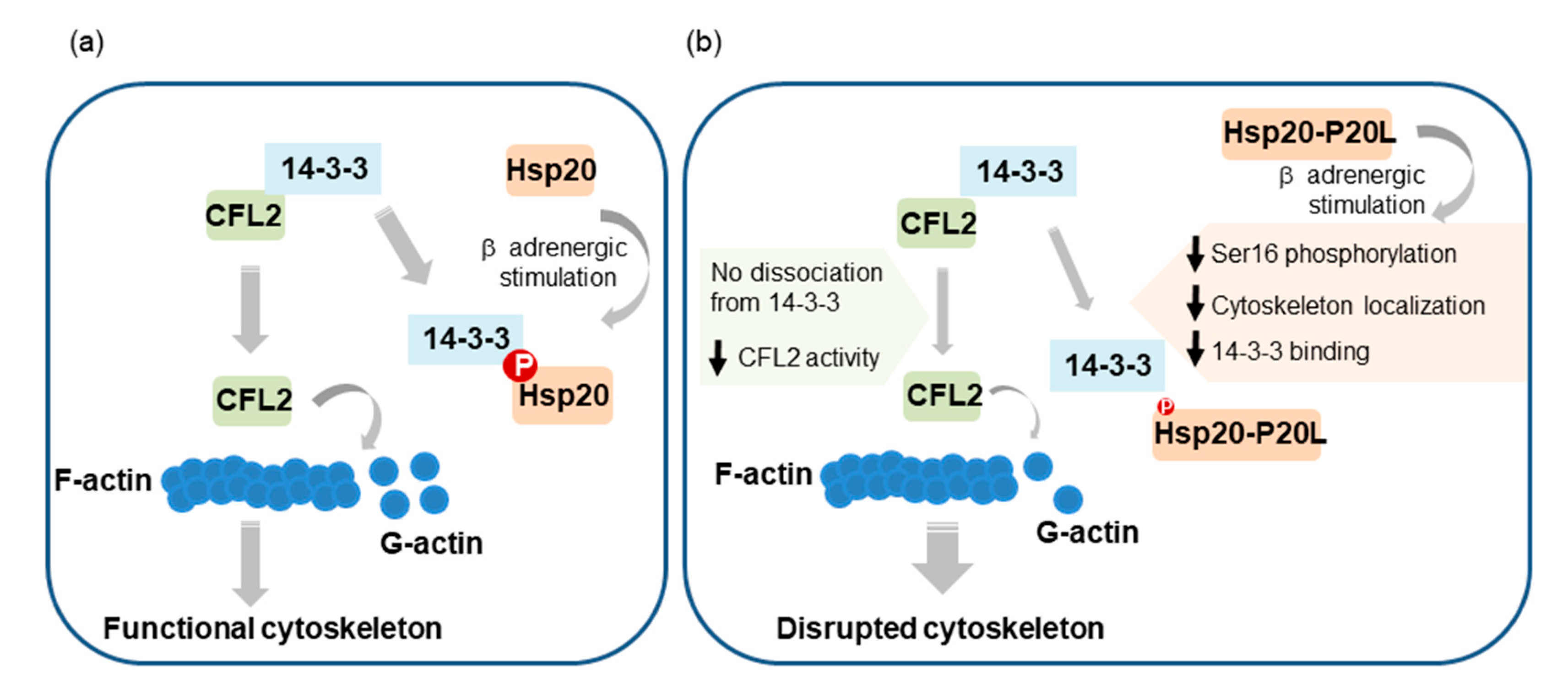
3. Discussion
4. Materials and Methods
4.1. Generation of Recombinant Proteins
4.2. Pull down Assays
4.3. Protein Phosphorylation
4.4. Immunoprecipitations
4.5. Blot Overlay Assays
4.6. Actin Depolymerization Assay
4.7. Cell Culture, Transfections and Immunofluorescence Studies
4.8. PCR Mutagenesis
4.9. Cell Fractionation
4.10. Protein Binding Competition Experiments
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Hsp20 | Heat shock protein 20 |
| CFL2 | cofilin-2 |
| DCM | dilated cardiomyopathy |
| Ser16 | Ser16 |
| PKA | cAMP-dependent protein kinase |
| ISO | isoproterenol |
References
- Chu, G.; Egnaczyk, G.F.; Zhao, W.; Jo, S.-H.; Fan, G.-C.; Maggio, J.E.; Xiao, R.-P.; Kranias, E.G. Phosphoproteome Analysis of Cardiomyocytes Subjected to β-Adrenergic Stimulation: Identification and characterization of a cardiac heat shock protein p20. Circ. Res. 2004, 94, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Goto, S.; Inaguma, Y.; Hasegawa, K.; Morishita, R.; Asano, T. Purification and characterization of a 20-kDa protein that is highly homologous to alpha B crystallin. J. Biol. Chem. 1994, 269, 15302–15309. [Google Scholar] [PubMed]
- Boluyt, M.O.; Brevick, J.L.; Rogers, D.S.; Randall, M.J.; Scalia, A.F.; Li, Z.B. Changes in the rat heart proteome induced by exercise training: Increased abundance of heat shock protein hsp20. Proteomics 2006, 6, 3154–3169. [Google Scholar] [CrossRef] [PubMed]
- Dohke, T.; Wada, A.; Isono, T.; Fujii, M.; Yamamoto, T.; Tsutamoto, T.; Horie, M. Proteomic Analysis Reveals Significant Alternations of Cardiac Small Heat Shock Protein Expression in Congestive Heart Failure. J. Card. Fail. 2006, 12, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.-C.; Chu, G.; Mitton, B.; Song, Q.; Yuan, Q.; Kranias, E.G. Small Heat-Shock Protein Hsp20 Phosphorylation Inhibits β-Agonist-Induced Cardiac Apoptosis. Circ. Res. 2004, 94, 1474–1482. [Google Scholar] [CrossRef]
- Qian, J.; Ren, X.; Wang, X.; Zhang, P.; Jones, W.K.; Molkentin, J.D.; Fan, G.-C.; Kranias, E.G. Blockade of Hsp20 phosphorylation exacerbates cardiac ischemia/reperfusion injury by suppressed autophagy and increased cell death. Circ. Res. 2009, 105, 1223–1231. [Google Scholar] [CrossRef]
- Fan, G.-C.; Ren, X.; Qian, J.; Yuan, Q.; Nicolaou, P.; Wang, Y.; Jones, W.K.; Chu, G.; Kranias, E.G. Novel Cardioprotective Role of a Small Heat-Shock Protein, Hsp20, Against Ischemia/Reperfusion Injury. Circulation 2005, 111, 1792–1799. [Google Scholar] [CrossRef]
- Fan, G.-C.; Yuan, Q.; Song, G.; Wang, Y.; Chen, G.; Qian, J.; Zhou, X.; Lee, Y.J.; Ashraf, M.; Kranias, E.G. Small Heat-Shock Protein Hsp20 Attenuates β-Agonist—Mediated Cardiac Remodeling Through Apoptosis Signal—Regulating Kinase 1. Circ. Res. 2006, 99, 1233–1242. [Google Scholar] [CrossRef]
- Ren, X.-P.; Wu, J.; Wang, X.; Sartor, M.A.; Qian, J.; Jones, K.; Nicolaou, P.; Pritchard, T.J.; Fan, G.-C. MicroRNA-320 Is Involved in the Regulation of Cardiac Ischemia/Reperfusion Injury by Targeting Heat-Shock Protein 20. Circulation 2009, 119, 2357–2366. [Google Scholar] [CrossRef]
- Edwards, H.V.; Scott, J.D.; Baillie, G.S. PKA phosphorylation of the small heat-shock protein Hsp20 enhances its cardioprotective effects. Biochem. Soc. Trans. 2012, 40, 210–214. [Google Scholar] [CrossRef]
- Fan, G.-C.; Chu, G.; Kranias, E.G. Hsp20 and Its Cardioprotection. Trends Cardiovasc. Med. 2005, 15, 138–141. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-S.; Zhu, H.; Cai, W.-F.; Wang, X.; Jiang, M.; Essandoh, K.; Vafiadaki, E.; Haghighi, K.; Lam, C.K.; Gardner, G.; et al. Regulation of BECN1-mediated autophagy by HSPB6: Insights from a human HSPB6S10F mutant. Autophagy 2018, 14, 80–97. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, P.; Knöll, R.; Haghighi, K.; Fan, G.-C.; Dorn, G.W.; Hasenfuß, G.; Kranias, E.G. Human Mutation in the Anti-apoptotic Heat Shock Protein 20 Abrogates Its Cardioprotective Effects. J. Biol. Chem. 2008, 283, 33465–33471. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-S.; Gardner, G.; Adly, G.; Jiang, M.; Cai, W.-F.; Lam, C.K.; Alogaili, F.; Robbins, N.; Rubinstein, J.; Kranias, E.G. A novel human S10F-Hsp20 mutation induces lethal peripartum cardiomyopathy. J. Cell. Mol. Med. 2018, 22, 3911–3919. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.-C.; Zhou, X.; Wang, X.; Song, G.; Qian, J.; Nicolaou, P.; Chen, G.; Ren, X.; Kranias, E.G. Heat Shock Protein 20 Interacting with Phosphorylated Akt Reduces Doxorubicin-Triggered Oxidative Stress and Cardiotoxicity. Circ. Res. 2008, 103, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Vafiadaki, E.; Florea, S.M.; Singh, V.P.; Song, W.; Lam, C.K.; Wang, Y.; Yuan, Q.; Pritchard, T.J.; Cai, W.; et al. Small Heat Shock Protein 20 Interacts with Protein Phosphatase-1 and Enhances Sarcoplasmic Reticulum Calcium Cycling. Circ. Res. 2011, 108, 1429–1438. [Google Scholar] [CrossRef]
- Sin, Y.Y.; Baillie, G.S. Heat shock protein 20 (HSP20) is a novel substrate for protein kinase D1 (PKD1). Cell Biochem. Funct. 2015, 33, 421–426. [Google Scholar] [CrossRef]
- Sin, Y.Y.; Edwards, H.V.; Li, X.; Day, J.P.; Christian, F.; Dunlop, A.J.; Adams, D.; Zaccolo, M.; Houslay, M.D.; Baillie, G.S. Disruption of the cyclic AMP phosphodiesterase-4 (PDE4)–HSP20 complex attenuates the β-agonist induced hypertrophic response in cardiac myocytes. J. Mol. Cell. Cardiol. 2011, 50, 872–883. [Google Scholar] [CrossRef]
- Sun, X.; Fontaine, J.-M.; Rest, J.S.; Shelden, E.A.; Welsh, M.J.; Benndorf, R. Interaction of Human HSP22 (HSPB8) with Other Small Heat Shock Proteins. J. Biol. Chem. 2003, 279, 2394–2402. [Google Scholar] [CrossRef]
- Chernik, I.S.; Seit-Nebi, A.; Marston, S.B.; Gusev, N.B. Small heat shock protein Hsp20 (HspB6) as a partner of 14-3-3γ. Mol. Cell. Biochem. 2007, 295, 9–17. [Google Scholar] [CrossRef]
- Dreiza, C.M.; Brophy, C.M.; Komalavilas, P.; Furnish, E.J.; Joshi, L.; Pallero, M.A.; Murphy-Ullrich, J.E.; Von Rechenberg, M.; Ho, Y.-S.J.; Richardson, B.; et al. Transducible heat shock protein 20 (HSP20) phosphopeptide alters cytoskeletal dynamics. FASEB J. 2004, 19, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sluchanko, N.N.; Artemova, N.V.; Sudnitsyna, M.V.; Safenkova, I.V.; Antson, A.A.; Levitsky, D.I.; Gusev, N.B. Monomeric 14-3-3ζ Has a Chaperone-Like Activity and Is Stabilized by Phosphorylated HspB6. Biochemistry 2012, 51, 6127–6138. [Google Scholar] [CrossRef] [PubMed]
- Sudnitsyna, M.V.; Seit-Nebi, A.; Gusev, N.B. Cofilin weakly interacts with 14-3-3 and therefore can only indirectly participate in regulation of cell motility by small heat shock protein HspB6 (Hsp20). Arch. Biochem. Biophys. 2012, 521, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Tessier, D.J.; Komalavilas, P.; Panitch, A.; Joshi, L.; Brophy, C.M. The small heat shock protein (HSP)20 is dynamically associated with the actin cross-linking protein actinin. J. Surg. Res. 2003, 111, 152–157. [Google Scholar] [CrossRef]
- Martin, T.P.; Currie, S.; Baillie, G.S. The cardioprotective role of small heat-shock protein 20. Biochem. Soc. Trans. 2014, 42, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Kanellos, G.; Frame, M.C. Cellular functions of the ADF/cofilin family at a glance. J. Cell Sci. 2016, 129, 3211–3218. [Google Scholar] [CrossRef]
- Pipkin, W.; Johnson, J.A.; Creazzo, T.L.; Burch, J.; Komalavilas, P.; Brophy, C.M. Localization, macromolecular associations, and function of the small heat shock-related protein HSP20 in rat heart. Circulation 2003, 107, 469–476. [Google Scholar] [CrossRef]
- Van De Klundert, F.A.; De Jong, W.W. The small heat shock proteins Hsp20 and αB-crystallin in cultured cardiac myocytes: Differences in cellular localization and solubilization after heat stress. Eur. J. Cell Biol. 1999, 78, 567–572. [Google Scholar] [CrossRef]
- Aitken, A. 14-3-3 proteins: A historic overview. Semin. Cancer Biol. 2006, 16, 162–172. [Google Scholar] [CrossRef]
- Muslin, A.J.; Tanner, J.; Allen, P.M.; Shaw, A.S. Interaction of 14-3-3 with Signaling Proteins Is Mediated by the Recognition of Phosphoserine. Cell 1996, 84, 889–897. [Google Scholar] [CrossRef]
- Sluchanko, N.N.; Gusev, N.B. 14-3-3 Proteins and regulation of cytoskeleton. Biochemistry 2010, 75, 1528–1546. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, M.B.; Rittinger, K.; Volinia, S.; Caron, P.R.; Aitken, A.; Leffers, H.; Gamblin, S.J.; Smerdon, S.J.; Cantley, L.C. The Structural Basis for 14-3-3: Phosphopeptide Binding Specificity. Cell 1997, 91, 961–971. [Google Scholar] [CrossRef]
- Sluchanko, N.N.; Bustos, D.M. Intrinsic disorder associated with 14-3-3 proteins and their partners. Prog. Mol. Biol. Transl. Sci. 2019, 166, 19–61. [Google Scholar] [CrossRef] [PubMed]
- Tinti, M.; Madeira, F.; Murugesan, G.; Hoxhaj, G.; Toth, R.; Mackintosh, C. ANIA: ANnotation and Integrated Analysis of the 14-3-3 interactome. Database 2014, 2014, bat085. [Google Scholar] [CrossRef] [PubMed]
- Ballone, A.; Centorrino, F.; Ottmann, C. 14-3-3: A Case Study in PPI Modulation. Molecules 2018, 23, 1386. [Google Scholar] [CrossRef] [PubMed]
- Obsilová, V.; Silhan, J.; Boura, E.; Teisinger, J.; Obsil, T. 14-3-3 proteins: A family of versatile molecular regulators. Physiol. Res. 2008, 57, 11–21. [Google Scholar]
- Bridges, D.; Moorhead, G.B.G. 14-3-3 Proteins: A Number of Functions for a Numbered Protein. Sci. STKE 2005, 2005, re10. [Google Scholar] [CrossRef]
- Hocking, K.M.; Evans, B.C.; Komalavilas, P.; Cheung-Flynn, J.; Duvall, C.L.; Brophy, C.M. Nanotechnology Enabled Modulation of Signaling Pathways Affects Physiologic Responses in Intact Vascular Tissue. Tissue Eng. Part A 2019, 25, 416–426. [Google Scholar] [CrossRef]
- Golenhofen, N.; Der Perng, M.; Quinlan, R.A.; Drenckhahn, D. Comparison of the small heat shock proteins αB-crystallin, MKBP, HSP25, HSP20, and cvHSP in heart and skeletal muscle. Histochem. Cell Biol. 2000, 122, 415–425. [Google Scholar] [CrossRef]
- Buja, L.M. Myocardial ischemia and reperfusion injury. Cardiovasc. Pathol. 2005, 14, 170–175. [Google Scholar] [CrossRef]
- Ndozangue-Touriguine, O.; Hamelin, J.; Bréard, J. Cytoskeleton and apoptosis. Biochem. Pharmacol. 2008, 76, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Franklin-Tong, V.E.; Gourlay, C.W. A role for actin in regulating apoptosis/programmed cell death: Evidence spanning yeast, plants and animals. Biochem. J. 2008, 413, 389–404. [Google Scholar] [CrossRef]
- Gourlay, C.W.; Ayscough, K.R. The actin cytoskeleton: A key regulator of apoptosis and ageing? Nat. Rev. Mol. Cell Biol. 2005, 6, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Perricone, A.J.; Heide, R.S.V. Novel therapeutic strategies for ischemic heart disease. Pharmacol. Res. 2014, 89, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Moulton, V.R.; Lapchak, P.H.; Deng, G.-M.; Lucca, J.J.D.; Tsokos, G.C. Ischemia-mediated aggregation of the actin cytoskeleton is one of the major initial events resulting in ischemia-reperfusion injury. Am. J. Physiol. Liver Physiol. 2009, 296, G339–G347. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haslbeck, M.; Vierling, E. A First Line of Stress Defense: Small Heat Shock Proteins and Their Function in Protein Homeostasis. J. Mol. Biol. 2015, 427, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Gusev, N.B.; Bogatcheva, N.V.; Marston, S.B. Structure and Properties of Small Heat Shock Proteins (sHsp) and Their Interaction with Cytoskeleton Proteins. Biochemistry 2002, 67, 511–519. [Google Scholar] [CrossRef]
- Mounier, N.; Arrigo, A.P. Actin cytoskeleton and small heat shock proteins: How do they interact? Cell Stress Chaperon 2002, 7, 167–176. [Google Scholar] [CrossRef]
- Chaudhuri, S.; Smith, P.G. Cyclic Strain–Induced HSP27 Phosphorylation Modulates Actin Filaments in Airway Smooth Muscle Cells. Am. J. Respir. Cell Mol. Biol. 2008, 39, 270–278. [Google Scholar] [CrossRef]
- García-Arguinzonis, M.; Padro, T.; Lugano, R.; Llorente-Cortes, V.; Badimon, L. Low-Density Lipoproteins Induce Heat Shock Protein 27 Dephosphorylation, Oligomerization, and Subcellular Relocalization in Human Vascular Smooth Muscle Cells. Arter. Thromb. Vasc. Biol. 2010, 30, 1212–1219. [Google Scholar] [CrossRef]
- Robinson, A.A.; Dunn, M.J.; McCormack, A.; Dos Remedios, C.; Rose, M.L. Protective effect of phosphorylated Hsp27 in coronary arteries through actin stabilization. J. Mol. Cell. Cardiol. 2010, 49, 370–379. [Google Scholar] [CrossRef]
- Wang, Y.; Xiong, Z.; Gong, W.; Zhou, P.; Xie, Q.; Zhou, Z.; Lu, G. Expression of heat shock protein 27 correlates with actin cytoskeletal dynamics and contractility of cultured human bladder smooth muscle cells. Exp. Cell Res. 2015, 338, 39–44. [Google Scholar] [CrossRef]
- Vafiadaki, E.; Arvanitis, D.A.; Sanoudou, D.; Kranias, E.G. Identification of a Protein Phosphatase-1/Phospholamban Complex That Is Regulated by cAMP-Dependent Phosphorylation. PLoS ONE 2013, 8, e80867. [Google Scholar] [CrossRef]
- Agrawal, P.B.; Greenleaf, R.S.; Tomczak, K.K.; Lehtokari, V.-L.; Wallgren-Pettersson, C.; Wallefeld, W.; Laing, N.G.; Darras, B.T.; Maciver, S.K.; Dormitzer, P.R.; et al. Nemaline Myopathy with Minicores Caused by Mutation of the CFL2 Gene Encoding the Skeletal Muscle Actin—Binding Protein, Cofilin-2. Am. J. Hum. Genet. 2007, 80, 162–167. [Google Scholar] [CrossRef]
- Agrawal, P.B.; Joshi, M.; Savic, T.; Chen, Z.; Beggs, A.H. Normal myofibrillar development followed by progressive sarcomeric disruption with actin accumulations in a mouse Cfl2 knockout demonstrates requirement of cofilin-2 for muscle maintenance. Hum. Mol. Genet. 2012, 21, 2341–2356. [Google Scholar] [CrossRef] [PubMed]
- Gurniak, C.B.; Chevessier, F.; Jokwitz, M.; Jönsson, F.; Perlas, E.; Richter, H.; Matern, G.; Boyl, P.P.; Chaponnier, C.; Fürst, D.; et al. Severe protein aggregate myopathy in a knockout mouse model points to an essential role of cofilin2 in sarcomeric actin exchange and muscle maintenance. Eur. J. Cell Biol. 2014, 93, 252–266. [Google Scholar] [CrossRef] [PubMed]
- Papalouka, V.; Arvanitis, D.A.; Vafiadaki, E.; Mavroidis, M.; Papadodima, S.A.; Spiliopoulou, C.A.; Kremastinos, D.T.; Kranias, E.G.; Sanoudou, D. Muscle Lim Protein Interacts with Cofilin 2 and Regulates F-Actin Dynamics in Cardiac and Skeletal Muscle. Mol. Cell. Biol. 2009, 29, 6046–6058. [Google Scholar] [CrossRef]
- Vafiadaki, E.; Sanoudou, D.; Arvanitis, D.A.; Catino, D.H.; Kranias, E.G.; Kontrogianni-Konstantopoulos, A. Phospholamban Interacts with HAX-1, a Mitochondrial Protein with Anti-apoptotic Function. J. Mol. Biol. 2007, 367, 65–79. [Google Scholar] [CrossRef]
- Vafiadaki, E.; Arvanitis, D.A.; Pagakis, S.N.; Papalouka, V.; Sanoudou, D.; Kontrogianni-Konstantopoulos, A.; Kranias, E.G. The Anti-apoptotic Protein HAX-1 Interacts with SERCA2 and Regulates Its Protein Levels to Promote Cell Survival. Mol. Biol. Cell 2009, 20, 306–318. [Google Scholar] [CrossRef]
- Arvanitis, D.A.; Vafiadaki, E.; Papalouka, V.; Sanoudou, D. Muscle Lim Protein and myosin binding protein C form a complex regulating muscle differentiation. Biochim. Biophys. Acta BBA Bioenerg. 2017, 1864, 2308–2321. [Google Scholar] [CrossRef]
- Vafiadaki, E.; Arvanitis, D.A.; Papalouka, V.; Terzis, G.; Roumeliotis, T.I.; Spengos, K.; Garbis, S.D.; Manta, P.; Kranias, E.G.; Sanoudou, D. Muscle lim protein isoform negatively regulates striated muscle actin dynamics and differentiation. FEBS J. 2014, 281, 3261–3279. [Google Scholar] [CrossRef]
- Lam, C.K.; Zhao, W.; Cai, W.; Vafiadaki, E.; Florea, S.M.; Ren, X.; Liu, Y.; Robbins, N.; Zhang, Z.; Zhou, X.; et al. Novel role of HAX-1 in ischemic injury protection involvement of heat shock protein 90. Circ. Res. 2013, 112, 79–89. [Google Scholar] [CrossRef]
- Liu, G.-S.; Morales, A.; Vafiadaki, E.; Lam, C.K.; Cai, W.-F.; Haghighi, K.; Adly, G.; Hershberger, R.E.; Kranias, E.G. A novel human R25C-phospholamban mutation is associated with super-inhibition of calcium cycling and ventricular arrhythmia. Cardiovasc. Res. 2015, 107, 164–174. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vafiadaki, E.; Arvanitis, D.A.; Eliopoulos, A.G.; Kranias, E.G.; Sanoudou, D. The Cardioprotective PKA-Mediated Hsp20 Phosphorylation Modulates Protein Associations Regulating Cytoskeletal Dynamics. Int. J. Mol. Sci. 2020, 21, 9572. https://doi.org/10.3390/ijms21249572
Vafiadaki E, Arvanitis DA, Eliopoulos AG, Kranias EG, Sanoudou D. The Cardioprotective PKA-Mediated Hsp20 Phosphorylation Modulates Protein Associations Regulating Cytoskeletal Dynamics. International Journal of Molecular Sciences. 2020; 21(24):9572. https://doi.org/10.3390/ijms21249572
Chicago/Turabian StyleVafiadaki, Elizabeth, Demetrios A. Arvanitis, Aristides G. Eliopoulos, Evangelia G. Kranias, and Despina Sanoudou. 2020. "The Cardioprotective PKA-Mediated Hsp20 Phosphorylation Modulates Protein Associations Regulating Cytoskeletal Dynamics" International Journal of Molecular Sciences 21, no. 24: 9572. https://doi.org/10.3390/ijms21249572
APA StyleVafiadaki, E., Arvanitis, D. A., Eliopoulos, A. G., Kranias, E. G., & Sanoudou, D. (2020). The Cardioprotective PKA-Mediated Hsp20 Phosphorylation Modulates Protein Associations Regulating Cytoskeletal Dynamics. International Journal of Molecular Sciences, 21(24), 9572. https://doi.org/10.3390/ijms21249572








