Flow Cytometry: From Experimental Design to Its Application in the Diagnosis and Monitoring of Respiratory Diseases
Abstract
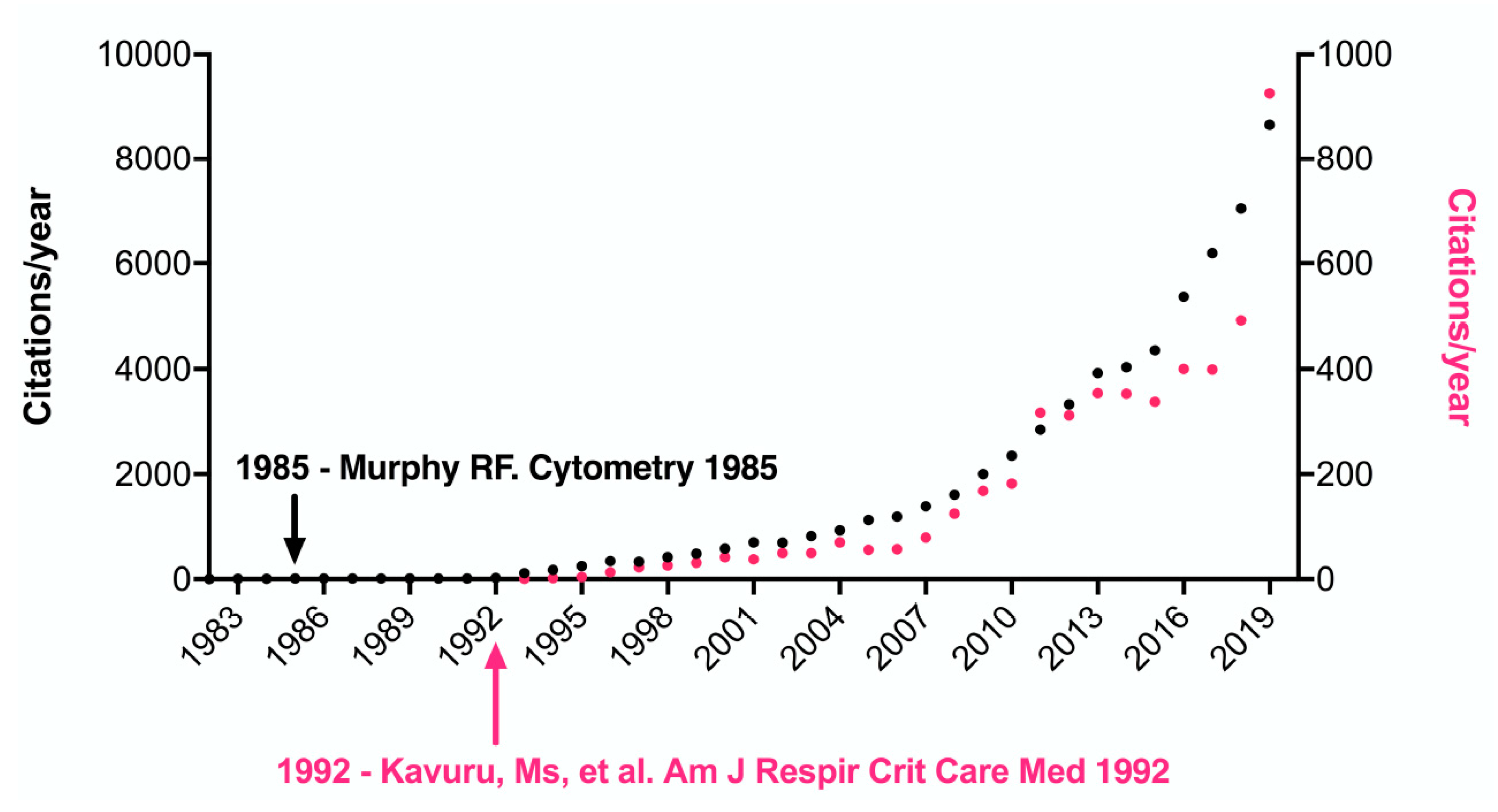
1. Introduction
2. Basic Systems for the Operation of the Flow Cytometer
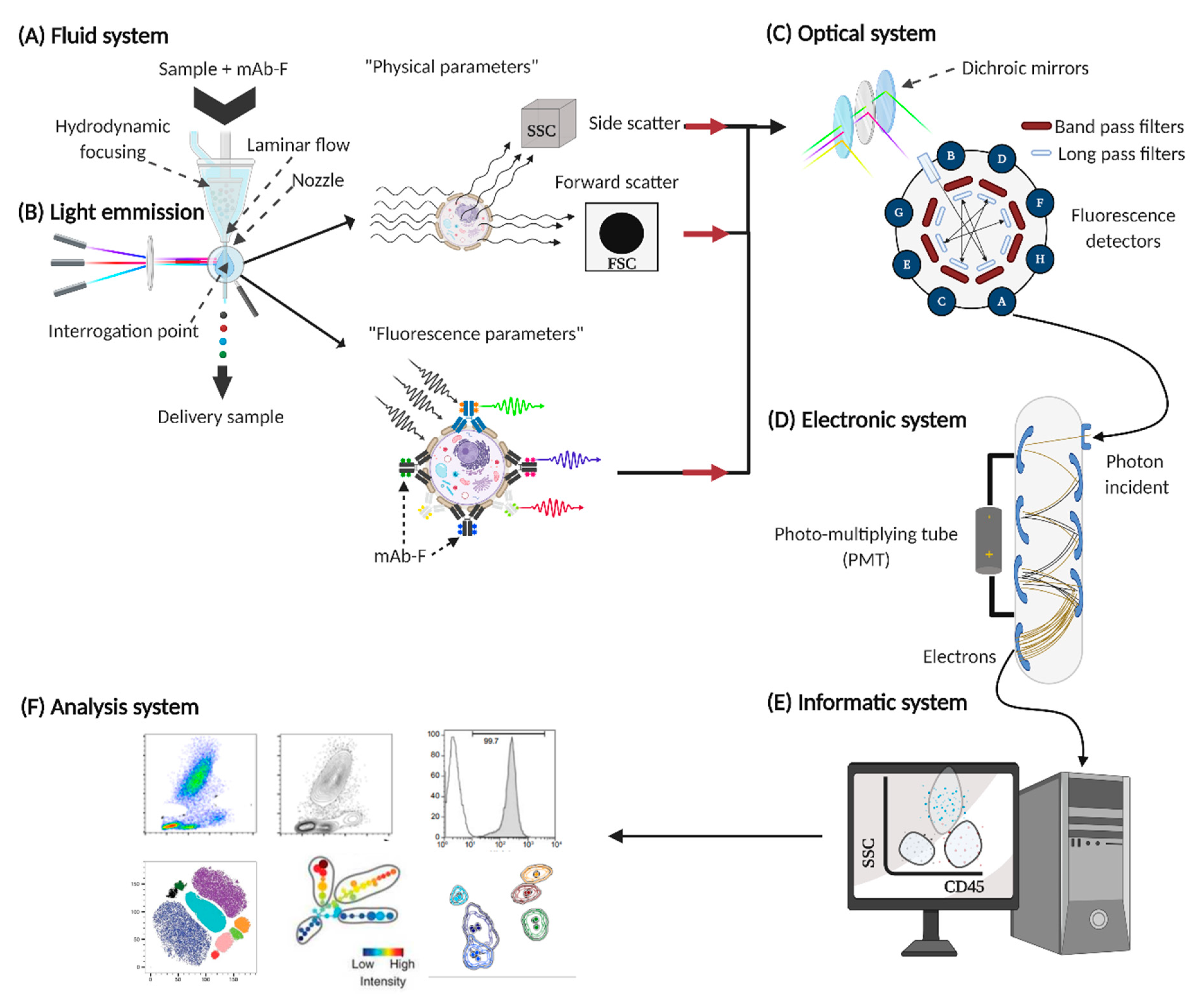
2.1. Fluid System
2.2. Source of Light Emission
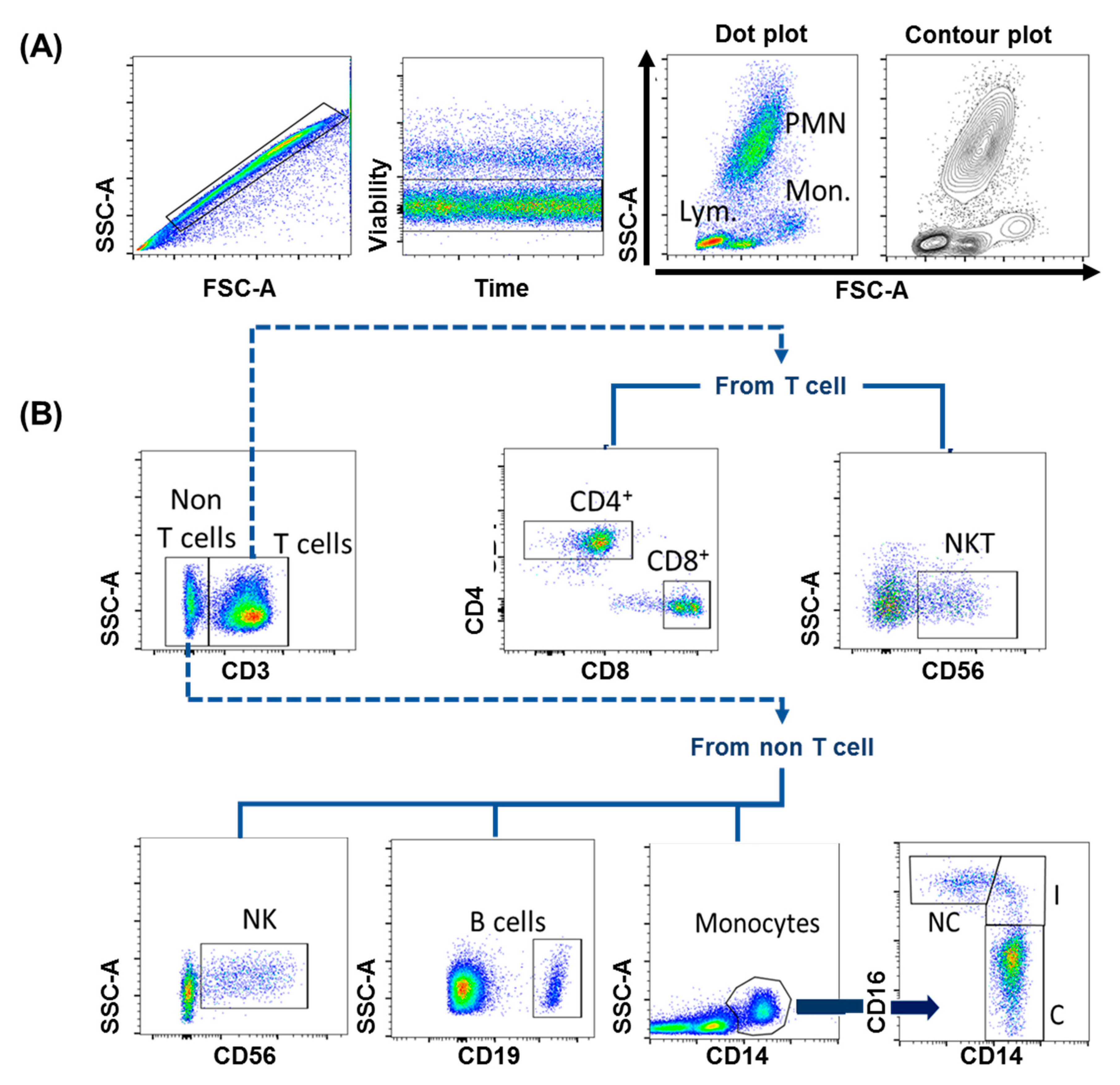
- Physical parameters, such as size (FSC, forward scatter), this is associated with interference in the horizontal path of visible light, as well as cell complexity (SSC, side scatter), which is associated with the change in the refractive index in every interface of the analyzed event. With these basic parameters, the region of lymphocytes, monocytes and polymorphonuclear cells can be generally identified (Figure 3A).
- Fluorescence parameters, quantify “n” fluorescences (depends on the capacity of each cytometer), allowing to obtain the percentage of positive events for a molecule (cell frequencies), and also to obtain the arbitrary unit called mean fluorescence intensity (MFI), which indirectly indicates the number of molecules expressed on the surface of an event [23].
2.3. Optic System
- Long pass (LP) transmits photons with a wavelength greater than that specified, for example, the LP 600 filter transmits light signals with a wavelength equal to or greater than 600 nm.
- Short pass (SP) transmit photons with a wavelength shorter than specified, for example, SP 600 filter transmits signals with a wavelength shorter than 600 nm [24].
- Band pass (BP) transmits light signals within the specified wavelength range, for example, the BP 525/50 filter, which allows light signals to pass between 500–550 nm.
- Dichroic mirrors (LP or SP), placed at a 45° angle to the incident light, reflecting non-transmitted light at a 90° angle to improve the transmission of wavelengths to specific detectors [25].
2.4. Electronic System
2.5. Informatic System
2.6. Analysis System
3. Basic Considerations to Perform Flow Cytometry
- Related to the object of study: Will cells, soluble components, functional processes (in vitro activation) be evaluated?
- Related to the staining protocol: What type of sample is it? Do I need to identify surface marks, intracellular, phosphoproteins, etc? Will the acquisition of the samples be immediate or do I need to preserve them?
- Equipment related: What equipment is available? How many types of lasers do you have attached? What detectors does it have?
- Related to fluorescence: How much do the emission signals of the selected fluorochromes overlap?
- Related to the analysis system: What analysis program is available?
4. Quality Management Systems in Flow Cytometry Assays
4.1. Pre-Analytical Phase
4.2. Analytical Phase
- Internal quality control: useful for monitoring the variations of the variables over time using Leving–Jennings graphs; this establishes the standard deviation of the signal from each fluorescence detector.
- Autofluorescence control: allows to know the natural fluorescence pattern of the analyzed events; the sample is subjected to all the processes, except the staining protocol. The result will be the inherent characteristic of the excitation and emission of the molecules that make up the analyzed event (background fluorescence).
- Fluorescence minus one (FMO): These controls contain all the mAb-F used in the panel, except for one, which is relevant to the molecular markers to be studied. It helps to explain propagation error in the empty detector by other interfering fluorescence signals (compensation problems) [42].
- Isotype control: This control allows to differentiate the non-specific binding of the fragment crystallizable region of the antibodies and the aggregation of fluorescent molecules by excess. Due to the variability of the manufacturing processes, it is impossible to have the ideal isotype control [42]; currently, its use is no longer widely recommended.
- Staining control: refers to the titration of the mAb-F to know the optimal concentration of mAb-F that saturates the available sites of the antigen to be detected, thereby avoiding the presence of false positives due to excess mAb-F [42].
- Background noise: phenomenon attributed to photons with random emission from cell fragments or inadequate compensation. The value of the signal that is considered a “normal” event (threshold) is limited, based on the size and complexity of the cellular event of interest.
4.3. Post-Analytical Phase
5. Use of Flow Cytometry in the Diagnosis of Respiratory Diseases
5.1. COPD
5.2. Asthma
5.3. Acute Infections of the Lower Respiratory Tract
5.4. Tuberculosis
5.5. Lung Cancer
6. Advantages and Limitations of Conventional Methods, Compared to FCM, in the Diagnosis of Respiratory Diseases
7. Conclusions
- Quick and specific diagnosis.
- Identification of clinical phases.
- Follow-up biomarkers to evaluate adequate response to treatments.
- Cost–benefit ratio.
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AILRT | Acute lower respiratory tract infections |
| ATS | American Thoracic Society |
| COPD | Chronic obstructive pulmonary disease |
| ERS | European Respiratory Society |
| EVs | Extracellular vesicles |
| FCM | Flow Cytometry |
| FCS | Flow cytometry standard |
| FIRS | Forum of International Respiratory Societies |
| FMO | Fluorescence minus one |
| FSC | Forward scatter |
| GINA | Global Initiative for Asthma |
| GOLD | Chronic Obstructive Lung Disease |
| ICCS | International Clinical Cytometry Society |
| IL | Interleukin |
| ISAC | International Society for Advancement of Cytometry |
| LAM | Lipoarabinomannan |
| mAB | Monoclonal antibodies |
| mAB-F | Monoclonal antibodies coupled to fluorochromes |
| MFI | Mean fluorescence intensity |
| Mtb | Mycobacterium tuberculosis |
| NKG2D | Natural killer group 2 member D |
| NSCLC | Non-small cell type |
| PBMC | Peripheral blood mononuclear cells |
| SSC | Side scatter |
| STRA | Therapy-resistant asthma |
| TB | Tuberculosis |
| WHO | World Health Organization |
References
- Malassez, L. De la Numération des Globules Rouges du Sang; Delahaye: Paris, France, 1873. [Google Scholar]
- Moldavan, A. Photo-electric technique for the counting of microscopical cells. Science 1934, 80, 188–189. [Google Scholar] [CrossRef] [PubMed]
- Gucker, F.T.; O’Konski, C.T.; Pickard, H.B.; Pitts, J.N. A photoelectronic counter for colloidal particles. J. Am. Chem. Soc. 1947, 69, 2422–2431. [Google Scholar] [CrossRef] [PubMed]
- Coulter, W.H. Means for Counting Particles in a Suspended Fluid. U.S. Patent 2,656,508, 20 October 1953. [Google Scholar]
- Crosland-Taylor, P.J. A device for counting small particles suspended in a fluid through a tube. Nature 1953, 171, 37–38. [Google Scholar] [CrossRef] [PubMed]
- Kamentsky, L.A.; Melamed, M.R.; Derman, H. Spectrophotometer: New instrument for ultrarapid cell analysis. Science 1965, 150, 630–631. [Google Scholar] [CrossRef]
- Sweet, R.G. High frequency recording with electrostatically deflected ink jets. Rev. Sci. Instrum. 1965, 36, 131–136. [Google Scholar] [CrossRef]
- Fulwyler, M.J. Electronic separation of biological cells by volume. Science 1965, 150, 910–911. [Google Scholar] [CrossRef]
- Oi, V.T.; Glazer, A.N.; Stryer, L. Fluorescent phycobiliprotein conjugates for analyses of cells and molecules. J. Cell Biol. 1982, 93, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Köhler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Herzenberg, L.A.; Parks, D.; Sahaf, B.; Perez, O.; Roederer, M.; Herzenberg, L.A. The history and future of the Fluorescence Activated Cell Sorter and flow cytometry: A view from Stanford. Clin. Chem. 2002, 48, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Bajgelman, M.C. Principles and applications of flow cytometry. In Data Processing Handbook for Complex Biological Data Sources; Elsevier: Amsterdam, The Netherlands, 2019; pp. 119–124. ISBN 978-0-12-816548-5. [Google Scholar]
- Yang, R.-J.; Fu, L.-M.; Hou, H.-H. Review and perspectives on microfluidic flow cytometers. Sens. Actuators B Chem. 2018, 266, 26–45. [Google Scholar] [CrossRef]
- Kormelink, T.G.; Arkesteijn, G.J.A.; Nauwelaers, F.A.; van den Engh, G.; Nolte-’t Hoen, E.N.M.; Wauben, M.H.M. Prerequisites for the analysis and sorting of extracellular vesicle subpopulations by high-resolution flow cytometry: Sorting extracellular vesicle subpopulations. Cytometry 2016, 89, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Montermini, L.; Jeong, H.; Sharma, S.; Meehan, B.; Rak, J. Mapping subpopulations of cancer cell-derived extracellular vesicles and particles by Nano-flow cytometry. ACS Nano 2019, 13, 10499–10511. [Google Scholar] [CrossRef] [PubMed]
- Ender, F.; Zamzow, P.; von Bubnoff, N.; Gieseler, F. Detection and quantification of extracellular vesicles via FACS: Membrane labeling matters! Int. J. Mol. Sci. 2019, 21, 291. [Google Scholar] [CrossRef] [PubMed]
- Cossarizza, A.; Chang, H.-D.; Radbruch, A.; Akdis, M.; Andrä, I.; Annunziato, F.; Bacher, P.; Barnaba, V.; Battistini, L.; Bauer, W.M.; et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies: Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur. J. Immunol. 2017, 47, 1584–1797. [Google Scholar] [CrossRef] [PubMed]
- Cossarizza, A.; Chang, H.; Radbruch, A.; Acs, A.; Adam, D.; Adam-Klages, S.; Agace, W.W.; Aghaeepour, N.; Akdis, M.; Allez, M.; et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur. J. Immunol. 2019, 49, 1457–1973. [Google Scholar] [CrossRef]
- Graves, S.W.; Nolan, J.P.; Jett, J.H.; Martin, J.C.; Sklar, L.A. Nozzle design parameters and their effects on rapid sample delivery in flow cytometry. Cytometry 2002, 47, 127–137. [Google Scholar] [CrossRef]
- Romanenko, S.; Begley, R.; Harvey, A.R.; Hool, L.; Wallace, V.P. The interaction between electromagnetic fields at megahertz, gigahertz and terahertz frequencies with cells, tissues and organisms: Risks and potential. J. R. Soc. Interface 2017, 14, 20170585. [Google Scholar] [CrossRef]
- Telford, W.G. Near infrared lasers in flow cytometry. Methods 2015, 82, 12–20. [Google Scholar] [CrossRef]
- Telford, W.; Stickland, L.; Koschorreck, M. Ultraviolet 320 nm laser excitation for flow cytometry: UV 320 nm laser for flow cytometry. Cytom. Part A 2017, 91, 314–325. [Google Scholar] [CrossRef]
- Flores-Montero, J.; Kalina, T.; Corral-Mateos, A.; Sanoja-Flores, L.; Pérez-Andrés, M.; Martin-Ayuso, M.; Sedek, L.; Rejlova, K.; Mayado, A.; Fernández, P.; et al. Fluorochrome choices for multi-color flow cytometry. J. Immunol. Methods 2019, 475, 112618. [Google Scholar] [CrossRef]
- Waggoner, A. Optical filter sets for multiparameter flow cytometry. Curr. Protoc. Cytom 1997, 00, 1.5.1–1.5.8. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Vakhare, D.; Chaphalkar, S.R. Flow cytometry: An overview in optical system and application in biological studies. Eur. J. Biol. Res. 2016, 6, 186–192. [Google Scholar] [CrossRef]
- Perfetto, S.P.; Ambrozak, D.; Nguyen, R.; Chattopadhyay, P.K.; Roederer, M. Quality assurance for polychromatic flow cytometry using a suite of calibration beads. Nat. Protoc. 2012, 7, 2067–2079. [Google Scholar] [CrossRef] [PubMed]
- How Flow Cytometry Converts Photons to Digital Data. Available online: https://expert.cheekyscientist.com/flow-cytometry-converts-photons-to-digital-data/ (accessed on 22 October 2020).
- Murphy, R.F.; Chused, T.M. A proposal for a flow cytometric data file standard. Cytometry 1984, 5, 553–555. [Google Scholar] [CrossRef]
- Verschoor, C.P.; Lelic, A.; Bramson, J.L.; Bowdish, D.M.E. An introduction to automated flow cytometry gating tools and their implementation. Front. Immunol. 2015, 6, 6. [Google Scholar] [CrossRef]
- Matarraz, S.; Almeida, J.; Flores-Montero, J.; Lécrevisse, Q.; Guerri, V.; López, A.; Bárrena, S.; Van Der Velden, V.H.J.; Te Marvelde, J.G.; Van Dongen, J.J.M.; et al. Introduction to the diagnosis and classification of monocytic-lineage leukemias by flow cytometry. Cytom. Part B Clin. Cytom. 2017, 92, 218–227. [Google Scholar] [CrossRef]
- Herzenberg, L.A.; Tung, J.; Moore, W.A.; Herzenberg, L.A.; Parks, D.R. Interpreting flow cytometry data: A guide for the perplexed. Nat. Immunol. 2006, 7, 681–685. [Google Scholar] [CrossRef]
- Khowawisetsut, L.; Sukapirom, K.; Pattanapanyasat, K. Data analysis and presentation in flow cytometry. ScienceAsia 2018, 44S, 19. [Google Scholar] [CrossRef]
- Novo, D.; Wood, J. Flow cytometry histograms: Transformations, resolution, and display. Cytom. Part A 2008, 73A, 685–692. [Google Scholar] [CrossRef]
- Tinnevelt, G.H.; Kokla, M.; Hilvering, B.; van Staveren, S.; Folcarelli, R.; Xue, L.; Bloem, A.C.; Koenderman, L.; Buydens, L.M.C.; Jansen, J.J. Novel data analysis method for multicolour flow cytometry links variability of multiple markers on single cells to a clinical phenotype. Sci. Rep. 2017, 7, 5471. [Google Scholar] [CrossRef]
- Home. Available online: http://www.hcdm.org/ (accessed on 22 October 2020).
- Ferrer-Font, L.; Pellefigues, C.; Mayer, J.U.; Small, S.J.; Jaimes, M.C.; Price, K.M. Panel design and optimization for high-dimensional immunophenotyping assays using spectral flow cytometry. Curr. Protoc. Cytom. 2020, 92, e70. [Google Scholar] [CrossRef] [PubMed]
- Khordad, M.; Mercer, R.E. Identifying genotype-phenotype relationships in biomedical text. J. Biomed. Semant. 2017, 8, 57. [Google Scholar] [CrossRef] [PubMed]
- Vázquez Rodríguez, S.; Pizano, L.A.A.; Servitje, E.L.; Ramirez, J.M.; Méndez, O.L.P.; Alcazar, G.V.; Cepeda, M.L.G.; Pelaez, M.G.H.; Gonzalez, G.C.; Villanueva, R.A.; et al. Multiparameter flow cytometry analysis of leukocyte markers for diagnosis in preterm neonatal sepsis. J. Matern. Neonatal. Med. 2019, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ellyard, J.I.; Tunningley, R.; Lorenzo, A.M.; Jiang, S.H.; Cook, A.; Chand, R.; Talaulikar, D.; Hatch, A.-M.; Wilson, A.; Vinuesa, C.G.; et al. Non-parametric heat map representation of flow cytometry data: Identifying cellular changes associated with genetic immunodeficiency disorders. Front. Immunol. 2019, 10, 2134. [Google Scholar] [CrossRef] [PubMed]
- ISO 9001:2015(en) Quality Management Systems—Requirements. Available online: https://www.iso.org/obp/ui/#iso:std:iso:9001:ed-5:v1:en (accessed on 22 October 2020).
- Campbell, J.D.M.; Fraser, A.R. Flow cytometric assays for identity, safety and potency of cellular therapies: Use of flow cytometric assays. Cytom. Part B Clin. Cytom. 2018, 94, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Spurgeon, B.E.J.; Naseem, K.M. Platelet flow cytometry: Instrument setup, controls, and panel performance. Cytom. Part B Clin. Cytom. 2020, 98, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 2015, 111, A3.B.1–A3.B.3. [Google Scholar] [CrossRef] [PubMed]
- Clinical Guidelines—Australasian Cytometry Society. Available online: https://cytometry.org.au/resources/clinical-guidelines/ (accessed on 22 October 2020).
- Wang, H.; Naghavi, M.; Allen, C.; Barber, R.M.; Bhutta, Z.A.; Carter, A.; Casey, D.C.; Charlson, F.J.; Chen, A.Z.; Coates, M.M.; et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [Google Scholar] [CrossRef]
- Forum of International Respiratory Societies. The Global Impact of Respiratory Disease, 2nd ed.; European Respiratory Society: Sheffield, UK, 2017; ISBN 978-1-84984-087-3. [Google Scholar]
- Oetjen, K.A.; Lindblad, K.E.; Goswami, M.; Gui, G.; Dagur, P.K.; Lai, C.; Dillon, L.W.; McCoy, J.P.; Hourigan, C.S. Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight 2018, 3, e124928. [Google Scholar] [CrossRef]
- Watza, D.; Lusk, C.M.; Dyson, G.; Purrington, K.S.; Wenzlaff, A.S.; Neslund-Dudas, C.; Soubani, A.O.; Gadgeel, S.M.; Schwartz, A.G. COPD-dependent effects of genetic variation in key inflammation pathway genes on lung cancer risk. Int. J. Cancer 2020, 147, 747–756. [Google Scholar] [CrossRef]
- Li, Q.; Hu, Y.; Chen, Y.; Lv, Z.; Wang, J.; An, G.; Du, X.; Wang, H.; Corrigan, C.J.; Wang, W.; et al. IL-33 induces production of autoantibody against autologous respiratory epithelial cells: A potential mechanism for the pathogenesis of COPD. Immunology 2019, 157, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Kadota, T.; Fujita, Y.; Yoshioka, Y.; Araya, J.; Kuwano, K.; Ochiya, T. Extracellular vesicles in chronic obstructive pulmonary disease. Int. J. Mol. Sci. 2016, 17, 1801. [Google Scholar] [CrossRef]
- Falcon-Rodriguez, C.I.; De Vizcaya-Ruiz, A.; Rosas-Pérez, I.A.; Osornio-Vargas, Á.R.; Segura-Medina, P. Inhalation of concentrated PM 2.5 from Mexico City acts as an adjuvant in a guinea pig model of allergic asthma. Environ. Pollut. 2017, 228, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Leffler, J.; Read, J.F.; Jones, A.C.; Mok, D.; Hollams, E.M.; Laing, I.A.; Le Souef, P.N.; Sly, P.D.; Kusel, M.M.H.; Klerk, N.H.; et al. Progressive increase of FcεRI expression across several PBMC subsets is associated with atopy and atopic asthma within school-aged children. Pediatr. Allergy Immunol. 2019, 30, 646–653. [Google Scholar] [CrossRef]
- Wiest, M.; Upchurch, K.; Hasan, M.M.; Cardenas, J.; Lanier, B.; Millard, M.; Turner, J.; Oh, S.; Joo, H. Phenotypic and functional alterations of regulatory B cell subsets in adult allergic asthma patients. Clin. Exp. Allerg 2019, 49, 1214–1224. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.; de Souza, A.P.D.; de Araújo, P.D.; Pinto, L.A.; Jones, M.H.; Stein, R.T.; Pitrez, P.M. iNKT cells are increased in children with severe therapy-resistant asthma. Allergol. Immunopathol. (Madr.) 2018, 46, 175–180. [Google Scholar] [CrossRef]
- Pasanen, A.; Karjalainen, M.K.; Kummola, L.; Waage, J.; Bønnelykke, K.; Ruotsalainen, M.; Piippo-Savolainen, E.; Goksör, E.; Nuolivirta, K.; Chawes, B.; et al. NKG2D gene variation and susceptibility to viral bronchiolitis in childhood. Pediatr. Res. 2018, 84, 451–457. [Google Scholar] [CrossRef]
- Schedel, M.; Jia, Y.; Michel, S.; Takeda, K.; Domenico, J.; Joetham, A.; Ning, F.; Strand, M.; Han, J.; Wang, M.; et al. 1,25D3 prevents CD8+Tc2 skewing and asthma development through VDR binding changes to the Cyp11a1 promoter. Nat. Commun. 2016, 7, 10213. [Google Scholar] [CrossRef]
- Massoud, A.H.; Charbonnier, L.-M.; Lopez, D.; Pellegrini, M.; Phipatanakul, W.; Chatila, T.A. An asthma-associated IL4R variant exacerbates airway inflammation by promoting conversion of regulatory T cells to TH17-like cells. Nat. Med. 2016, 22, 1013–1022. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Z.; Wang, L.; Wang, J.; Chen, L.; Xie, H.; Zhang, H.; He, S. Altered expression of IL-18 binding protein and IL-18 receptor in basophils and mast cells of asthma patients. Scand. J. Immunol. 2018, 87, e12658. [Google Scholar] [CrossRef]
- Feldman, C.; Shaddock, E. Epidemiology of lower respiratory tract infections in adults. Expert Rev. Respir. Med. 2019, 13, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.-Y.; Wu, W.-T.; Chang, C.-Y.; Wong, Y.-C.; Lai, C.-C.; Chan, Y.-J.; Wu, K.-G.; Hung, M.-C. Viral etiologies of acute respiratory tract infections among hospitalized children—A comparison between single and multiple viral infections. J. Microbiol. Immunol. Infect. 2019, 52, 902–910. [Google Scholar] [CrossRef]
- Pérez-López, F.R.; Tajada, M.; Savirón-Cornudella, R.; Sánchez-Prieto, M.; Chedraui, P.; Terán, E. Coronavirus disease 2019 and gender-related mortality in European countries: A meta-analysis. Maturitas 2020, 141, 59–62. [Google Scholar] [CrossRef]
- Swieboda, D.; Guo, Y.; Sagawe, S.; Thwaites, R.S.; Nadel, S.; Openshaw, P.J.M.; Culley, F.J. OMIP-062: A 14-color, 16-antibody panel for immunophenotyping human innate lymphoid, myeloid and t cells in small volumes of whole blood and pediatric airway samples. Cytom. Part A 2019, 95, 1231–1235. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Schielke, E.G.; Grace, K.M.; Hassell, C.; Marrone, B.L.; Nolan, J.P. Microsphere-based duplexed immunoassay for influenza virus typing by flow cytometry. J. Immunol. Methods 2004, 284, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Hobbie, S.N.; Viswanathan, K.; Bachelet, I.; Aich, U.; Shriver, Z.; Subramanian, V.; Raman, R.; Sasisekharan, R. Modular glycosphere assays for high-throughput functional characterization of influenza viruses. BMC Biotechnol. 2013, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Hartley, G.E.; Edwards, E.S.J.; Bosco, J.J.; Ojaimi, S.; Stirling, R.G.; Cameron, P.U.; Flanagan, K.; Plebanski, M.; Hogarth, P.M.; O’Hehir, R.E.; et al. Influenza-specific IgG1+ memory B-cell numbers increase upon booster vaccination in healthy adults but not in patients with predominantly antibody deficiency. Clin. Transl. Immunol. 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Cossarizza, A.; De Biasi, S.; Guaraldi, G.; Girardis, M.; Mussini, C.; for the Modena Covid-19 Working Group (MoCo19). SARS-CoV-2, the virus that causes COVID-19: Cytometry and the new challenge for global health. Cytom. Part. A 2020, 97, 340–343. [Google Scholar] [CrossRef]
- Mathew, D.; Giles, J.R.; Baxter, A.E.; Oldridge, D.A.; Greenplate, A.R.; Wu, J.E.; Alanio, C.; Kuri-Cervantes, L.; Pampena, M.B.; D’Andrea, K.; et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 2020, 369, eabc8511. [Google Scholar] [CrossRef]
- Fong, C.H.-Y.; Cai, J.-P.; Dissanayake, T.K.; Chen, L.-L.; Choi, C.Y.-K.; Wong, L.-H.; Ng, A.C.-K.; Pang, P.K.P.; Ho, D.T.-Y.; Poon, R.W.-S.; et al. Improved detection of antibodies against SARS-CoV-2 by microsphere-based antibody assay. Int. J. Mol. Sci. 2020, 21, 6595. [Google Scholar] [CrossRef]
- World Health Organization. Global Tuberculosis Report 2019; World Health Organization: Geneva, Switzerland, 2019; ISBN 978-92-4-156571-4. [Google Scholar]
- Chávez-Galán, L.; Ocaña-Guzmán, R.; Torre-Bouscoulet, L.; García-de-Alba, C.; Sada-Ovalle, I. Exposure of monocytes to lipoarabinomannan promotes their differentiation into functionally and phenotypically immature macrophages. J. Immunol. Res. 2015, 2015, 1–16. [Google Scholar] [CrossRef]
- Chávez-Galán, L.; Ramon-Luing, L.; Carranza, C.; Garcia, I.; Sada-Ovalle, I. Lipoarabinomannan decreases Galectin-9 expression and tumor necrosis factor pathway in macrophages favoring mycobacterium tuberculosis intracellular growth. Front. Immunol. 2017, 8, 1659. [Google Scholar] [CrossRef]
- Chávez-Galán, L.; Illescas-Eugenio, J.; Alvarez-Sekely, M.; Baez-Saldaña, R.; Chávez, R.; Lascurain, R. Tuberculosis patients display a high proportion of CD8+ T cells with a high cytotoxic potential. Microbiol. Immunol. 2019, 63, 316–327. [Google Scholar] [CrossRef]
- Beham, A.W.; Puellmann, K.; Laird, R.; Fuchs, T.; Streich, R.; Breysach, C.; Raddatz, D.; Oniga, S.; Peccerella, T.; Findeisen, P.; et al. A TNF-regulated recombinatorial macrophage immune receptor implicated in granuloma formation in tuberculosis. PLoS Pathog. 2011, 7, e1002375. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Galan, L.; Vesin, D.; Blaser, G.; Uysal, H.; Benmerzoug, S.; Rose, S.; Ryffel, B.; Quesniaux, V.F.J.; Garcia, I. Myeloid cell TNFR1 signaling dependent liver injury and inflammation upon BCG infection. Sci. Rep. 2019, 9, 5297. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Cruz, A.; Vesin, D.; Ramon-Luing, L.; Zuñiga, J.; Quesniaux, V.F.J.; Ryffel, B.; Lascurain, R.; Garcia, I.; Chávez-Galán, L. CD3+ macrophages deliver proinflammatory cytokines by a CD3− and transmembrane TNF-dependent pathway and are increased at the BCG-infection site. Front. Immunol. 2019, 10, 2550. [Google Scholar] [CrossRef] [PubMed]
- Estévez, O.; Anibarro, L.; Garet, E.; Martínez, A.; Pena, A.; Barcia, L.; Peleteiro, M.; González-Fernández, Á. Multi-parameter flow cytometry immunophenotyping distinguishes different stages of tuberculosis infection. J. Infect. 2020, 81, 57–71. [Google Scholar] [CrossRef]
- Yang, Q.; Xu, Q.; Chen, Q.; Li, J.; Zhang, M.; Cai, Y.; Liu, H.; Zhou, Y.; Deng, G.; Deng, Q.; et al. Discriminating active tuberculosis from latent tuberculosis infection by flow cytometric measurement of CD161-expressing T cells. Sci. Rep. 2015, 5, 17918. [Google Scholar] [CrossRef]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non–small cell lung cancer: Epidemiology, screening, diagnosis, and treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef]
- Yabe, M.; Gao, Q.; Ozkaya, N.; Huet, S.; Lewis, N.; Pichardo, J.D.; Moskowitz, A.J.; Horwitz, S.M.; Dogan, A.; Roshal, M. Bright PD-1 expression by flow cytometry is a powerful tool for diagnosis and monitoring of angioimmunoblastic T-cell lymphoma. Blood Cancer J. 2020, 10, 32. [Google Scholar] [CrossRef]
- Khattak, M.A.; Reid, A.; Freeman, J.; Pereira, M.; McEvoy, A.; Lo, J.; Frank, M.H.; Meniawy, T.; Didan, A.; Spencer, I.; et al. PD-L1 expression on circulating tumor cells may be predictive of response to pembrolizumab in advanced melanoma: Results from a pilot study. Oncologist 2020, 25. [Google Scholar] [CrossRef] [PubMed]
- Heim, L.; Kachler, K.; Siegmund, R.; Trufa, D.I.; Mittler, S.; Geppert, C.-I.; Friedrich, J.; Rieker, R.J.; Sirbu, H.; Finotto, S. Increased expression of the immunosuppressive interleukin-35 in patients with non-small cell lung cancer. Br. J. Cancer 2019, 120, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Kotsakis, A.; Koinis, F.; Katsarou, A.; Gioulbasani, M.; Aggouraki, D.; Kentepozidis, N.; Georgoulias, V.; Vetsika, E.-K. Prognostic value of circulating regulatory T cell subsets in untreated non-small cell lung cancer patients. Sci. Rep. 2016, 6, 39247. [Google Scholar] [CrossRef] [PubMed]
- Islas-Vazquez, L.; Prado-Garcia, H.; Aguilar-Cazares, D.; Meneses-Flores, M.; Galicia-Velasco, M.; Romero-Garcia, S.; Camacho-Mendoza, C.; Lopez-Gonzalez, J.S. LAP TGF-beta subset of CD4+CD25+CD127- Treg cells is increased and overexpresses LAP TGF-beta in lung adenocarcinoma patients. Biomed. Res. Int. 2015, 2015, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Laheurte, C.; Dosset, M.; Vernerey, D.; Boullerot, L.; Gaugler, B.; Gravelin, E.; Kaulek, V.; Jacquin, M.; Cuche, L.; Eberst, G.; et al. Distinct prognostic value of circulating anti-telomerase CD4+ Th1 immunity and exhausted PD-1+/TIM-3+ T cells in lung cancer. Cell. Mol. Biol. 2019, 121, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Jing, W.; An, N.; Li, A.; Yan, W.; Zhu, H.; Yu, J. Prognostic significance of peripheral CD8+CD28+ and CD8+CD28− T cells in advanced non-small cell lung cancer patients treated with chemo(radio)therapy. J. Transl. Med. 2019, 17, 344. [Google Scholar] [CrossRef]
- Graham, B.L.; Steenbruggen, I.; Miller, M.R.; Barjaktarevic, I.Z.; Cooper, B.G.; Hall, G.L.; Hallstrand, T.S.; Kaminsky, D.A.; McCarthy, K.; McCormack, M.C.; et al. Standardization of spirometry 2019 update. An official American thoracic society and European respiratory society technical statement. Am. J. Respir. Crit. Care Med. 2019, 200, e70–e88. [Google Scholar] [CrossRef]
- American Thoracic Society Statements. Guidelines & Reports. Available online: https://www.thoracic.org/statements/index.php (accessed on 9 November 2020).
- Lopes, A.J. Advances in spirometry testing for lung function analysis. Expert Rev. Respir. Med. 2019, 13, 559–569. [Google Scholar] [CrossRef]
- Bednarek, M.; Grabicki, M.; Piorunek, T.; Batura-Gabryel, H. Current place of impulse oscillometry in the assessment of pulmonary diseases. Respir. Med. 2020, 170, 105952. [Google Scholar] [CrossRef]
- Halpin, D.M.G.; Criner, G.J.; Papi, A.; Singh, D.; Anzueto, A.; Martinez, F.J.; Agusti, A.A.; Vogelmeier, C.F.; on Behalf of the GOLD Science Committee. Global initiative for the diagnosis, management, and prevention of chronic obstructive lung disease: The 2020 GOLD science committee report on COVID-19 & COPD. Am. J. Respir. Crit. Care Med. 2020. [Google Scholar] [CrossRef]
- Bursle, E.; Robson, J. Non-culture methods for detecting infection. Aust. Prescr. 2016, 39, 171–175. [Google Scholar] [CrossRef] [PubMed]
- García-Arroyo, L.; Prim, N.; Martí, N.; Roig, M.C.; Navarro, F.; Rabella, N. Benefits and drawbacks of molecular techniques for diagnosis of viral respiratory infections. Experience with two multiplex PCR assays: Multiplex PCR Methods, Benefits, and Drawbacks. J. Med. Virol. 2016, 88, 45–50. [Google Scholar] [CrossRef] [PubMed]
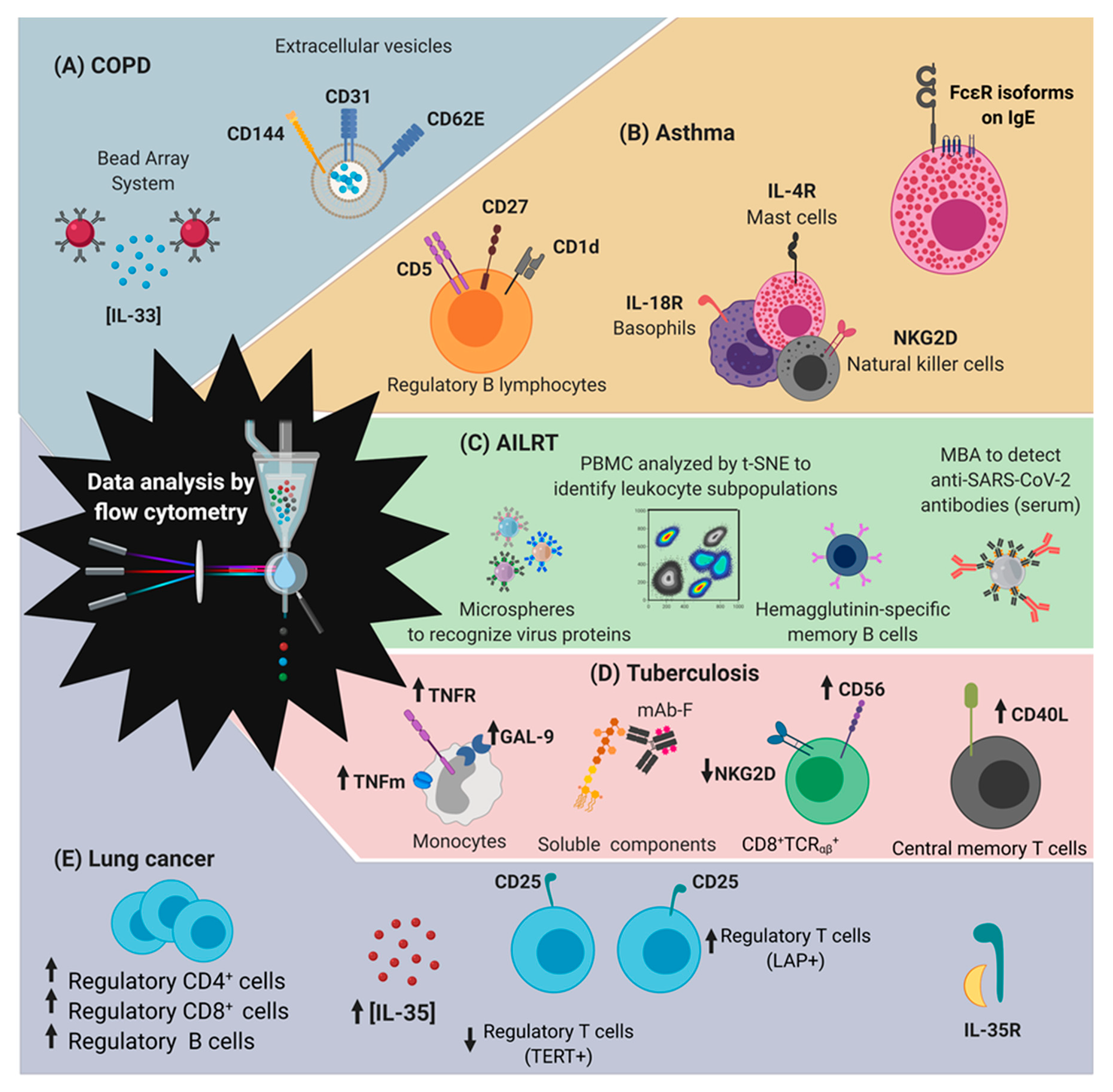
| Respiratory Disease | [A]Conventional Diagnostic and Monitoring Methods | Alternative to Evaluate by Flow Cytometry to Support the Diagnostics and Monitoring | References | |
|---|---|---|---|---|
| [B]COPD | Spirometry test | IL-33 (alarmin) | [49] | |
| Extracellular vesicles (CD144+CD31+CD62E+) | [50] | |||
| Asthma | Skin test [C]FeNO test Family history | IgE –FcεRIα expression on basophils, monocytes and plasmacytoid dendritic cells | [52] | |
| CD5, CD1d and CD27 expression on B cell | [53] | |||
| NKT invariant cells (Vα24Jα18) | [54] | |||
| NKG2D expression on NK cells | [55] | |||
| CD8+IL-13+ T cell | [56] | |||
| IL-4R expression on conversion from Tireg to Th17 | [57] | |||
| IL- 18R expression on basophils and mast cells | [58] | |||
| [D]AILRT | Influenza | [E]RT-PCR Lung exploration | Cell maps to identify variations of PBMC based on [F]t-SNE analysis | [62,66] |
| Microsphere-based antibody assay | [63,64] | |||
| Hemagglutinin-specific memory B cells | [65] | |||
| SARS-CoV-2 | [G]NAAT [E]RT-PCR Serological testing | Cell maps to identify variations of T cells based on [F]t-SNE analysis | [67] | |
| Microsphere-based antibody assay | [68] | |||
| TB | Clinical data [H]IGRA [I]TST [JI]AFB Smear Mycobacterial Cultures [F]NAAT | Galectin 9, TLR-2 and 4, CD68, CD33, and CD86 expression on macrophages and monocytes | [70,71] | |
| CD8+TCRαβ+NKG2DdimCD56high | [72] | |||
| CD3/TCRαβ monocytes | [73,74,75] | |||
| CD40L expression on T cells (central memory) | [76] | |||
| CD161 expression on T cells | [77] | |||
| Lung cancer | Imaging tests Biopsy Lung function tests | PD-1 expression | [79,80] | |
| IL-35 on NSCLC | [81] | |||
| TregCD4+CD25high | [82] | |||
| CD4+TERT+ / CD4+LAP-TGF-β+ | [83,84] | |||
| CD8+CD28+ | [85] | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flores-Gonzalez, J.; Cancino-Díaz, J.C.; Chavez-Galan, L. Flow Cytometry: From Experimental Design to Its Application in the Diagnosis and Monitoring of Respiratory Diseases. Int. J. Mol. Sci. 2020, 21, 8830. https://doi.org/10.3390/ijms21228830
Flores-Gonzalez J, Cancino-Díaz JC, Chavez-Galan L. Flow Cytometry: From Experimental Design to Its Application in the Diagnosis and Monitoring of Respiratory Diseases. International Journal of Molecular Sciences. 2020; 21(22):8830. https://doi.org/10.3390/ijms21228830
Chicago/Turabian StyleFlores-Gonzalez, Julio, Juan Carlos Cancino-Díaz, and Leslie Chavez-Galan. 2020. "Flow Cytometry: From Experimental Design to Its Application in the Diagnosis and Monitoring of Respiratory Diseases" International Journal of Molecular Sciences 21, no. 22: 8830. https://doi.org/10.3390/ijms21228830
APA StyleFlores-Gonzalez, J., Cancino-Díaz, J. C., & Chavez-Galan, L. (2020). Flow Cytometry: From Experimental Design to Its Application in the Diagnosis and Monitoring of Respiratory Diseases. International Journal of Molecular Sciences, 21(22), 8830. https://doi.org/10.3390/ijms21228830






