Long-Lasting Activity of ERK Kinase Depends on NFATc1 Induction and Is Involved in Cell Migration-Fusion in Murine Macrophages RAW264.7
Abstract
1. Introduction
2. Results
2.1. ERK Activation in RAW 264.7 Cells RANKL-Stimulated
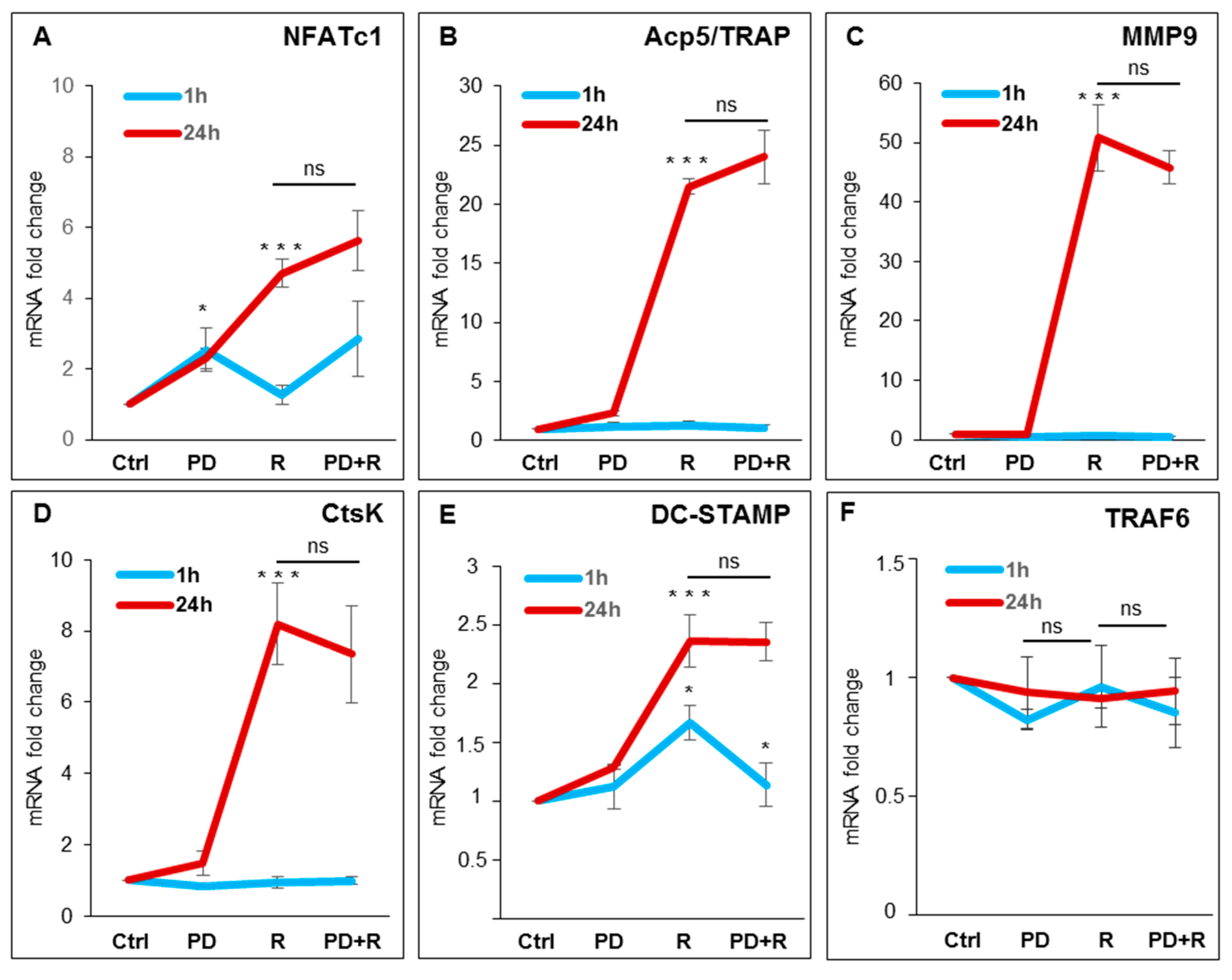
2.2. RANKL-Induced Osteoclast Hallmarks Expression at 24 h
2.3. Effects of ERK1/2 Inhibitors on Osteoclastogenesis
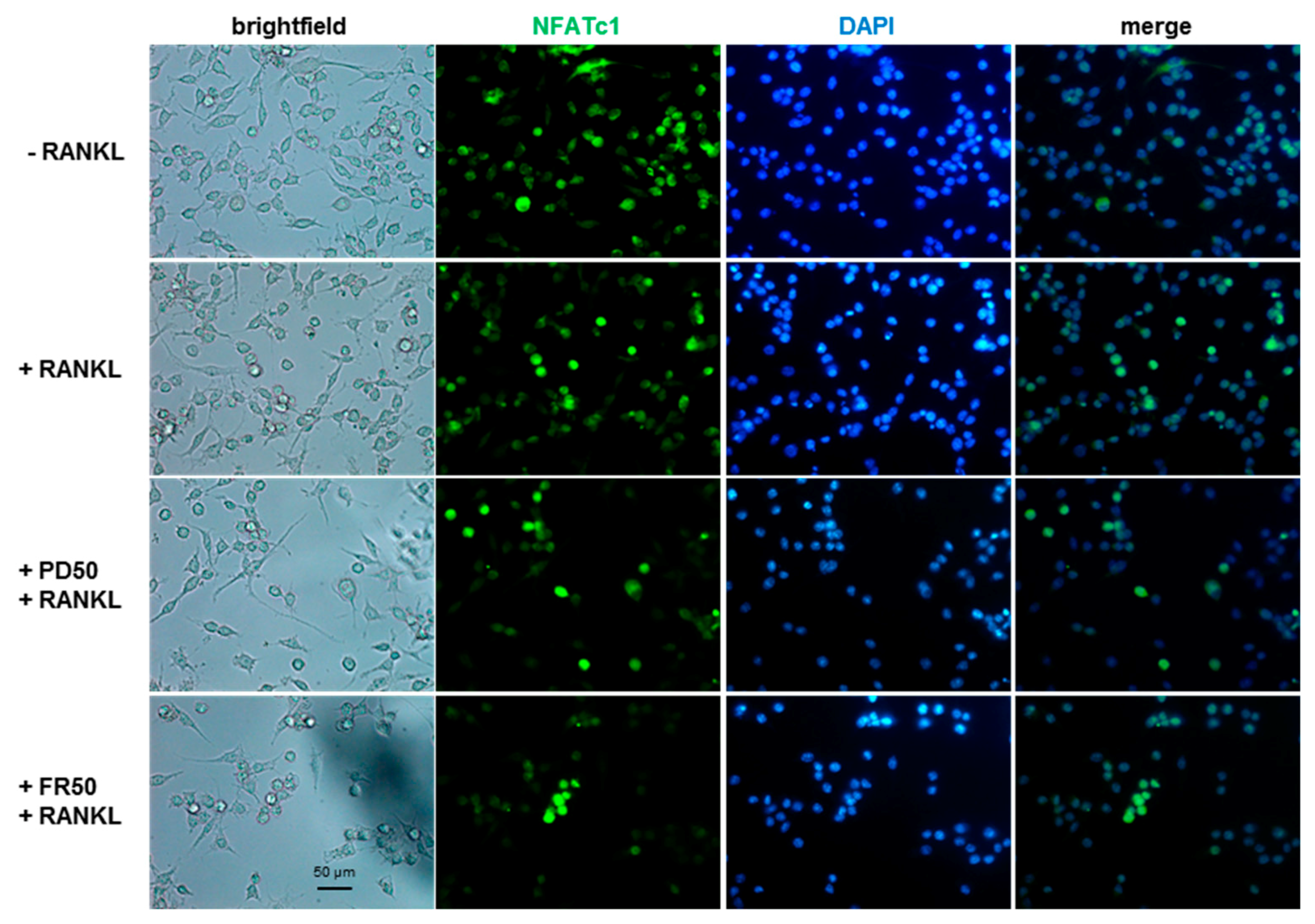
2.4. Effects of ERK1/2 Inhibitors on NFATc1 Nuclear Translocation
2.5. Effects of NFATc1 Depletion on Osteoclastogenesis
2.6. Functional Analysis of ERK Activation on Cell Migration and Cell-Cell Fusion
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. TRAP Staining
4.3. Cell Fusion
4.4. RNA Extraction, cDNA Synthesis, and Quantitative Polymerase Chain Reaction (qPCR)
4.5. Immunofluorescence Analysis
4.6. Western Blot
4.7. Small Interfering RNA (siRNA) Transfection
4.8. Migration Assay
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Xing, L. Regulation of osteoclast precursors in inflammatory bone loss. Curr. Opin. Investig. Drugs (London, England 2000) 2009, 10, 1195–1203. [Google Scholar]
- Yoshida, H.; Hayashi, S.-I.; Kunisada, T.; Ogawa, M.; Nishikawa, S.; Okamura, H.; Sudo, T.; Shultz, L.D.; Nishikawa, S.-I. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nat. Cell Biol. 1990, 345, 442–444. [Google Scholar] [CrossRef] [PubMed]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast Differentiation and Activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Segovia-Silvestre, T.; Neutzsky-Wulff, A.V.; Sorensen, M.G.; Christiansen, C.; Bollerslev, J.; Karsdal, M.A.; Henriksen, K. Advances in osteoclast biology resulting from the study of osteopetrotic mutations. Qual. Life Res. 2009, 124, 561–577. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Schwarz, E.M.; Boyce, B.F. Osteoclast precursors, RANKL/RANK, and immunology. Immunol. Rev. 2005, 208, 19–29. [Google Scholar] [CrossRef]
- Hofbauer, L.C.; Heufelder, A.E. Role of receptor activator of nuclear factor-κB ligand and osteoprotegerin in bone cell biology. J. Mol. Med. 2001, 79, 243–253. [Google Scholar] [CrossRef]
- Horowitz, M.C.; Xi, Y.; Wilson, K.; Kacena, M. Control of osteoclastogenesis and bone resorption by members of the TNF family of receptors and ligands. Cytokine Growth Factor Rev. 2001, 12, 9–18. [Google Scholar] [CrossRef]
- Wu, H.; Arron, J.R. TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. BioEssays 2003, 25, 1096–1105. [Google Scholar] [CrossRef]
- Teitelbaum, S.L.; Ross, F.P. Genetic regulation of osteoclast development and function. Nat. Rev. Genet. 2003, 4, 638–649. [Google Scholar] [CrossRef]
- Kawamura, N.; Kugimiya, F.; Oshima, Y.; Ohba, S.; Ikeda, T.; Saito, T.; Shinoda, Y.; Kawasaki, Y.; Ogata, N.; Hoshi, K.; et al. Akt1 in Osteoblasts and Osteoclasts Controls Bone Remodeling. PLoS ONE 2007, 2, e1058. [Google Scholar] [CrossRef]
- Asagiri, M.; Sato, K.; Usami, T.; Ochi, S.; Nishina, H.; Yoshida, H.; Morita, I.; Wagner, E.F.; Mak, T.W.; Serfling, E.; et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J. Exp. Med. 2005, 202, 1261–1269. [Google Scholar] [CrossRef]
- Takayanagi, H.; Kim, S.; Koga, T.; Nishina, H.; Isshiki, M.; Yoshida, H.; Saiura, A.; Isobe, M.; Yokochi, T.; Inoue, J.-I.; et al. Induction and Activation of the Transcription Factor NFATc1 (NFAT2) Integrate RANKL Signaling in Terminal Differentiation of Osteoclasts. Dev. Cell 2002, 3, 889–901. [Google Scholar] [CrossRef]
- Matsumoto, M.; Sudo, T.; Saito, T.; Osada, H.; Tsujimoto, M. Involvement of p38 mitogen-activated protein kinase signaling pathway in osteoclastogenesis mediated by receptor activator of NF-κB ligand (RANKL). J. Biol. Chem. 2000, 275, 31155–31161. [Google Scholar] [CrossRef]
- David, J.-P.; Sabapathy, K.; Hoffmann, O.; Idarraga, M.H.; Wagner, E.F. JNK1 modulates osteoclastogenesis through both c-Jun phosphorylation-dependent and -independent mechanisms. J. Cell Sci. 2002, 115, 4317–4325. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Staser, K.; Rhodes, S.D.; Liu, Y.; Wu, X.; Park, S.-J.; Yuan, J.; Yang, X.; Li, X.; Jiang, L.; et al. Erk1 Positively Regulates Osteoclast Differentiation and Bone Resorptive Activity. PLoS ONE 2011, 6, e24780. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Chung, Y.H.; Ahn, H.; Kim, H.; Rho, J.; Jeong, D. Selective Regulation of MAPK Signaling Mediates RANKL-dependent Osteoclast Differentiation. Int. J. Biol. Sci. 2016, 12, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Russo, R.; Mallia, S.; Zito, F.; Lampiasi, N. Gene Expression Profiling of NFATc1-Knockdown in RAW 264.7 Cells: An Alternative Pathway for Macrophage Differentiation. Cells 2019, 8, 131. [Google Scholar] [CrossRef]
- Ohori, M.; Kinoshita, T.; Okubo, M.; Sato, K.; Yamazaki, A.; Arakawa, H.; Nishimura, S.; Inamura, N.; Nakajima, H.; Neya, M.; et al. Identification of a selective ERK inhibitor and structural determination of the inhibitor–ERK2 complex. Biochem. Biophys. Res. Commun. 2005, 336, 357–363. [Google Scholar] [CrossRef]
- Qiao, H.; May, J.M. Macrophage differentiation increases expression of the ascorbate transporter (SVCT2). Free. Radic. Biol. Med. 2009, 46, 1221–1232. [Google Scholar] [CrossRef]
- Ross, F.P. M-CSF, c-Fms, and Signaling in Osteoclasts and their Precursors. Ann. N. Y. Acad. Sci. 2006, 1068, 110–116. [Google Scholar] [CrossRef]
- Wada, T.; Nakashima, T.; Hiroshi, N.; Penninger, J.M. RANKL–RANK signaling in osteoclastogenesis and bone disease. Trends Mol. Med. 2006, 12, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Chuderland, D.; Konson, A.; Seger, R. Article Identification and Characterization of a General Nuclear Translocation Signal in Signaling Proteins. Mol. Cell 2008, 31, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Asano, S.; Nakamura, T.; Adachi, M.; Yoshida, M.; Yanagida, M.; Nishida, E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nat. Cell Biol. 1997, 390, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Prowse, C.N.; Lew, J. Mechanism of Activation of ERK2 by Dual Phosphorylation. J. Biol. Chem. 2000, 276, 99–103. [Google Scholar] [CrossRef]
- Adachi, M.; Fukuda, M.; Nishida, E. Two co-existing mechanisms for nuclear import of MAP kinase: Passive diffusion of a monomer and active transport of a dimer. EMBO J. 1999, 18, 5347–5358. [Google Scholar] [CrossRef]
- Yazicioglu, M.N.; Goad, D.L.; Ranganathan, A.; Whitehurst, A.W.; Goldsmith, E.J.; Cobb, M.H. Mutations in ERK2 Binding Sites Affect Nuclear Entry. J. Biol. Chem. 2007, 282, 28759–28767. [Google Scholar] [CrossRef]
- Caunt, C.J.; McArdle, C.A. Stimulus-induced uncoupling of extracellular signal-regulated kinase phosphorylation from nuclear localization is dependent on docking domain interactions. J. Cell Sci. 2010, 123, 4310–4320. [Google Scholar] [CrossRef][Green Version]
- Lee, T.; Hoofnagle, A.N.; Kabuyama, Y.; Stroud, J.; Min, X.; Goldsmith, E.J.; Chen, L.; A Resing, K.; Ahn, N.G. Docking Motif Interactions in MAP Kinases Revealed by Hydrogen Exchange Mass Spectrometry. Mol. Cell 2004, 14, 43–55. [Google Scholar] [CrossRef]
- Wolf, I.; Rubinfeld, H.; Yoon, S.; Marmor, G.; Hanoch, T.; Seger, R. Involvement of the Activation Loop of ERK in the Detachment from Cytosolic Anchoring. J. Biol. Chem. 2001, 276, 24490–24497. [Google Scholar] [CrossRef]
- Tanoue, T.; Maeda, R.; Adachi, M.; Nishida, E. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 2001, 20, 466–479. [Google Scholar] [CrossRef]
- Jacobs, D.; Glossip, D.; Xing, H.; Muslin, A.J.; Kornfeld, K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999, 13, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Casar, B.; Pinto, A.; Crespo, P. Essential Role of ERK Dimers in the Activation of Cytoplasmic but Not Nuclear Substrates by ERK-Scaffold Complexes. Mol. Cell 2008, 31, 708–721. [Google Scholar] [CrossRef] [PubMed]
- Matsubayashi, Y.; Fukuda, M.; Nishida, E. Evidence for Existence of a Nuclear Pore Complex-mediated, Cytosol-independent Pathway of Nuclear Translocation of ERK MAP Kinase in Permeabilized Cells. J. Biol. Chem. 2001, 276, 41755–41760. [Google Scholar] [CrossRef] [PubMed]
- Tanoue, T.; Adachi, M.; Moriguchi, T.; Nishida, E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2000, 2, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Sharrocks, A.D.; Yang, S.-H.; Galanis, A. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 2000, 25, 448–453. [Google Scholar] [CrossRef]
- Bardwell, L.; Thorner, J. A conserved motif at the amino termini of MEKs might mediate high-affinity interaction with the cognate MAPKs. Trends Biochem. Sci. 1996, 21, 373–374. [Google Scholar] [CrossRef]
- Carlson, J.; Cui, W.; Zhang, Q.; Xu, X.; Mercan, F.; Bennett, A.M.; Vignery, A. Role of MKP-1 in Osteoclasts and Bone Homeostasis. Am. J. Pathol. 2009, 175, 1564–1573. [Google Scholar] [CrossRef]
- Sasaki, A.; Taketomi, T.; Kato, R.; Saeki, K.; Nonami, A.; Sasaki, M.; Kuriyama, M.; Saito, N.; Shibuya, M.; Yoshimura, A. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat. Cell Biol. 2003, 5, 427–432. [Google Scholar] [CrossRef]
- Sun, M.; Huang, F.; Yu, D.; Zhang, Y.; Xu, H.; Zhang, L.; Li, L.; Dong, L.; Guo, L.; Wang, S. Autoregulatory loop between TGF-β1/miR-411-5p/SPRY4 and MAPK pathway in rhabdomyosarcoma modulates proliferation and differentiation. Cell Death Dis. 2015, 6, e1859. [Google Scholar] [CrossRef]
- Li, Y.; Shi, Z.; Jules, J.; Chen, S.; Kesterson, R.A.; Zhao, D.; Zhang, P.; Feng, X. Specific RANK Cytoplasmic Motifs Drive Osteoclastogenesis. J. Bone Miner. Res. 2019, 34, 1938–1951. [Google Scholar] [CrossRef]
- Alessi, D.R.; Cuenda, A.; Cohen, P.; Dudley, D.T.; Saltiel, A.R. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 1995, 270, 27489–27494. [Google Scholar] [CrossRef] [PubMed]
- Hotokezaka, H.; Sakai, E.; Kanaoka, K.; Saito, K.; Matsuo, K.-I.; Kitaura, H.; Yoshida, N.; Nakayama, K. U0126 and PD98059, Specific Inhibitors of MEK, Accelerate Differentiation of RAW264.7 Cells into Osteoclast-like Cells. J. Biol. Chem. 2002, 277, 47366–47372. [Google Scholar] [CrossRef]
- Oh, J.H.; Lee, J.Y.; Park, J.H.; No, J.H.; Lee, N.K. Obatoclax Regulates the Proliferation and Fusion of Osteoclast Precursors through the Inhibition of ERK Activation by RANKL. Mol. Cells 2015, 38, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Zito, F.; Lampiasi, N.; Kireev, I.; Russo, R. United we stand: Adhesion and molecular mechanisms driving cell fusion across species. Eur. J. Cell Biol. 2016, 95, 552–562. [Google Scholar] [CrossRef]
- Lampiasi, N.; Russo, R.; Zito, F. The Alternative Faces of Macrophage Generate Osteoclasts. BioMed Res. Int. 2016, 2016, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tanimura, S.; Takeda, K. ERK signalling as a regulator of cell motility. J. Biochem. 2017, 162, 145–154. [Google Scholar] [CrossRef]
- Kim, B.H.; Oh, J.H.; Lee, N.K. The inactivation of ERK1/2, P38 and NF-kU is involved in the down-regulation of osteoclastogenesis and function by A2B adenosine receptor stimulation. Mol. Cells 2017, 40, 752–760. [Google Scholar]
- Sundaram, K.; Nishimura, R.; Senn, J.; Youssef, R.F.; London, S.D.; Reddy, S.V. RANK ligand signaling modulates the matrix metalloproteinase-9 gene expression during osteoclast differentiation. Exp. Cell Res. 2007, 313, 168–178. [Google Scholar] [CrossRef]
- Engsig, M.T.; Chen, Q.-J.; Vu, T.H.; Pedersen, A.-C.; Therkidsen, B.; Lund, L.R.; Henriksen, K.; Lenhard, T.; Foged, N.T.; Werb, Z.; et al. Matrix Metalloproteinase 9 and Vascular Endothelial Growth Factor Are Essential for Osteoclast Recruitment into Developing Long Bones. J. Cell Biol. 2000, 151, 879–890. [Google Scholar] [CrossRef]
- Moon, S.-H.; Choi, S.; Kim, S.H. In vitro anti-osteoclastogenic activity of p38 inhibitor doramapimod via inhibiting migration of pre-osteoclasts and NFATc1 activity. J. Pharmacol. Sci. 2015, 129, 135–142. [Google Scholar] [CrossRef]
- Wilson, S.R.; Peters, C.; Saftig, P.; Brömme, D. Cathepsin K Activity-dependent Regulation of Osteoclast Actin Ring Formation and Bone Resorption. J. Biol. Chem. 2009, 284, 2584–2592. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Jacobson, K.; Schaller, M.D. MAP kinases and cell migration. J. Cell Sci. 2004, 117, 4619–4628. [Google Scholar] [CrossRef] [PubMed]
- Bruzzaniti, A.; Neff, L.; Sanjay, A.; Horne, W.C.; De Camilli, P.; Baron, R. Dynamin Forms a Src Kinase–sensitive Complex with Cbl and Regulates Podosomes and Osteoclast Activity. Mol. Biol. Cell 2005, 16, 3301–3313. [Google Scholar] [CrossRef] [PubMed]
- Murillo, A.; Guerrero, C.A.; Acosta, O.; A Cardozo, C. Bone resorptive activity of osteoclast-like cells generated in vitro by PEG-induced macrophage fusion. Biol. Res. 2010, 43, 205–224. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
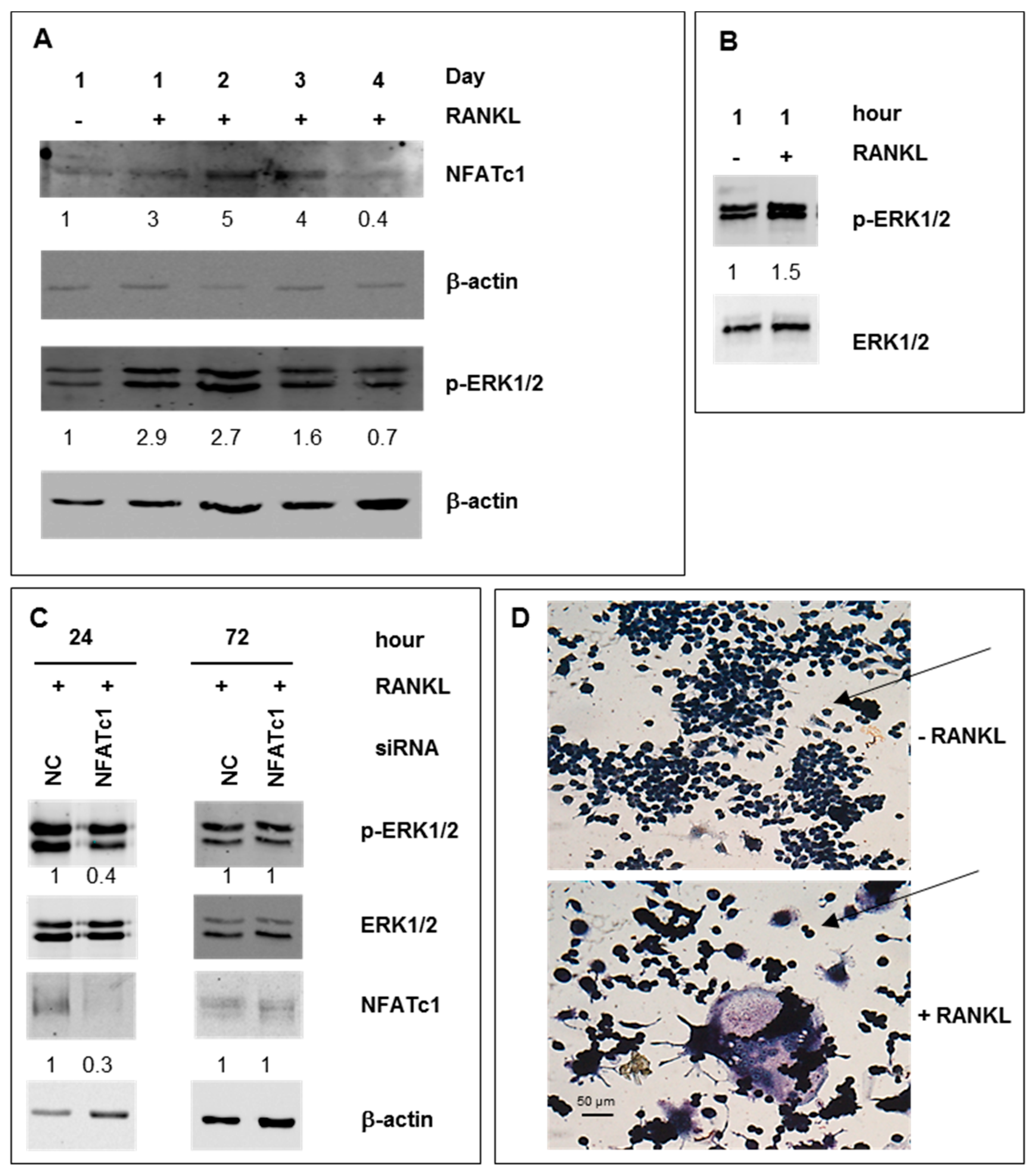

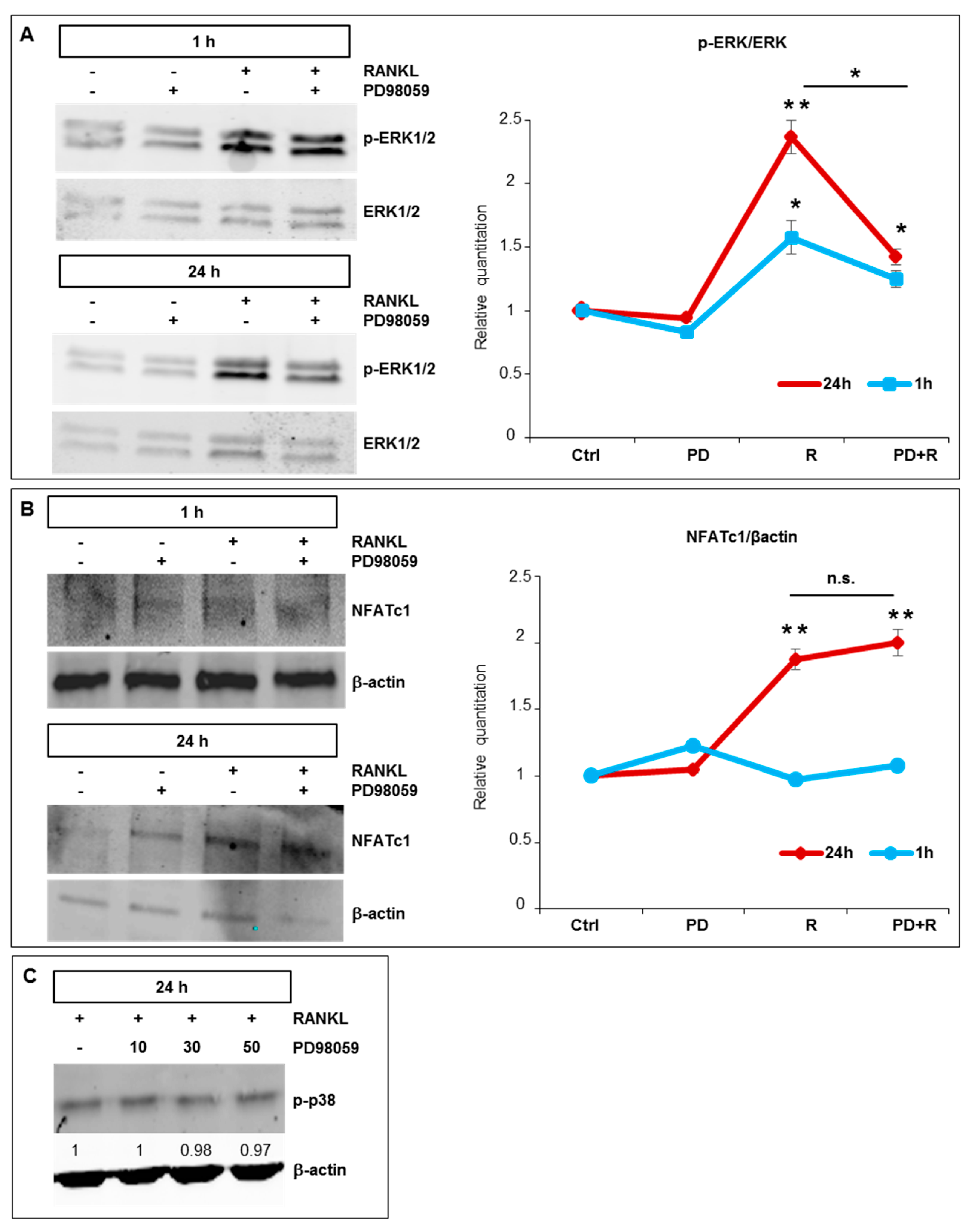
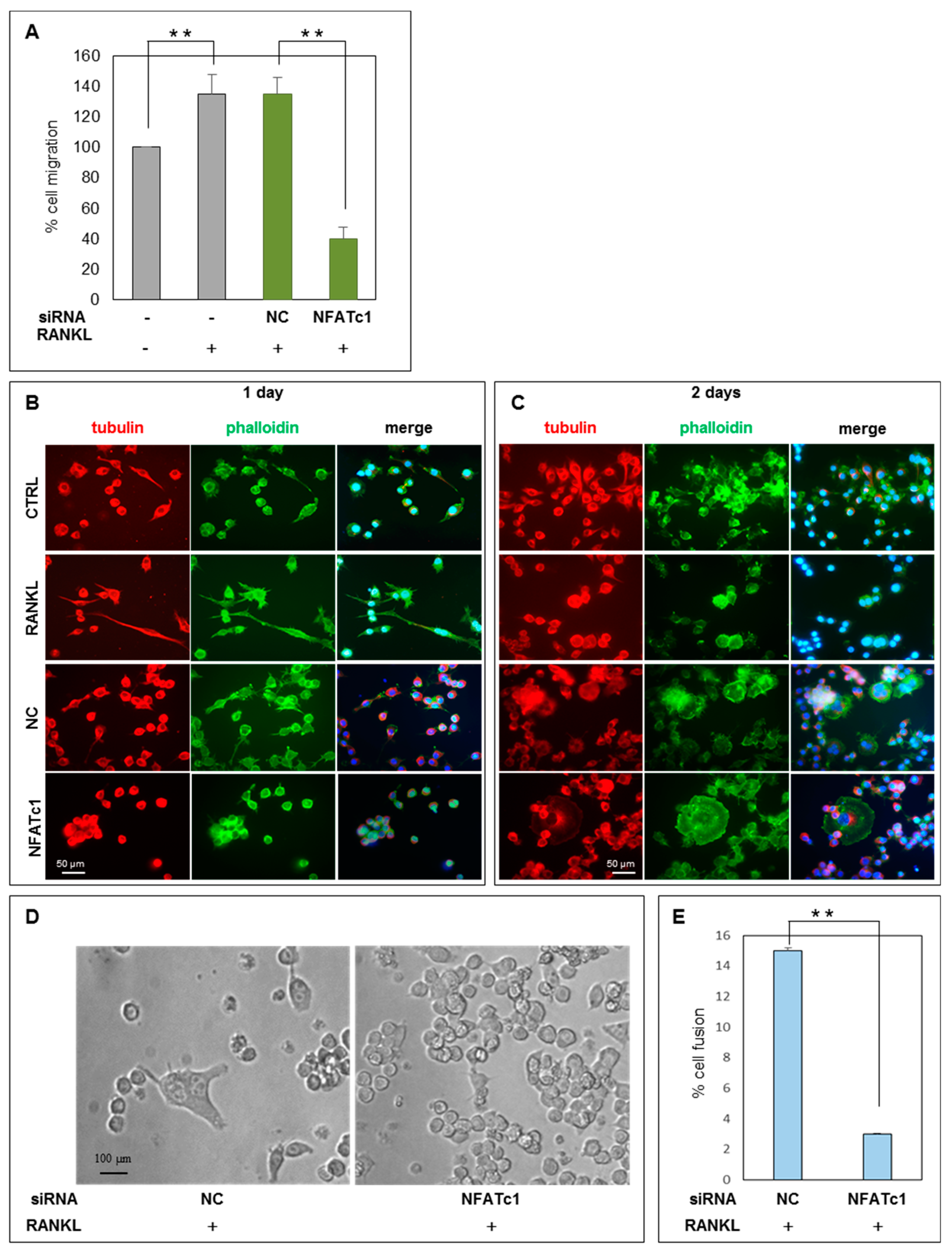

 , inhibition.
, inhibition.
 , inhibition.
, inhibition.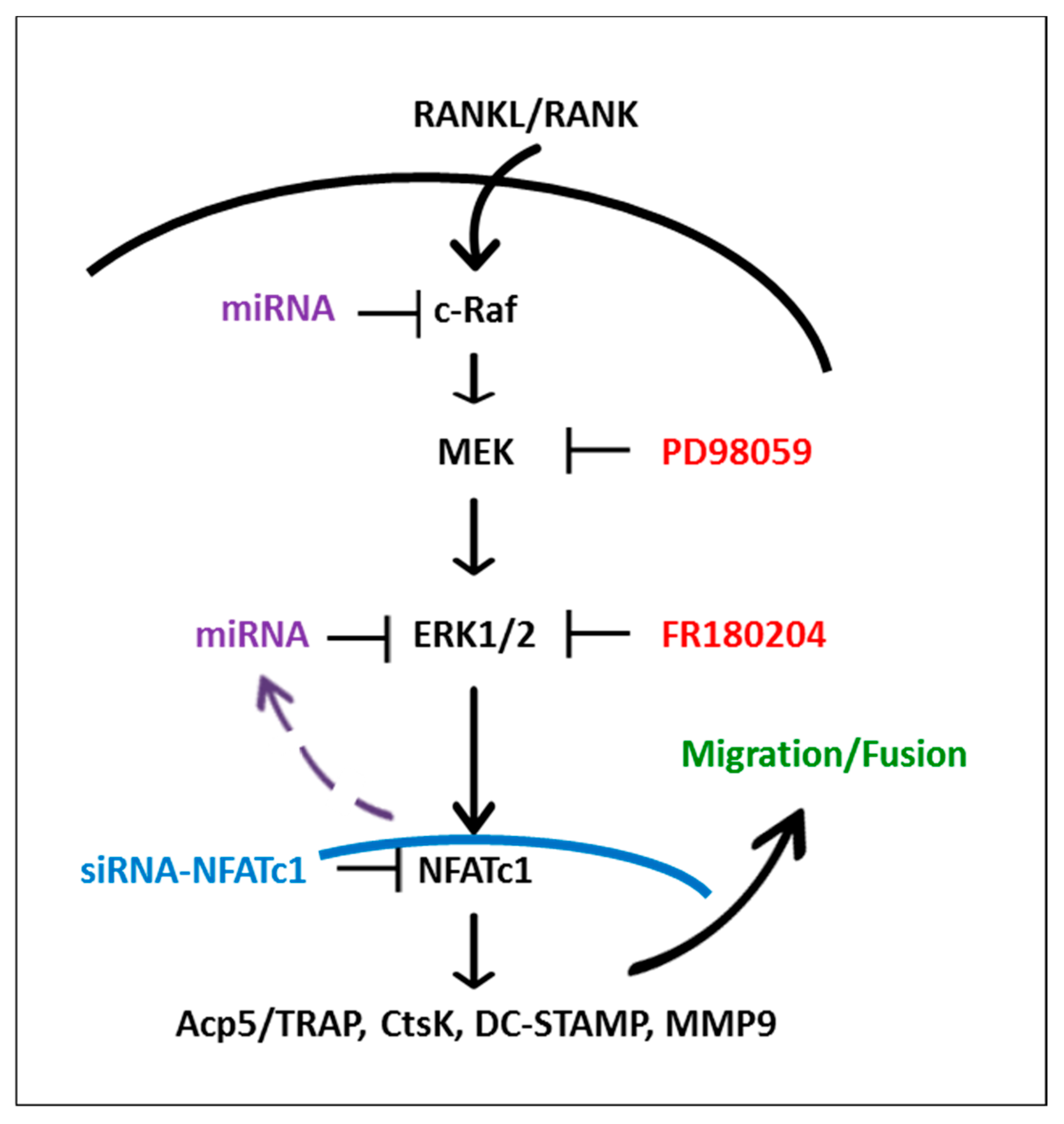
| Gene | 1 h | ±SD | 24 h | ±SD |
|---|---|---|---|---|
| NFATc1 | 1.3 | 0.6 | 3.6 | 1.6 |
| Acp5/TRAP | 1.2 | 0.4 | 18.6 | 3.5 |
| MMP9 | 0.8 | 0.1 | 46.5 | 7.0 |
| CtsK | 1.0 | 0.3 | 9.0 | 1.9 |
| DC-STAMP | 1.7 | 0.3 | 1.9 | 0.5 |
| TRAF6 | 0.9 | 0.3 | 1.1 | 0.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Russo, R.; Mallia, S.; Zito, F.; Lampiasi, N. Long-Lasting Activity of ERK Kinase Depends on NFATc1 Induction and Is Involved in Cell Migration-Fusion in Murine Macrophages RAW264.7. Int. J. Mol. Sci. 2020, 21, 8965. https://doi.org/10.3390/ijms21238965
Russo R, Mallia S, Zito F, Lampiasi N. Long-Lasting Activity of ERK Kinase Depends on NFATc1 Induction and Is Involved in Cell Migration-Fusion in Murine Macrophages RAW264.7. International Journal of Molecular Sciences. 2020; 21(23):8965. https://doi.org/10.3390/ijms21238965
Chicago/Turabian StyleRusso, Roberta, Selene Mallia, Francesca Zito, and Nadia Lampiasi. 2020. "Long-Lasting Activity of ERK Kinase Depends on NFATc1 Induction and Is Involved in Cell Migration-Fusion in Murine Macrophages RAW264.7" International Journal of Molecular Sciences 21, no. 23: 8965. https://doi.org/10.3390/ijms21238965
APA StyleRusso, R., Mallia, S., Zito, F., & Lampiasi, N. (2020). Long-Lasting Activity of ERK Kinase Depends on NFATc1 Induction and Is Involved in Cell Migration-Fusion in Murine Macrophages RAW264.7. International Journal of Molecular Sciences, 21(23), 8965. https://doi.org/10.3390/ijms21238965








