BIRC6 Is Associated with Vulnerability of Carotid Atherosclerotic Plaque
Abstract
1. Introduction
2. Results
2.1. Identification of RNA-Seq-Derived Single Nucleotide Polymorphisms (SNPs) and Indels Associated with Carotid Plaque Symptomatology
2.2. Synonymous Exonic BIRC6 SNP rs35286811 Is Associated with Vulnerable Carotid Plaque
2.3. Full-Length BIRC6 Expression Is Upregulated in Carotid Atheroma Plaques from Symptomatic Patients
2.4. rs35286811 Genotype Is a cis-QTL Regulating BIRC6 Expression Levels in Carotid Plaque
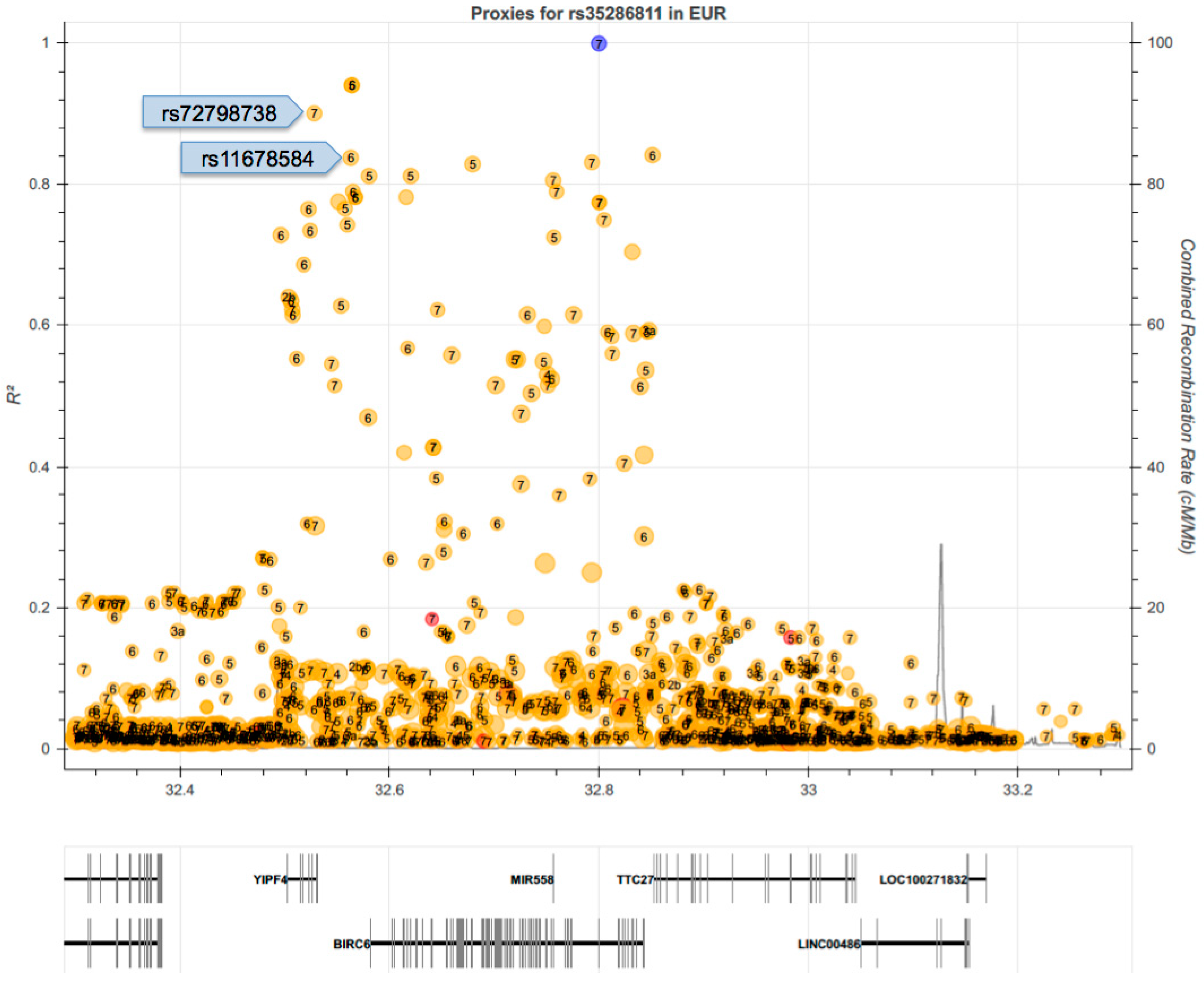
2.5. Linkage Disequilibrium (LD) of rs35286811 with GWAS SNPs Associated with Independent Traits Indicates Pleiotropy of the BIRC6 Locus
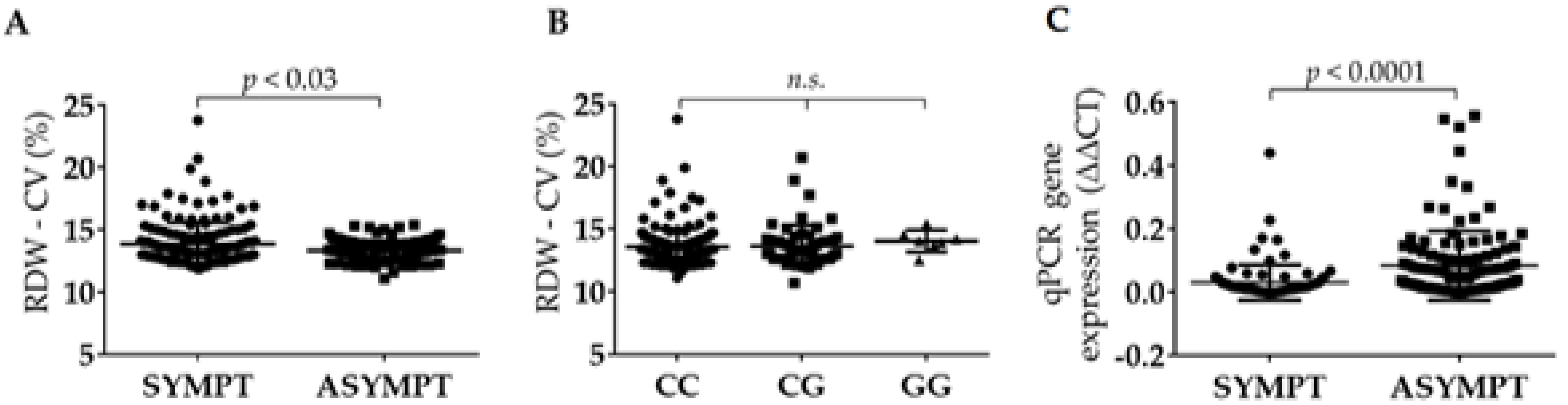
2.6. RDW Is Increased in Symptomatic Patients and Is Independent from BIRC6 rs35286811 Genotype
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. RNA-Seq Variant Detection Analysis
4.3. DNA and RNA Extraction from Carotid Atheroma Plaques
4.4. Selection and Genotyping of SNPs
4.5. Real-Time qPCR for Detection of BIRC6 and MAP1LC3B Expression in Carotid Atheroma RNA
4.6. Statistical Analysis
5. Patents
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BIRC6 | Baculoviral IAP Repeat Containing 6 |
| CI | Confidence interval |
| cIMT | Carotid intima media thickness |
| CVA | Cerebrovascular accident |
| LD | Linkage disequilibrium |
| OR | Odds ratio |
| QTL | Quantitative trait locus |
| RDW | Red cell distribution width |
| RDW-CV | RDW coefficient of variation |
| SNP | Single nucleotide polymorphism |
| VSMC | Vascular smooth muscle cells |
Appendix A
| Patient Characteristics | RNA-Seq Group | Asymp | Sympt |
|---|---|---|---|
| Number, n | 14 | 7 | 7 |
| Years | 68 ± 8 | 68 ± 8 | 68 ± 8 |
| Sex M/F, n | 11/3 | 4/3 | 7/0 |
| Treatment with statins | 14 | 7 | 7 |
| Risk Factors (%): | |||
| Diabetes Mellitus | 7 (50) | 3 (43) | 4 (57) |
| Dyslipidemia | 14 (100) | 7 (100) | 7 (100) |
| Arterial hypertension | 14 (100) | 7 (100) | 7 (100) |
| Tobacco | 7 (50) | 4 (57) | 3 (43) |
| Patient Characteristics | Validation Group | Asymp | Sympt |
|---|---|---|---|
| Number, n | 301 | 169 | 132 |
| Years | 70 ± 15 | 70 ± 15 | 70 ± 15 |
| Sex M/F, n | 263/38 | 147/22 | 116/16 |
| Treatment with statins | 61 | 110 | 78 |
| Risk Factors (%): | |||
| Diabetes Mellitus | 130 (43) | 69 (40) | 61 (41) |
| Dyslipidemia | 209 (68) | 122 (72) | 87 (66) |
| Arterial hypertension | 240 (79) | 137 (81) | 103 (78) |
| Tobacco | 101 (33) | 64 (37) | 37 (27) |
References
- Lubna, A.; Schnitzler, J.G.; Kroon, J. Metabolism: The road to inflammation and atherosclerosis. Curr. Opin. Lipidol. 2018, 29, 474–480. [Google Scholar]
- Goikuria, H.; Vandenbroeck, K.; Alloza, I. Inflammation in human carotid atheroma plaques. Cytokine Growth Factor Rev. 2018, 39, 62–70. [Google Scholar] [CrossRef] [PubMed]
- GBD 2015 DALYs and HALE Collaborators. Global, regional, and national disability-adjusted life-years (DALYs) for 315 diseases and injuries and healthy life expectancy (HALE), 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1603–1658. [Google Scholar] [CrossRef]
- Favate, A.S.; Younger, D.S. Epidemiology of ischemic stroke. Neurol. Clin. 2016, 34, 967–980. [Google Scholar] [CrossRef] [PubMed]
- Montaner, J.; Ramiro, L.; Simats, A.; Tiedt, S.; Makris, K.; Jickling, G.C.; Debette, S.; Sanchez, J.C.; Bustamante, A. Multilevel omics for the discovery of biomarkers and therapeutic targets for stroke. Nat. Rev. Neurol. 2020, 16, 247–264. [Google Scholar] [CrossRef] [PubMed]
- Chauan, G.; Debette, S. Genetic risk factors for ischemic and hemorrhagic stroke. Curr. Radiol. Rep. 2016, 18, 124. [Google Scholar]
- NINDS Stroke Genetics Network (SiGN); International Stroke Genetics Consortium (ISGC). The NINDS stroke genetics network: A genome-wide association study of ischemic stroke and its subtypes. Lancet Neurol. 2016, 15, 174–184. [Google Scholar] [CrossRef]
- Malik, R.; Chauhan, G.; Traylor, M.; Sargurupremraj, M.; Okada, Y.; Mishra, A.; Rutten-Jacobs, L.; Giese, A.K.; van der Laan, S.W.; Gretarsdottir, S. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018, 50, 524–537. [Google Scholar] [CrossRef]
- Mathiesen, E.B.; Johnsen, S.H.; Wilsgaard, T.; Bønaa, K.H.; Løchen, M.L.; Njølstad, I. Carotid plaque area and intima-media thickness in prediction of first-ever ischemic stroke. Stroke 2011, 42, 972–978. [Google Scholar] [CrossRef]
- Bis, J.C.; Kavousi, M.; Franceschini, N.; Iaacs, A.; Abecasis, G.R.; Schminke, U.; Post, W.Q.; Smith, A.V.; Cupples, L.A.; Markus, H.S. Meta-analysis of genome-wide association studies from the CHARGE consortium identifies common variants associated with carotid intima media thickness and plaque. Nat. Genet. 2011, 43, 940–947. [Google Scholar] [CrossRef]
- Franceschini, N.; Giambartolomei, C.; de Vries, P.S.; Finan, C.; Bis, J.C.; Huntley, R.P.; Lovering, R.C.; Tajuddin, S.M.; Winkler, T.W.; Graff, M.; et al. GWAS and colocalization analyses implicate carotid intima-media thickness and carotid plaque loci in cardiovascular outcomes. Nat. Commun. 2018, 9, 5141. [Google Scholar] [CrossRef] [PubMed]
- Traylor, M.; Mäkelä, K.M.; Kilarski, L.L.; Holliday, E.G.; Devan, W.J.; Nalls, M.A.; Wiggins, K.L.; Zhao, W.; Cheng, Y.C.; Achterberg, S.; et al. A novel MMP12 locus is associated with large artery atherosclerotic stroke using a genome-wide age-at-onset informed approach. PLoS Genet. 2014, 10, e1004469. [Google Scholar] [CrossRef] [PubMed]
- Collura, S.; Morsiani, C.; Vacirca, A.; Fronterrè, S.; Ciavarella, C.; Vasuri, F.; D’Errico, A.; Franceschi, C.; Pasquinelli, G.; Gargiulo, M.; et al. The carotid plaque as paradigmatic case of site-specific acceleration of aging process: The microRNAs and the inflammaging contribution. Aging Res. Rev. 2020, 61, 101090. [Google Scholar] [CrossRef] [PubMed]
- Moreno, P.R. Vulnerable plaque: Definition, diagnosis, and treatment. Cardiol. Clin. 2010, 28, 1–30. [Google Scholar] [CrossRef]
- Virmani, R.; Burke, A.P.; Farb, A.; Kolodgie, F. Pathology of the vulnerable plaque. J. Am. Coll. Cardiol. 2006, 47, C13–C18. [Google Scholar] [CrossRef]
- Golldedge, J.; Greenhalgh, R.M.; Davies, A.H. The symptomatic carotid plaque. Stroke 2000, 31, 774–781. [Google Scholar] [CrossRef]
- Silvestre-Roig, C.; de Winther, M.P.; Weber, C.; Daemen, M.J.; Lutgens, E.; Soehnlein, O. Atherosclerotic plaque destabilization: Mechanisms, models, and therapeutic strategies. Circ. Res. 2014, 114, 214–226. [Google Scholar] [CrossRef]
- Bennett, M.R.; Sinha, S.; Owens, G.K. Vascular smooth muscle cells in atherosclerosis. Circ. Res. 2016, 118, 692–702. [Google Scholar] [CrossRef]
- Goikuria, H.; Freijo, M.D.M.; Vega Manrique, R.; Sasztre, M.; Elizagaray, E.; Lorenzo, A.; Vandenbroeck, K.; Alloza, I. Characterization of carotid smooth muscle cells during phenotypic transition. Cells 2018, 7, 23. [Google Scholar] [CrossRef]
- Alloza, I.; Goikuria, H.; Idor, J.L.; Triviño, J.C.; Fernández Velasco, J.M.; Elizagary, E.; García-Barcina, M.; Montoya-Murillo, G.; Sarasola, E.; Vega Manrique, R.; et al. RNAseq based transcriptomics study of SMCs from carotid atherosclerotic plaque: BMP2 and IDs proteins are crucial regulators of plaque stability. Sci. Rep. 2017, 7, 3470. [Google Scholar] [CrossRef]
- GATK—Broad Institute. Available online: https://gatk.broadinstitute.org/hc/en-us (accessed on 3 July 2020).
- Ensembl Genome Browser 102. Available online: https://www.ensembl.org/index.html (accessed on 3 July 2020).
- GWAS Catalog—EMBL-EBI. Available online: https://www.ebi.ac.uk/gwas/downloads (accessed on 7 July 2020).
- LDlink—An Interactive Web Tool for Exploring Linkage Disequilibrium. Available online: https://ldlink.nci.nih.gov/?tab=home (accessed on 7 July 2020).
- Kichaev, G.; Bhatia, G.; Loh, P.R.; Gazal, S.; Burch, K.; Freund, M.K.; Schoech, A.; Pasaniuc, B.; Price, A.L. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 2019, 104, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Astle, W.J.; Elding, H.; Jiang, T.; Allen, D.; Ruklisa, D.; Mann, A.L.; Mead, D.; Bouman, H.; Riveros-Mckay, F.; Kostadima, M.A.; et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 2016, 167, 1415–1429. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.E.; Lane, J.M.; Wood, A.R.; van Hees, V.T.; Tyrrell, J.; Baumont, R.N.; Jeffries, A.R.; Dashti, H.S.; Hillsdon, M.; Ruth, K.S.; et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rythms. Nat. Commun. 2019, 10, 343. [Google Scholar] [CrossRef] [PubMed]
- Open Target Genetics. Available online: https://genetics.opentargets.org (accessed on 3 July 2020).
- Liu, H.; Cao, Y.; Tong, T.; Shi, J.; Zhang, Y.; Yang, Y.; Liu, C. Autophagy in atherosclerosis: A phenomenon found in human carotid atherosclerotic plaques. Chin. Med. J. 2015, 128, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, B.; Goikuria, H.; Vega, R.; Rodríguez-Antigüedad, A.; López Medina, A.; Freijo, M.; Vandenbroeck, K.; Alloza, I. Autophagic marker MAP1LC3B expression levels are associated with carotid atherosclerosis symptomatology. PLoS ONE 2014, 9, e115176. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, F. The anti-apoptotic ubiquitin-conjugating enzyme BIRC6/BRUCE regulates autophagosome-lysosome fusion. Autophagy 2018, 14, 1283–1284. [Google Scholar] [CrossRef]
- Ebner, P.; Poetsch, I.; Deszcz, L.; Hoffmann, T.; Zuber, J.; Ikeda, F. The IAP family member BRUCE regulates autophagosome-lysosome fusion. Nat. Commun. 2018, 9, 599. [Google Scholar] [CrossRef]
- Jia, R.; Bonifacino, J.S. Negative regulation of autophagy by UBA6-BIRC6-mediated ubiquitination of LC3B. Elife 2019, 8, e50034. [Google Scholar] [CrossRef]
- Jia, R.; Bonifacino, J.S. Regulation of LC3B levels by ubiquitination and proteasomal degradation. Autophagy 2020, 16, 382–384. [Google Scholar] [CrossRef]
- Jiang, T.X.; Zou, J.B.; Zhu, Q.Q.; Liu, C.H.; Wang, G.F.; Du, T.T.; Luo, Z.Y.; Guo, F.; Zhou, L.M.; Liu, J.J.; et al. SIP/CacyBP promotes autophagy by regulating levels of BRUCE/Apollon, which stimulates LC3-I degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 13404–13413. [Google Scholar] [CrossRef]
- Che, L.; Yang, X.; Chunmen, G.; El-Amouri, S.S.; Wang, Q.E.; Pan, D.; Herzog, T.; Du, C. Loss of BRUCE reduces cellular energy level and induces autophagy by driving activation of the AMPK-ULK1 autophagic initiating axis. PLoS ONE 2019, 14, e0216553. [Google Scholar] [CrossRef] [PubMed]
- Alloza, I.; Goikuria, H.; Freijo, M.D.M.; Vandenbroeck, K. A role for autophagy in carotid atherosclerosis. Eur. J. Stroke 2016, 1, 255–263. [Google Scholar] [CrossRef]
- Zhu, Y.N.; Fan, W.J.; Zhang, C.; Guo, F.; Li, W.; Wang, Y.F.; Jiang, Z.S.; Qu, S.L. Role of autophagy in advanced atherosclerosis. Mol. Med. Rep. 2017, 15, 2903–2908. [Google Scholar] [CrossRef] [PubMed]
- Grootaert, M.O.J.; Roth, L.; Schrijvers, D.M.; De Meyer, G.R.Y.; Martinet, W. Defective autophagy in atherosclerosis: To die or to senesce? Oxidative Med. Cell. Longev. 2018, 2018, 7687083. [Google Scholar] [CrossRef] [PubMed]
- Song, S.Y.; Hua, C.; Dornbors, D.; Kang, R.; Zhao, X.X.; Du, X.; He, W.; Ding, Y.; Meng, R. Baseline red blood cell distribution width as a predictor of stroke occurrence and outcome: A comprehensive meta-analysis of 31 studies. Front. Neurol. 2019, 10, 1237. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, S.; Liu, X.; Fang, J.; Zhuang, W. Association between red cell distribution width level and risk of stroke. Medicine 2020, 99, e19691. [Google Scholar] [CrossRef]
- Feng, G.H.; Li, H.P.; Li, Q.L.; Fu, Y.; Huang, R.B. Red blood cell distribution width and ischaemic stroke. Stroke Vasc. Neurol. 2017, 2, 172–175. [Google Scholar] [CrossRef]
- Lappegård, J.; Ellingsen, T.S.; Vik, A.; Skjelbakken, T.; Brox, J.; Mathiesen, E.B.; Johnsen, S.H.; Brækkan, S.K.; Hansen, J.B. Red cell distribution width and carotid atherosclerosis progression. The Tromsø study. Thromb. Haemost. 2015, 113, 649–654. [Google Scholar]
- Silva, C.M.; Giovani, P.; Viana, M.B. High reticulocyte count is an independent risk factor for cardiovascular disease in children with sickle cell anemia. Pediatr. Blood Cancer 2011, 5, 116–121. [Google Scholar] [CrossRef]
- Fan, M.; Sun, D.; Zhou, T.; Heianza, Y.; Lv, J.; Li, L.; Qi, L. Sleep patterns, genetic susceptibility, and incident of cardiovascular disease: A prospective study of 385,292 UK biobank participants. Eur. Heart J. 2020, 41, 1182–1189. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, K.; Xiao, X.; Liao, J.; Hu, Q.; Chen, H.; Liu, J.; An, X. Autophagy as a regulatory component of erythropoiesis. Int. J. Mol. Sci. 2015, 16, 4083–4093. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Panda, S.; Lin, J.D. Temporal orchestration of circadian autophagy rhythm by C/EBPbeta. EMBO J. 2011, 30, 4642–4651. [Google Scholar] [CrossRef] [PubMed]
- Maiese, K. Moving to the rhythm with clock (circadian) genes, autophagy, mTOR, and SIRT1 in degenerative disease and cancer. Curr. Neurovasc. Res. 2017, 14, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, E.; Wilterdink, J.L.; Kosinski, A.; Lynn, M.; Chimowitz, M.I.; Sarafin, J.; Smith, H.H.; Nichols, F.; Rogg, J.; Cloft, H.J.; et al. The stroke outcomes and neuroimaging of intracranial atherosclerosis (SONIA) trial. Neurology 2007, 68, 2099–2106. [Google Scholar] [CrossRef] [PubMed]
- Seqgene Download. Available online: https://sourceforge.net/projects/seqgene/ (accessed on 5 July 2020).
- D3 Assay Design—Fluidigm. Available online: https://d3.fluidigm.com/account/login (accessed on 5 July 2020).


| Effects by Type | Counts (%) | Effects by Impact | Counts (%) |
|---|---|---|---|
| 3′UTR | 154 (19.5) | HIGH | 12 (1.5) |
| 5′UTR | 26 (3.3) | MODERATE | 40 (5) |
| downstream_gene | 164 (20.8) | LOW | 31 (4) |
| frameshift | 11 (1.4) | MODIFIER | 706 (89.5) |
| intron | 168 (21.3) | ||
| missense | 40 (5) | ||
| non_coding_exon | 81 (10.3) | ||
| regulatory_region | 40 (5) | ||
| splice_region | 1 (0.13) | ||
| synonymous | 31 (4) | ||
| upstream_gene | 73 (9.2) |
| Gene | Chr | SNP Position | Ref./Var. Alleles 1 | Variant Effect | rs Number | Carrier Counts (S/A) 2 | p | Cis-QTL p |
|---|---|---|---|---|---|---|---|---|
| PDLIM4 | 5 | 132272249 | G/C | synonymous | rs9895 | 5/0 | 0.021 | 0.03 |
| ATL3 | 11 | 63625771 | A/G | 3_prime_UTR | rs79429913 | 0/5 | 0.021 | 0.04 |
| BIRC6 | 2 | 32575355 | C/G | synonymous | rs35286811 | 5/0 | 0.021 | 0.07 |
| TMEM167A | 5 | 83054823 | C/T | 3_prime_UTR | rs13162274 | 0/5 | 0.021 | 0.05 |
| HTT | 4 | 3135947 | G/A | missense | rs363075 | 5/0 | 0.021 | n.s. 3 |
| BDH2 | 4 | 103998912 | T/C | intron | rs6825519 | 5/0 | 0.021 | n.s. |
| SLK | 10 | 62885307 | A/G | synonymous | rs10883960 | 5/0 | 0.021 | n.s. |
| SLC3A2 | 11 | 55828523 | C/T | synonymous | rs4726 | 5/0 | 0.021 | n.s. |
| NEDD4 | 15 | 55828523 | C/T | 3_prime_UTR | rs2899593 | 5/0 | 0.021 | n.s. |
| MGLL | 3 | 127691190 | C/G | 3_prime_UTR | rs76232599 | 0/6 | 0.005 | n.s. |
| RMND1 | 6 | 151405165 | G/C | 3_prime_UTR | rs1065310 | 0/6 | 0.005 | n.s. |
| ZNF664 | 12 | 124015292 | C/T | 3_prime_UTR | rs3768 | 0/6 | 0.005 | n.s. |
| Gene | SNP | Risk Allele | RAF 1 Asympt | RAF Sympt | Other Allele | p | OR 2 (95% CI 3) |
|---|---|---|---|---|---|---|---|
| ATL3 | rs79429913 | A | 0.85 | 0.86 | G | 0.72 | 1.09 (0.641.72) |
| BDH2 | rs6825519 | C | 0.18 | 0.22 | T | 0.18 | 1.33 (0.88–1.98) |
| BIRC6 | rs35286811 | G | 0.07 | 0.15 | C | 0.002 | 2.24 (1.27–3.93) |
| HTT | rs363075 | A | 0.09 | 0.10 | G | 0.63 | 1.14 (0.66–1.98) |
| MGLL | rs76232599 | G | 0.29 | 0.31 | C | 0.61 | 1.07 (0.75–1.53) |
| NEDD4 | rs2899593 | T | 0.09 | 0.13 | C | 0.16 | 1.47 (0.84–2.55) |
| PDLIM4 | rs9895 | C | 0.06 | 0.09 | G | 0.22 | 1.47 (0.478–2.48) |
| RNMD1 | rs1065310 | G | 0.71 | 0.77 | C | 0.12 | 1.35 (0.92–1.94) |
| SLC3A2 | rs4726 | T | 0.20 | 0.27 | C | 0.08 | 1.41 (0.94–2.13) |
| SLK | rs10883960 | A | 0.73 | 0.77 | G | 0.31 | 1.21 (0.83–1.77) |
| TMEM167A | rs10883960 | C | 0.74 | 0.76 | T | 0.41 | 1.12 (0.77–1.64) |
| ZNF664 | rs13162274 | C | 0.72 | 0.78 | T | 0.06 | 1.45 (0.97–2.15) |
| SNP | Alleles (Risk Allele) | RAF 1 | Linkage Disequilibrium (LD) 2 | p for Assoc. with Trait | Trait 3 | Ref. | ||
|---|---|---|---|---|---|---|---|---|
| Allele Correlation | D’ | r2 | ||||||
| rs72798738 | T/C (T) | 0.12 | rs72798738*T—rs35286811*G | 0.96 | 0.90 | 7 × 10−18 | Red cell distribution width | [25] |
| rs11678584 | T/A (T) | 0.13 | rs11678584*T—rs35286811*G | 0.96 | 0.87 | 2 × 10−11 | Reticulocyte fraction of red cells | [26] |
| 1 × 10−10 | Reticulocyte count | [26] | ||||||
| 4 × 10−9 | Chronotype | [27] | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alloza, I.; Salegi, A.; Mena, J.; Navarro, R.T.; Martin, C.; Aspichueta, P.; Salazar, L.M.; Carpio, J.U.; Cagigal, P.D.-l.-H.; Vega, R.; et al. BIRC6 Is Associated with Vulnerability of Carotid Atherosclerotic Plaque. Int. J. Mol. Sci. 2020, 21, 9387. https://doi.org/10.3390/ijms21249387
Alloza I, Salegi A, Mena J, Navarro RT, Martin C, Aspichueta P, Salazar LM, Carpio JU, Cagigal PD-l-H, Vega R, et al. BIRC6 Is Associated with Vulnerability of Carotid Atherosclerotic Plaque. International Journal of Molecular Sciences. 2020; 21(24):9387. https://doi.org/10.3390/ijms21249387
Chicago/Turabian StyleAlloza, Iraide, Andrea Salegi, Jorge Mena, Raquel Tulloch Navarro, César Martin, Patricia Aspichueta, Lucía Martínez Salazar, Jon Uriarte Carpio, Patricia De-la-Hera Cagigal, Reyes Vega, and et al. 2020. "BIRC6 Is Associated with Vulnerability of Carotid Atherosclerotic Plaque" International Journal of Molecular Sciences 21, no. 24: 9387. https://doi.org/10.3390/ijms21249387
APA StyleAlloza, I., Salegi, A., Mena, J., Navarro, R. T., Martin, C., Aspichueta, P., Salazar, L. M., Carpio, J. U., Cagigal, P. D.-l.-H., Vega, R., Triviño, J. C., Freijo, M. d. M., & Vandenbroeck, K. (2020). BIRC6 Is Associated with Vulnerability of Carotid Atherosclerotic Plaque. International Journal of Molecular Sciences, 21(24), 9387. https://doi.org/10.3390/ijms21249387






