Connection between the Gut Microbiome, Systemic Inflammation, Gut Permeability and FOXP3 Expression in Patients with Primary Sjögren’s Syndrome
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics of the pSS and the Healthy Control Groups
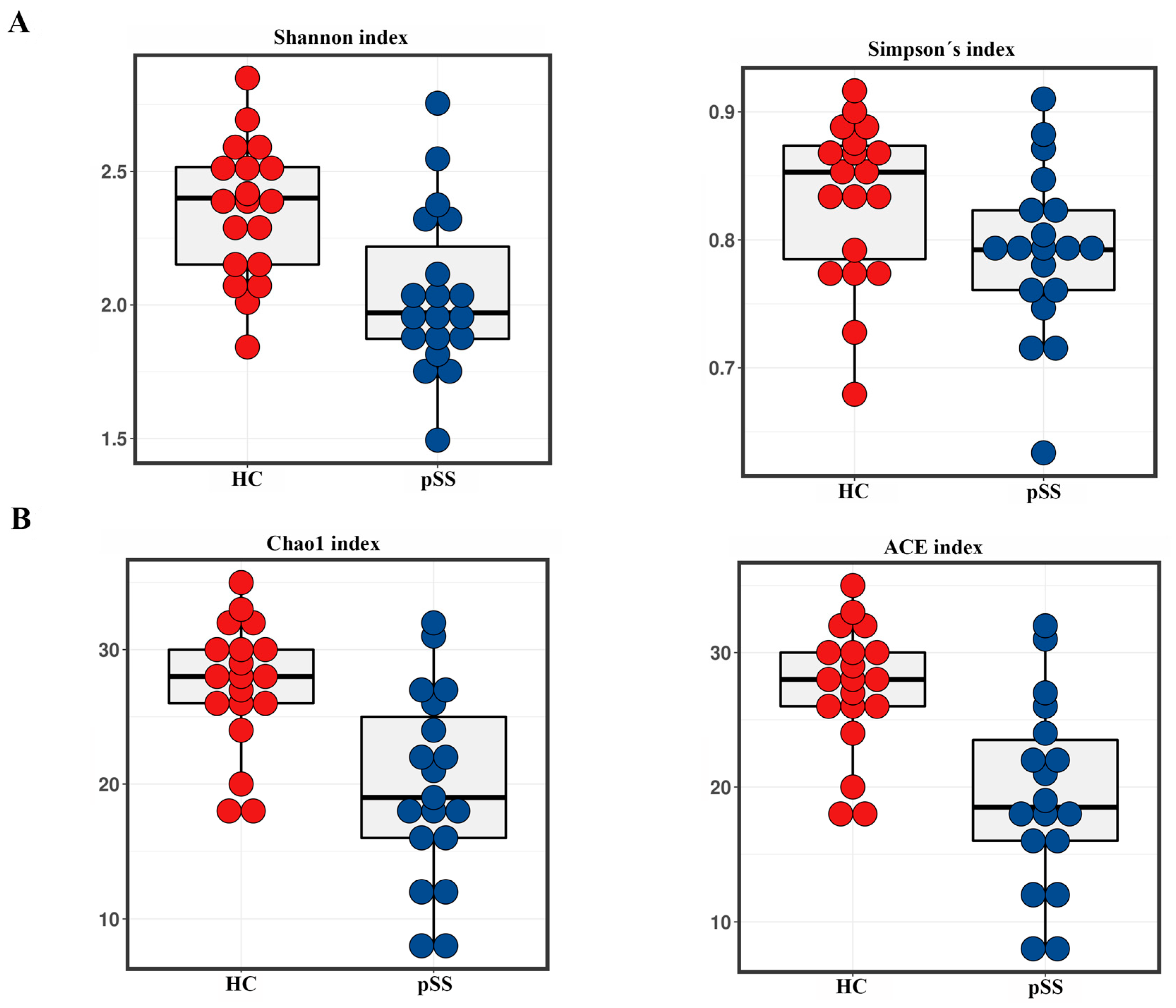
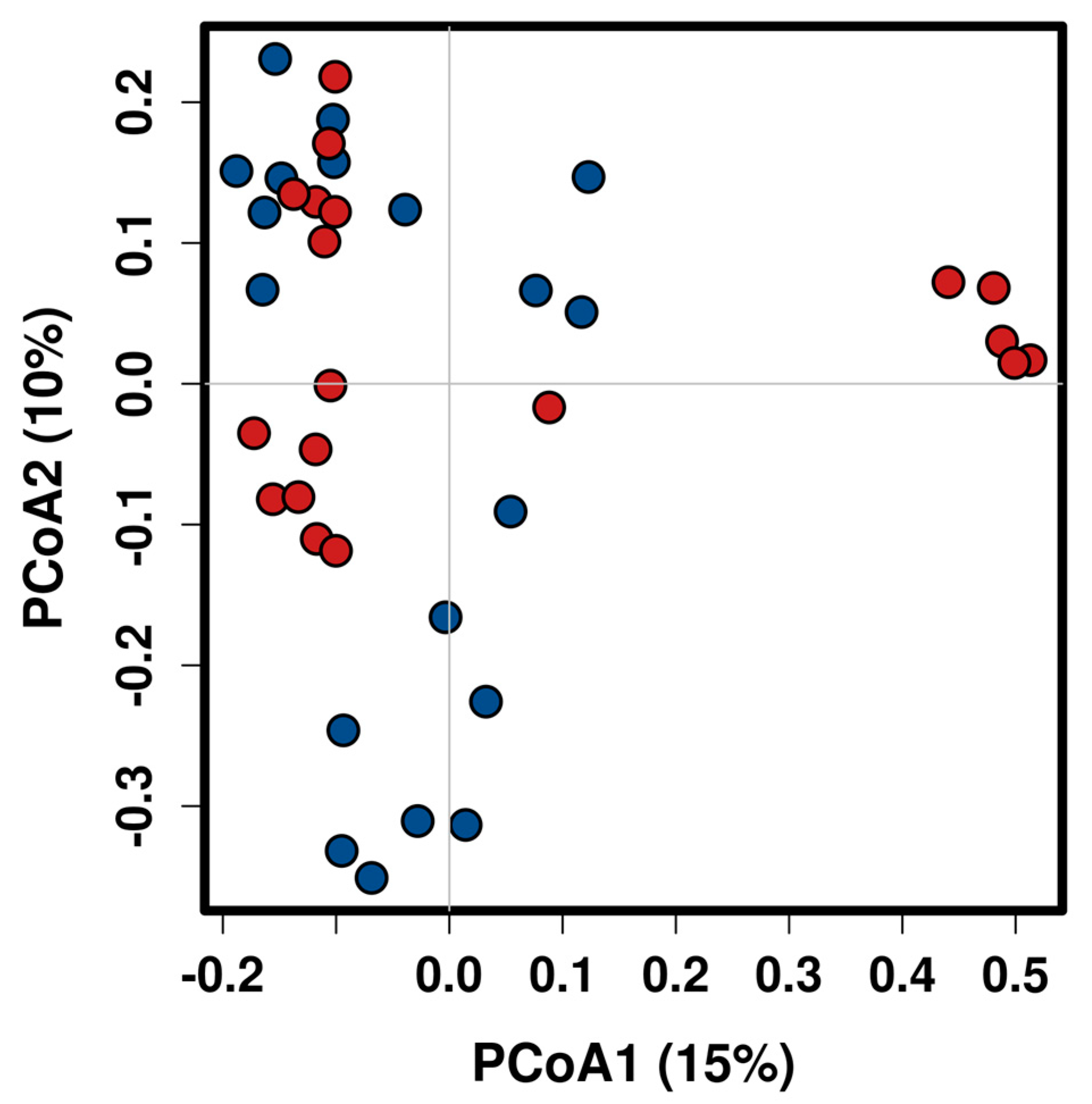
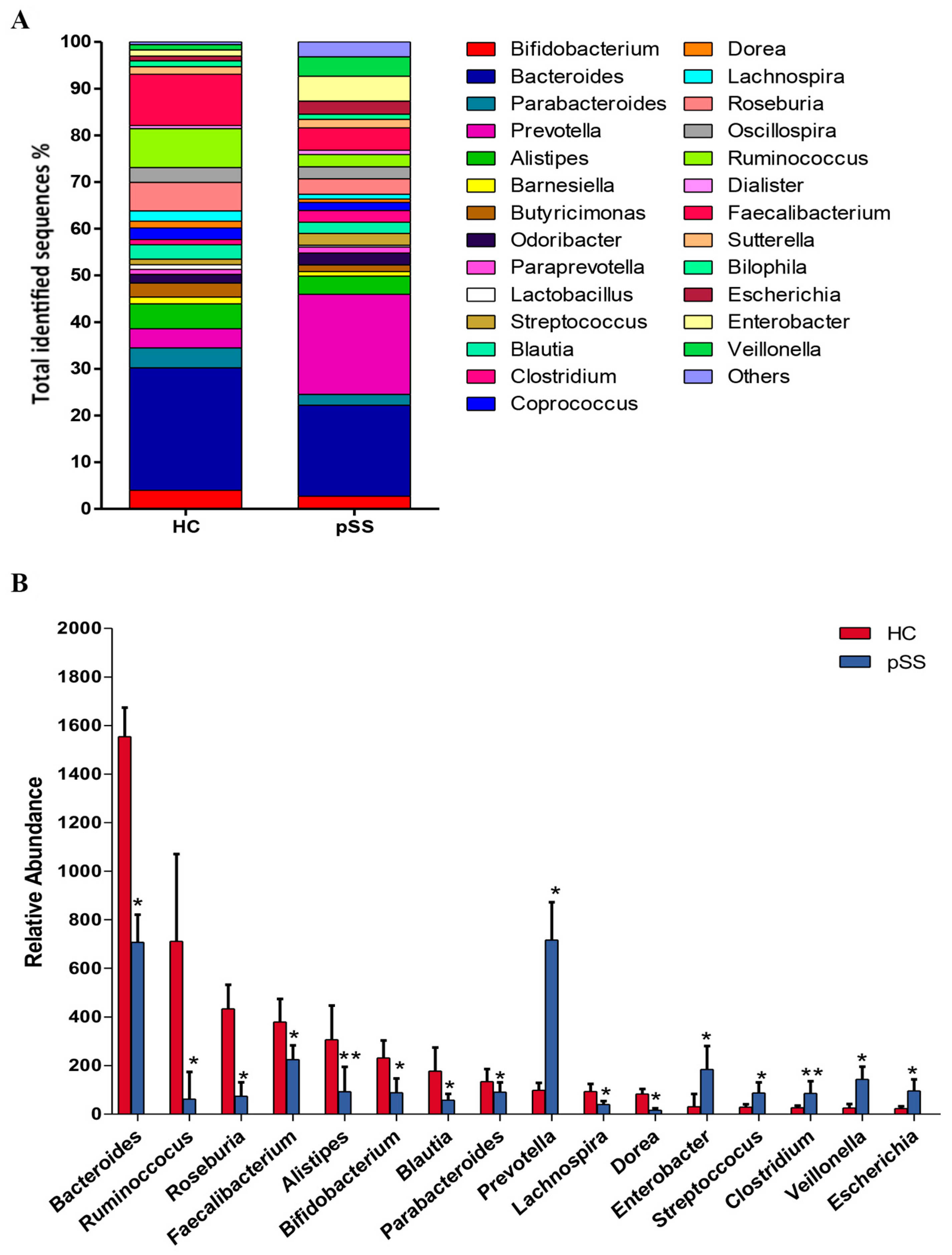
2.2. Taxonomic Composition and Diversity of Gut Microbiota
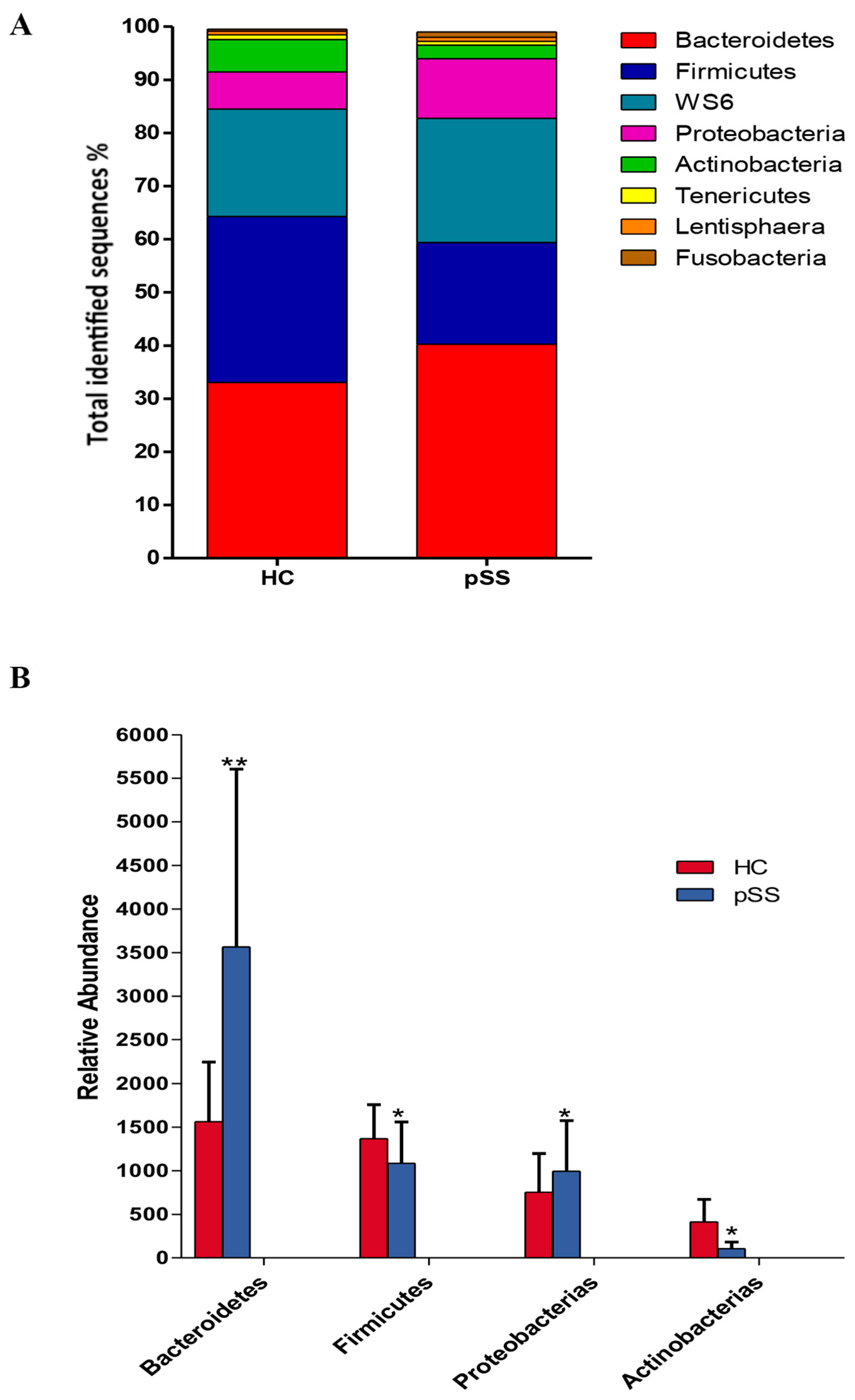
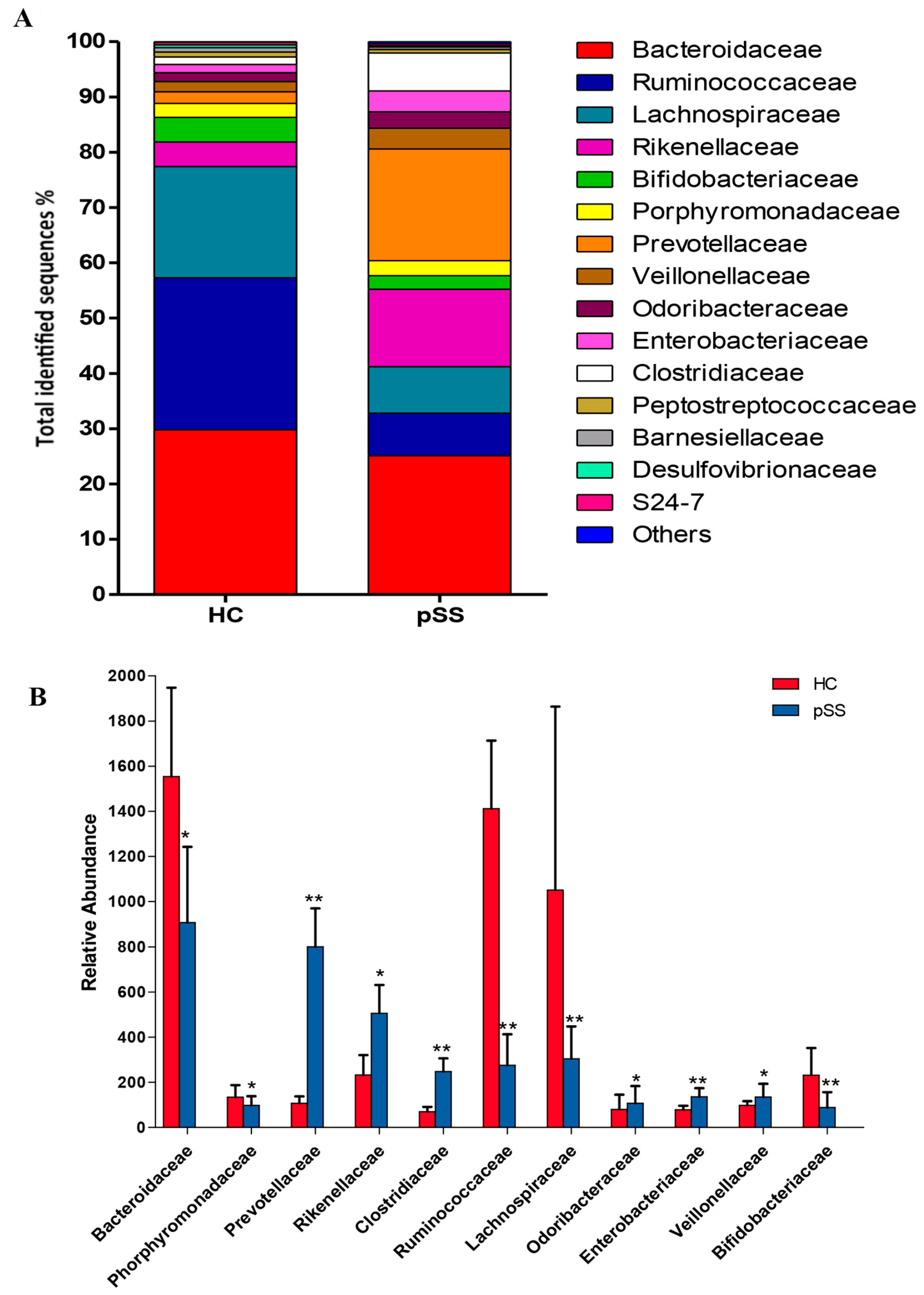
2.3. Comparative Analysis of the Gut Microbial Composition of Patients with pSS and Healthy Controls
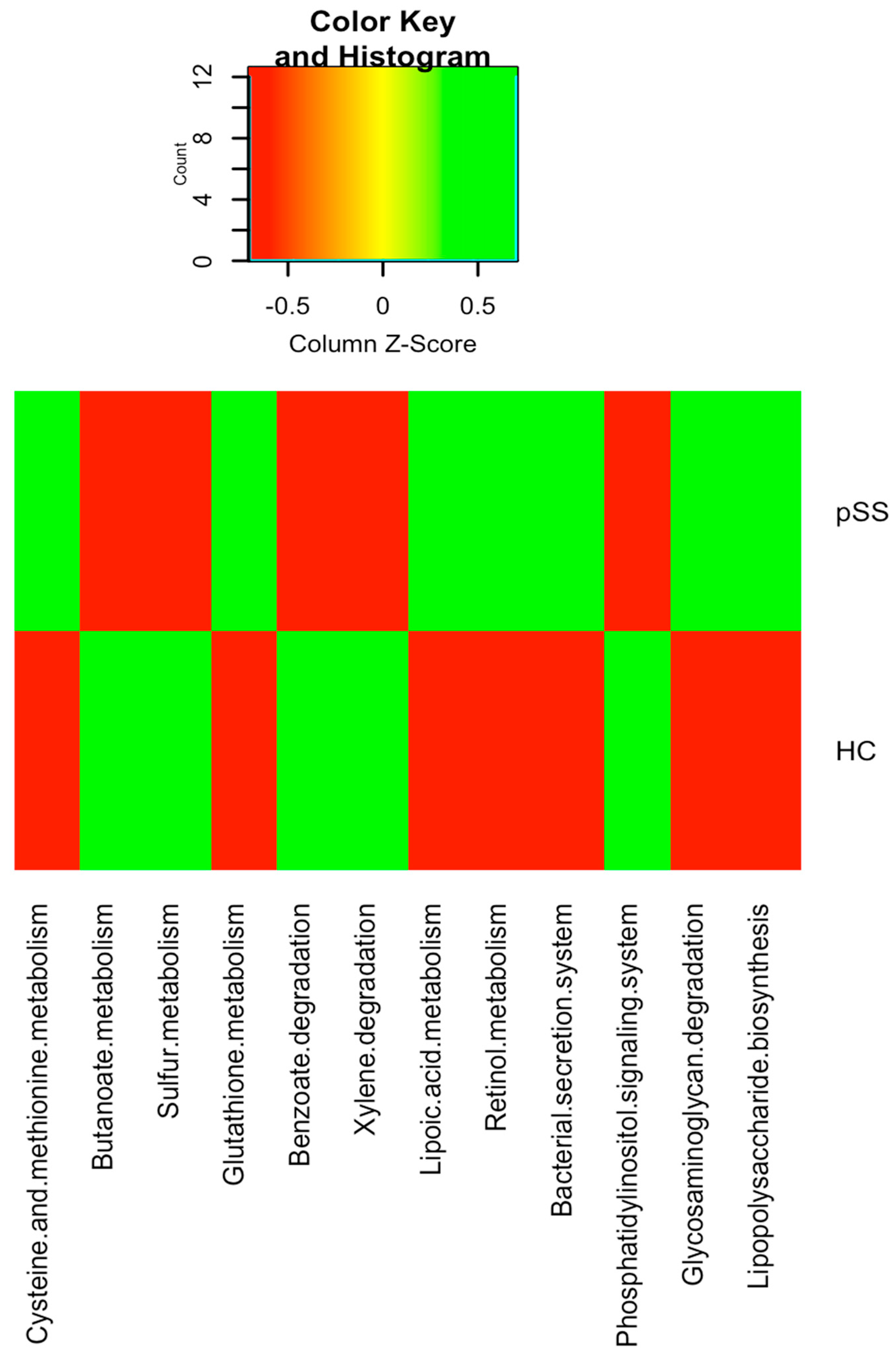
2.4. Functional Differences in Gut Microbiota between pSS Patients and Healthy Individuals
2.5. Differences in the Serum Levels of Inflammatory Mediators and Zonulin and the Relative Expression of FOXP3 in pSS Patients Compared to Healthy Controls
2.6. Gut Microbiota was Significantly Associated with Serum Levels of Inflammatory Mediators and Zonulin and the Relative Expression of FOXP3 in pSS Patients
3. Discussion
4. Materials and Methods
4.1. Study Subjects
4.2. Laboratory Measurements
4.3. DNA Extraction and Gut Microbiota Sequencing
4.4. Bioinformatics Analysis
4.5. Intestinal Permeability Analysis
4.6. Measurement of Serum Cytokine Levels
4.7. Real-Time Quantitative PCR for Treg Cells
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACR EULAR | American College of Rheumatology/European League Against Rheumatism |
| ANA | Antinuclear antibody |
| BMI | Body mass index |
| C3/C4 | Complement C3/C4 |
| CRP | c-reactive protein |
| ENA | Extractable nuclear antigen |
| FDR | False discovery rate |
| FoxP3 | Transcription factor Forkhead box protein P3 |
| HDL | High density lipoprotein |
| IFN | gamma Interferon-gamma |
| IL | Interleukin |
| KEGG | Kyoto encyclopedia of genes and genomes |
| La/SSB | Anti La/Sjögren’s syndrome |
| LDL | Low density lipoprotein |
| OTU | Operational taxonomic unit |
| PCoA | Principal Coordinates Analysis |
| PICRUSt | Phylogenetic investigation of communities of unobserved states |
| pSS | Primary Sjögren’s syndrome |
| QIIME | Quantitative Insights Into Microbial Ecology |
| Ro/SSA | Anti-Ro/Sjögren’s syndrome autoantibody A |
| SD | standard deviation |
| TNF | alpha Tumor necrosis factor-alpha |
| Tregs | Regulatory T cells |
References
- Mariette, X.; Criswell, L.A. Primary Sjögren’s Syndrome. N. Engl. J. Med. 2018, 378, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Carsons, S. A review and update of Sjögren’s syndrome: Manifestations, diagnosis, and treatment. Am. J. Manag. Care 2001, 7, S433–S443. [Google Scholar] [PubMed]
- Kuo, C.; Grainge, M.J.; Valdes, A.M.; See, L.; Luo, S.; Yu, K.; Zhang, W.; Doherty, M. Familial Risk of Sjögren’s Syndrome and Co-aggregation of Autoimmune Diseases in Affected Families: A Nationwide Population Study. Arthr. Rheumatol. 2015, 67, 1904–1912. [Google Scholar] [CrossRef] [PubMed]
- Breban, M.; Beaufrère, M.; Glatigny, S. The microbiome in spondyloarthritis. Best Pr. Res. Clin. Rheumatol. 2019, 33, 101495. [Google Scholar] [CrossRef] [PubMed]
- Scher, J.U.; Littman, D.R.; Abramson, S.B. Review: Microbiome in Inflammatory Arthritis and Human Rheumatic Diseases. Arthr. Rheumatol. 2016, 68, 35–45. [Google Scholar] [CrossRef]
- Hevia, A.; Milani, C.; López, P.; Cuervo, A.; Arboleya, S.; Duranti, S.; Turroni, F.; González, S.; Suárez, A.; Gueimonde, M.; et al. Intestinal Dysbiosis Associated with Systemic Lupus Erythematosus. mBio 2014, 5, e01548-14. [Google Scholar] [CrossRef]
- De Paiva, C.S.; Jones, D.B.; Stern, M.E.; Bian, F.; Moore, Q.L.; Corbiere, S.; Streckfus, C.F.; Hutchinson, D.S.; Ajami, N.J.; Petrosino, J.F.; et al. Altered Mucosal Microbiome Diversity and Disease Severity in Sjögren Syndrome. Sci. Rep. 2016, 6, 23561. [Google Scholar] [CrossRef]
- Mandl, T.; Marsal, J.; Olsson, P.; Ohlsson, B.; Andréasson, K. Severe intestinal dysbiosis is prevalent in primary Sjögren’s syndrome and is associated with systemic disease activity. Arthr. Res. 2017, 19, 237. [Google Scholar] [CrossRef]
- Zhong, D.; Wu, C.; Zeng, X.; Wang, Q. The role of gut microbiota in the pathogenesis of rheumatic diseases. Clin. Rheumatol. 2017, 37, 25–34. [Google Scholar] [CrossRef]
- Kim, J.; Lahl, K.; Hori, S.; Loddenkemper, C.; Chaudhry, A.; DeRoos, P.; Rudensky, A.; Sparwasser, T. Cutting Edge: Depletion of Foxp3+ Cells Leads to Induction of Autoimmunity by Specific Ablation of Regulatory T Cells in Genetically Targeted Mice. J. Immunol. 2009, 183, 7631–7634. [Google Scholar] [CrossRef]
- Honda, K.; Littman, D.R. The microbiota in adaptive immune homeostasis and disease. Nat. Cell Biol. 2016, 535, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Forbes, J.D.; Van Domselaar, G.; Bernstein, C.N. The Gut Microbiota in Immune-Mediated Inflammatory Diseases. Front. Microbiol. 2016, 7, 1081. [Google Scholar] [CrossRef] [PubMed]
- Koskenpato, K.; Ainola, M.; Przybyla, B.; Kouri, V.-P.; Virkki, L.; Koskenpato, J.; Ristimäki, A.; Konttinen, Y. Diminished salivary epidermal growth factor secretion: A link between Sjögren’s syndrome and autoimmune gastritis? Scand. J. Rheumatol. 2016, 45, 118–121. [Google Scholar] [CrossRef]
- Ebert, E.C. Gastrointestinal and Hepatic Manifestations of Sjogren Syndrome. J. Clin. Gastroenterol. 2012, 46, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Li, D.; Shi, G.; Zhang, X.; Pan, Y.; Dou, H.; Wang, T.; Yao, G.; Hou, Y. Interleukin-12 exacerbates Sjögren’s syndrome through induction of myeloid-derived suppressor cells. Mol. Med. Rep. 2019, 20, 1131–1138. [Google Scholar] [CrossRef]
- Ferreira, C.M.; Vieira, A.T.; Vinolo, M.A.R.; Oliveira, F.A.; Curi, R.; Martins, F.D.S. The Central Role of the Gut Microbiota in Chronic Inflammatory Diseases. J. Immunol. Res. 2014, 2014, 1–12. [Google Scholar] [CrossRef]
- Mendez, R.; Watane, A.; Farhangi, M.; Cavuoto, K.M.; Leith, T.; Budree, S.; Galor, A.; Banerjee, S. Gut microbial dysbiosis in individuals with Sjögren’s syndrome. Microb. Cell Fact. 2020, 19, 1–13. [Google Scholar] [CrossRef]
- Van Der Meulen, T.A.; Harmsen, H.J.; Vila, A.V.; Kurilshikov, A.; Liefers, S.C.; Zhernakova, A.; Fu, J.; Wijmenga, C.; Weersma, R.K.; De Leeuw, K.; et al. Shared gut, but distinct oral microbiota composition in primary Sjögren’s syndrome and systemic lupus erythematosus. J. Autoimmun. 2019, 97, 77–87. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nat. Cell Biol. 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Pianta, A.; Arvikar, S.; Strle, K.; Drouin, E.E.; Wang, Q.; Costello, C.E.; Steere, A.C. Evidence of the Immune Relevance of Prevotella copri, a Gut Microbe, in Patients With Rheumatoid Arthritis. Arthr. Rheumatol. 2017, 69, 964–975. [Google Scholar] [CrossRef]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2013, 2, e01202. [Google Scholar] [CrossRef] [PubMed]
- Eomenetti, S.; Pizarro, T.T. The Treg/Th17 Axis: A Dynamic Balance Regulated by the Gut Microbiome. Front. Immunol. 2015, 6, 639. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Choi, S.H.; Yoon, C.H.; Kim, M.K. Gut dysbiosis is prevailing in Sjögren’s syndrome and is related to dry eye severity. PLoS ONE 2020, 15, e0229029. [Google Scholar] [CrossRef] [PubMed]
- Favier, C.; Neut, C.; Mizon, C.; Cortot, A.; Colombel, J.F.; Mizon, J. Fecal beta-D-galactosidase production and Bifidobacteria are decreased in Crohn’s disease. Dig. Dis Sci. 1997, 42, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Radjabzadeh, D.; Boer, C.G.; Beth, S.A.; Van Der Wal, P.; Jong, J.C.K.-D.; Jansen, M.A.E.; Konstantinov, S.R.; Peppelenbosch, M.P.; Hays, J.P.; Jaddoe, V.W.; et al. Diversity, compositional and functional differences between gut microbiota of children and adults. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Takagi, T.; Naito, Y.; Inoue, R.; Kashiwagi, S.; Uchiyama, K.; Mizushima, K.; Tsuchiya, S.; Dohi, O.; Yoshida, N.; Kamada, K.; et al. Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects. J. Gastroenterol. 2019, 54, 53–63. [Google Scholar] [CrossRef]
- De La Cuesta-Zuluaga, J.; Kelley, S.T.; Chen, Y.; Escobar, J.S.; Mueller, N.T.; Ley, R.E.; McDonald, D.; Huang, S.; Swafford, A.D.; Knight, R.; et al. Age- and Sex-Dependent Patterns of Gut Microbial Diversity in Human Adults. mSystems 2019, 4, e00261-19. [Google Scholar] [CrossRef]
- Fasano, A. Zonulin and Its Regulation of Intestinal Barrier Function: The Biological Door to Inflammation, Autoimmunity, and Cancer. Physiol. Rev. 2011, 91, 151–175. [Google Scholar] [CrossRef]
- Sturgeon, C.; Fasano, A. Zonulin, a regulator of epithelial and endothelial barrier functions, and its involvement in chronic inflammatory diseases. Tissue Barriers 2016, 4, e1251384. [Google Scholar] [CrossRef]
- Wright, D.P.; Rosendale, D.; Roberton, A.M. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol. Lett. 2000, 190, 73–79. [Google Scholar] [CrossRef]
- Su, T.; Liu, R.; Lee, A.; Long, Y.; Du, L.; Lai, S.; Chen, X.; Wang, L.; Si, J.; Owyang, C.; et al. Altered Intestinal Microbiota with Increased Abundance of Prevotella Is Associated with High Risk of Diarrhea-Predominant Irritable Bowel Syndrome. Gastroenterol. Res. Pr. 2018, 2018, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.R. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009, 9, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Iljazovic, A.; Roy, U.; Gálvez, E.J.C.; Lesker, T.R.; Zhao, B.; Gronow, A.; Amend, L.; Will, S.E.; Hofmann, J.D.; Pils, M.C.; et al. Perturbation of the gut microbiome by Prevotella spp. enhances host susceptibility to mucosal inflammation. Mucosal Immunol. 2020, 1–12. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed]
- 35. Tamanai-Shacoori, Z.; Smida, I.; Bousarghin, L.; Loreal, O.; Meuric, V.; Fong, S.B.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Roseburia spp.: A marker of health? Future Microbiol. 2017, 12, 157–170. [Google Scholar] [CrossRef]
- Lewis, K.; Lutgendorff, F.; Phan, V.; Söderholm, J.D.; Sherman, P.M.; McKay, D.M. Enhanced translocation of bacteria across metabolically stressed epithelia is reduced by butyrate†. Inflamm. Bowel Dis. 2010, 16, 1138–1148. [Google Scholar] [CrossRef]
- Masui, R.; Sasaki, M.; Funaki, Y.; Ogasawara, N.; Mizuno, M.; Iida, A.; Izawa, S.; Kondo, Y.; Ito, Y.; Tamura, Y.; et al. G Protein-Coupled Receptor 43 Moderates Gut Inflammation Through Cytokine Regulation from Mononuclear Cells. Inflamm. Bowel Dis. 2013, 19, 2848–2856. [Google Scholar] [CrossRef]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, Receptor for Niacin and the Commensal Metabolite Butyrate, Suppresses Colonic Inflammation and Carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef]
- Fernando, M.R.; Saxena, A.; Reyes, J.L.; McKay, D.M. Butyrate enhances antibacterial effects while suppressing other features of alternative activation in IL-4-induced macrophages. Am. J. Physiol. Liver Physiol. 2016, 310, G822–G831. [Google Scholar] [CrossRef]
- Saemann, M.D.; Böhmig, G.A.; Österreicher, C.H.; Burtscher, H.; Parolini, O.; Diakos, C.; Stöckl, J.; Hörl, W.H.; Zlabinger, G.J. Anti-inflammatory effects of sodium butyrate on human monocytes: Potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J. 2000, 14, 2380–2382. [Google Scholar] [CrossRef]
- Chen, J.; Vitetta, L. The Role of Butyrate in Attenuating Pathobiont-Induced Hyperinflammation. Immune Netw. 2020, 20, e15. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Esquivel-Elizondo, S.; Ilhan, Z.E.; Garcia-Peña, E.I.; Krajmalnik-Brown, R. Insights into Butyrate Production in a Controlled Fermentation System via Gene Predictions. mSystems 2017, 2, e00051-17. [Google Scholar] [CrossRef]
- De Oliveira, G.L.V.; Leite, A.Z.; Higuchi, B.S.; Gonzaga, M.I.; Mariano, V.S. Intestinal dysbiosis and probiotic applications in autoimmune diseases. Immunology 2017, 152, 1–12. [Google Scholar] [CrossRef]
- Shah, D.; Sah, S.; Nath, S.K. Interaction between glutathione and apoptosis in systemic lupus erythematosus. Autoimmun. Rev. 2013, 12, 741–751. [Google Scholar] [CrossRef]
- Spalding, M.D.; Prigge, S.T. Lipoic Acid Metabolism in Microbial Pathogens. Microbiol. Mol. Biol. Rev. 2010, 74, 200–228. [Google Scholar] [CrossRef]
- Winslet, M.C.; Poxon, V.; Allan, A.; Keighley, M.R.B. Mucosal glucosamine synthetase activity in inflammatory bowel disease. Dig. Dis. Sci. 1994, 39, 540–544. [Google Scholar] [CrossRef]
- Abdelhamid, L.; Luo, X.M. Retinoic Acid, Leaky Gut, and Autoimmune Diseases. Nutrients 2018, 10, 1016. [Google Scholar] [CrossRef]
- Shiboski, C.H.; Shiboski, S.C.; Seror, R.; Criswell, L.A.; Labetoulle, M.; Lietman, T.M.; Rasmussen, A.; Scofield, H.; Vitali, C.; Bowman, S.J.; et al. 2016 American College of Rheumatology/European League Against Rheumatism Classification Criteria for Primary Sjögren’s Syndrome: A Consensus and Data-Driven Methodology Involving Three International Patient Cohorts. Arthr. Rheumatol. 2017, 69, 35–45. [Google Scholar] [CrossRef]
- Martínez-González, M.-A.; García-Arellano, A.; Toledo, E.; Salas-Salvadó, J.; Buil-Cosiales, P.; Corella, D.; Covas, M.I.; Schröder, H.; Arós, F.; Gómez-Gracia, E.; et al. A 14-Item Mediterranean Diet Assessment Tool and Obesity Indexes among High-Risk Subjects: The PREDIMED Trial. PLoS ONE 2012, 7, e43134. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Alcoholado, L.; Ordóñez, R.; Otero, A.; Plaza-Andrade, I.; Laborda-Illanes, A.; Medina, J.A.; Ramos-Molina, B.; Gómez-Millán, J.; Queipo-Ortuño, M.I. Gut Microbiota-Mediated Inflammation and Gut Permeability in Patients with Obesity and Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 6782. [Google Scholar] [CrossRef] [PubMed]
- Bhute, S.; Pande, P.; Shetty, S.A.; Shelar, R.; Mane, S.; Kumbhare, S.V.; Gawali, A.; Makhani, H.; Navandar, M.; Dhotre, D.; et al. Molecular Characterization and Meta-Analysis of Gut Microbial Communities Illustrate Enrichment of Prevotella and Megasphaera in Indian Subjects. Front. Microbiol. 2016, 7, 660. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| pSS (n = 19) | Healthy Controls (n = 19) | p | |
|---|---|---|---|
| Age in years, mean ± SD | 56.43 ± 8.74 | 54.70 ± 8.42 | 0.545 |
| Female sex, n (%) | 19 (100) | 19 (100) | 0.589 |
| BMI, mean ±SD | 26.17 ± 6.27 | 26.25 ± 4.15 | 0.963 |
| Smoking, n (%) | 7 (36.84) | 8 (42.10) | 0.736 |
| Caucasian, n (%) | 19 (100) | 19 (100) | 0.589 |
| Fulfilling ACR EULAR 2016 criteria for SS, n (%) | 19 (100) | NA | - |
| Serum biochemistry | |||
| Glucose (mg/dl) | 93.44 ± 22.14 | 92.16 ± 18.24 | 0.847 |
| Insulin (mg/dl) | 8.81 ± 3.63 | 6.82 ± 3.72 | 0.104 |
| Triglycerides (mg/dl) | 90.41 ± 29.65 | 97.92 ± 27.51 | 0.398 |
| Cholesterol (mg/dl) | 189.38 ± 22.29 | 176.3 ± 34.6 | 0.175 |
| LDL-cholesterol (mg/dl) | 123.76 ± 31.0 | 118.20 ± 33.75 | 0.600 |
| HDL-cholesterol (mg/dl) | 63.16 ± 14.73 | 59.84 ± 15.60 | 0.504 |
| CRP (ng/mL) | 3.80 ± 0.49 | 2.77 ± 0.63 | 0.001 |
| ANA positive, n (%) | 15 (78.94) | NA | - |
| ENA positive, n (%) | 6 (31.57%) | NA | - |
| Ro/SSA positive, n (%) | 11 (57.89) | NA | - |
| La/SSB positive, n (%) | 12 (63.15) | NA | - |
| Rheumatoid factor positive, n (%) | 13 (68.42) | NA | - |
| C3 low at inclusion (<0.90 g/L), n (%) | 7 (36.84) | NA | - |
| C4 low at inclusion (<0.10 g/L), n (%) | 7 (36.84) | NA | - |
| Other symptoms reported | |||
| Hypothyroidism, n (%) | 3(15.78) | 0 (0) | - |
| Fibromyalgia, n (%) | 2 (10.52) | 0 (0) | - |
| Oral ulcers, n (%) | 1 (5.26) | 0 (0) | - |
| Arthralgia, n (%) | 3 (15.78) | 0 (0) | - |
| Type 2 diabetes, n (%) | 1 (5.26) | 0 (0) | - |
| pSS | Healthy Controls | pSS | Healthy Controls | pSS | Healthy Controls | |
|---|---|---|---|---|---|---|
| IL-6 | IL-12 | IL-17 | ||||
| Butyciromonas | −0.598 (p = 0.030) | |||||
| Roseburia | −0.718 (p = 0.010) | −0.767 (p = 0.020) | −0.511 (p = 0.045) | −0.675 (p = 0.047) | ||
| Ruminococcus | −0.527 (p = 0.020) | −0.533 (p = 0.029) | −0.631 (p = 0.025) | |||
| Blautia | −0.635 (p = 0.007) | |||||
| Enterobacter | 0.873 (p = 0.008) | 0.549 (p = 0.039) | ||||
| Streptococcus | 0.466 (p = 0.044) | |||||
| Faecalibacterium prausnitzii | −0.587 (p = 0.028) | −0.508 (p = 0.027) | −0.550 (p= 0.025) | |||
| Escherichia coli | 0.723 (p = 0.009) | |||||
| Bifidobacterium | −0.573 (p = 0.030) | −0.508 (p = 0.018) | −0.507 (p = 0.027) | −0.789 (p = 0.002) | −0.459 (p = 0.041) | |
| Bacteroides fragilis | −0.473 (p = 0.041) | |||||
| Parabacteroides distasonis | −0.767 (p = 0.004) | |||||
| Lachnospira | −0.514 (p = 0.014) | |||||
| Prevotella | 0.465 (p = 0.045) | |||||
| Clostridium | 0.462 (p = 0.046) | 0.412 (p = 0.043) | ||||
| pSS | Healthy Controls | pSS | Healthy Controls | pSS | Healthy Controls | pSS | Healthy Controls | |
|---|---|---|---|---|---|---|---|---|
| TNF-Alpha | IL-10 | Zonulin | FOXP3 mRNA Expression | |||||
| Ruminococcus | −0.496 (p = 0.031) | 0.563 (p = 0.035) | 0.494 (p = 0.032) | −0.496 (p = 0.031) | −0.665 (p = 0.005) | 0.573 (p = 0.034) | ||
| Butyciromonas | −0.560 (p = 0.033) | 0.525 (p = 0.021) | −0.542 (p = 0.027) | 0.598 (p = 0.043) | ||||
| Blautia | 0.516 (p = 0.024) | 0.576 (p = 0.005) | ||||||
| Roseburia | −0.458 (p = 0.048) | 0.501 (p = 0.042) | ||||||
| Prevotella copri | 0.645 (p = 0.003) | |||||||
| Clostridium | 0.550 (p = 0.025) | −0.651 (p = 0.033) | ||||||
| Bifidobacterium | −0.580 (p = 0.009) | −0.631 (p = 0.010) | −0.584 (p = 0.017) | |||||
| Bacteroides fragilis | −0.567 (p = 0.041) | 0.631 (p = 0.002) | 0.563 (p = 0.009) | |||||
| Faecalibacterium prausnitzii | 0.651 (p = 0.010) | 0.597 (p = 0.025) | −0.502 (p = 0.029) | |||||
| Parabacteroides distasonis | −0.559 (p = 0.009) | −0.700 (p = 0.035) | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cano-Ortiz, A.; Laborda-Illanes, A.; Plaza-Andrades, I.; Membrillo del Pozo, A.; Villarrubia Cuadrado, A.; Rodríguez Calvo de Mora, M.; Leiva-Gea, I.; Sanchez-Alcoholado, L.; Queipo-Ortuño, M.I. Connection between the Gut Microbiome, Systemic Inflammation, Gut Permeability and FOXP3 Expression in Patients with Primary Sjögren’s Syndrome. Int. J. Mol. Sci. 2020, 21, 8733. https://doi.org/10.3390/ijms21228733
Cano-Ortiz A, Laborda-Illanes A, Plaza-Andrades I, Membrillo del Pozo A, Villarrubia Cuadrado A, Rodríguez Calvo de Mora M, Leiva-Gea I, Sanchez-Alcoholado L, Queipo-Ortuño MI. Connection between the Gut Microbiome, Systemic Inflammation, Gut Permeability and FOXP3 Expression in Patients with Primary Sjögren’s Syndrome. International Journal of Molecular Sciences. 2020; 21(22):8733. https://doi.org/10.3390/ijms21228733
Chicago/Turabian StyleCano-Ortiz, Antonio, Aurora Laborda-Illanes, Isaac Plaza-Andrades, Alberto Membrillo del Pozo, Alberto Villarrubia Cuadrado, Marina Rodríguez Calvo de Mora, Isabel Leiva-Gea, Lidia Sanchez-Alcoholado, and María Isabel Queipo-Ortuño. 2020. "Connection between the Gut Microbiome, Systemic Inflammation, Gut Permeability and FOXP3 Expression in Patients with Primary Sjögren’s Syndrome" International Journal of Molecular Sciences 21, no. 22: 8733. https://doi.org/10.3390/ijms21228733
APA StyleCano-Ortiz, A., Laborda-Illanes, A., Plaza-Andrades, I., Membrillo del Pozo, A., Villarrubia Cuadrado, A., Rodríguez Calvo de Mora, M., Leiva-Gea, I., Sanchez-Alcoholado, L., & Queipo-Ortuño, M. I. (2020). Connection between the Gut Microbiome, Systemic Inflammation, Gut Permeability and FOXP3 Expression in Patients with Primary Sjögren’s Syndrome. International Journal of Molecular Sciences, 21(22), 8733. https://doi.org/10.3390/ijms21228733





