In-Silico Identified New Natural Sortase A Inhibitors Disrupt S. aureus Biofilm Formation
Abstract
1. Introduction
2. Results
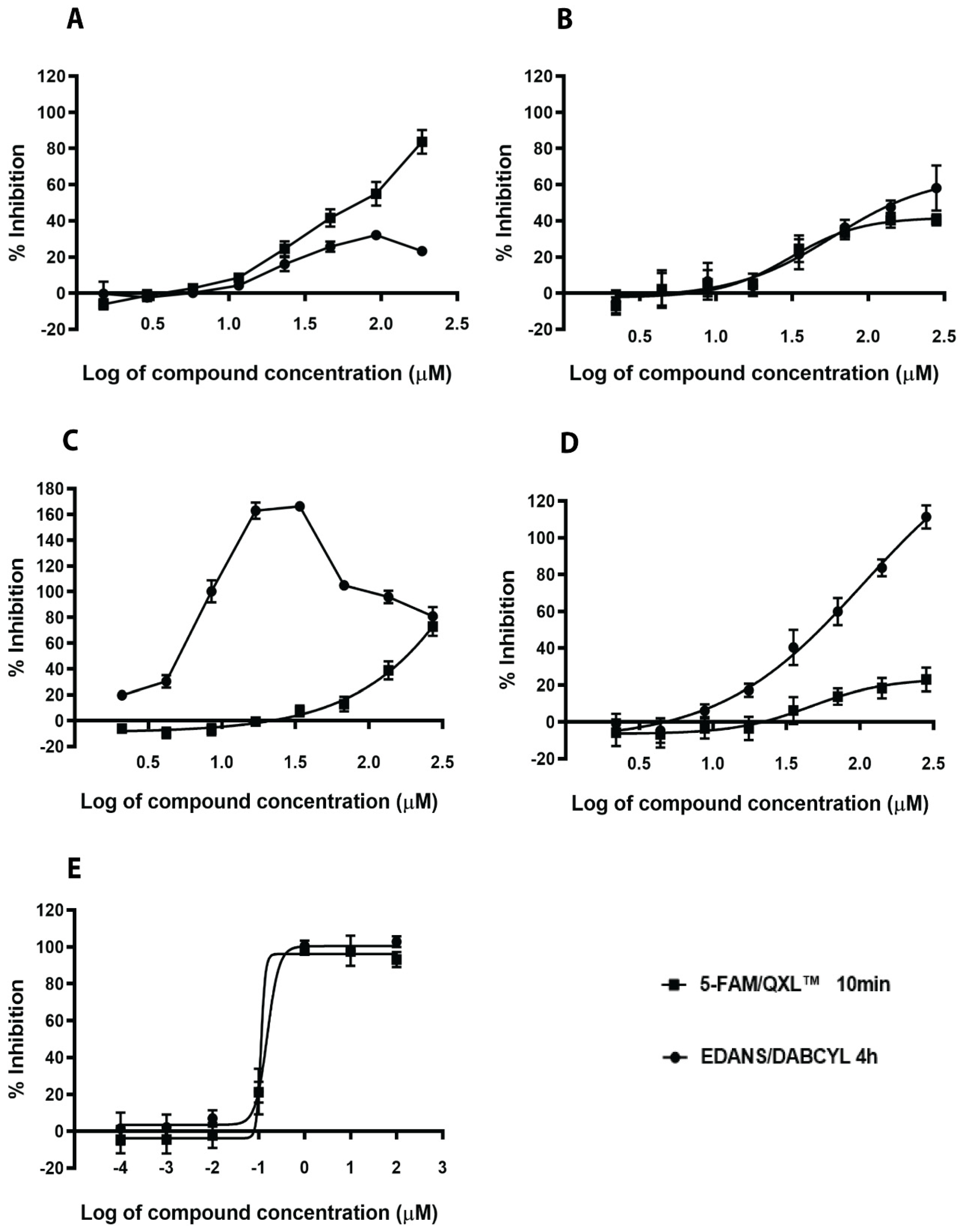
2.1. Virtual Screening and Validation of SrtA Inhibitors
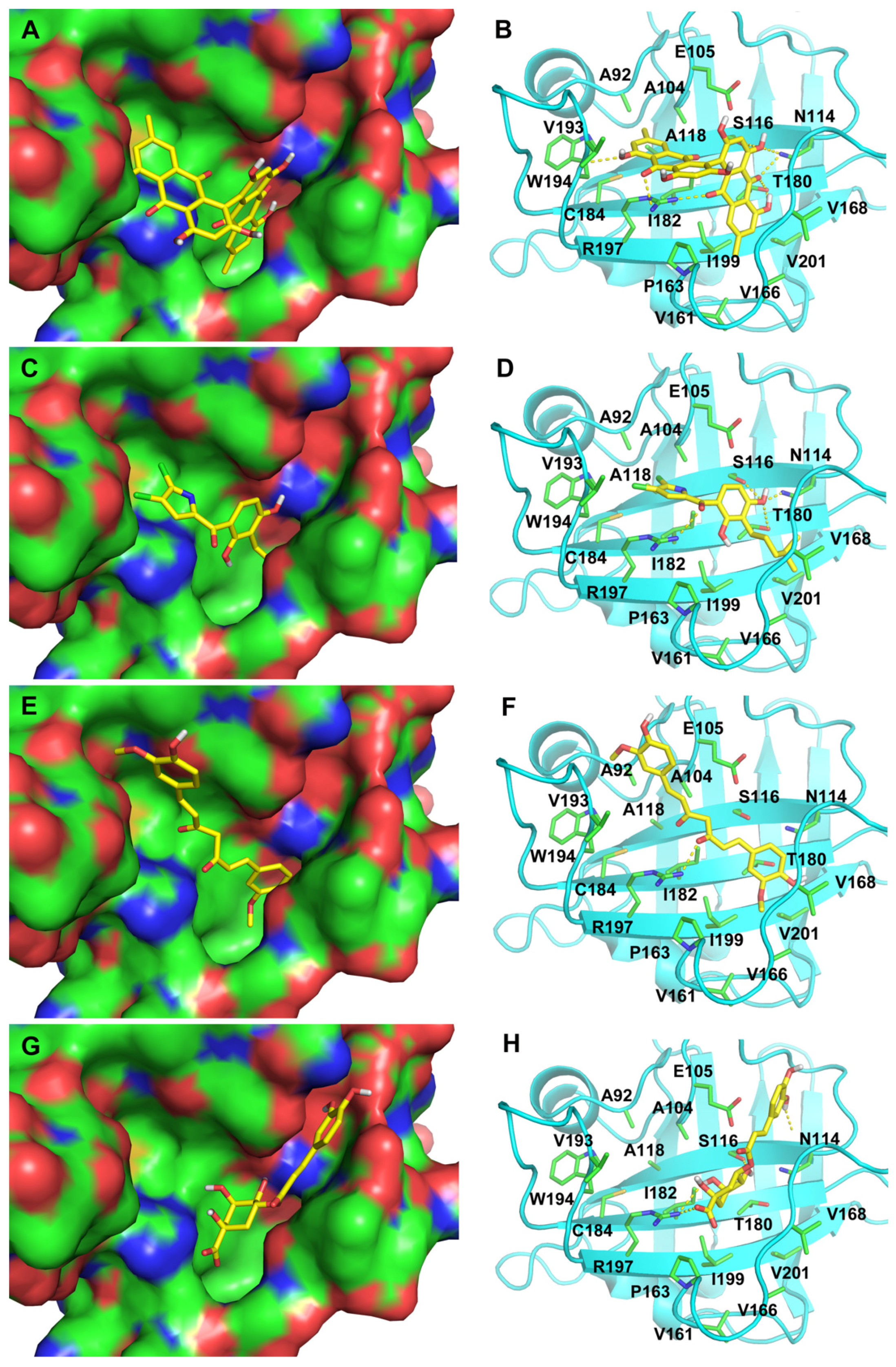
2.2. Structure Analysis of SrtA-Ligand Complexes
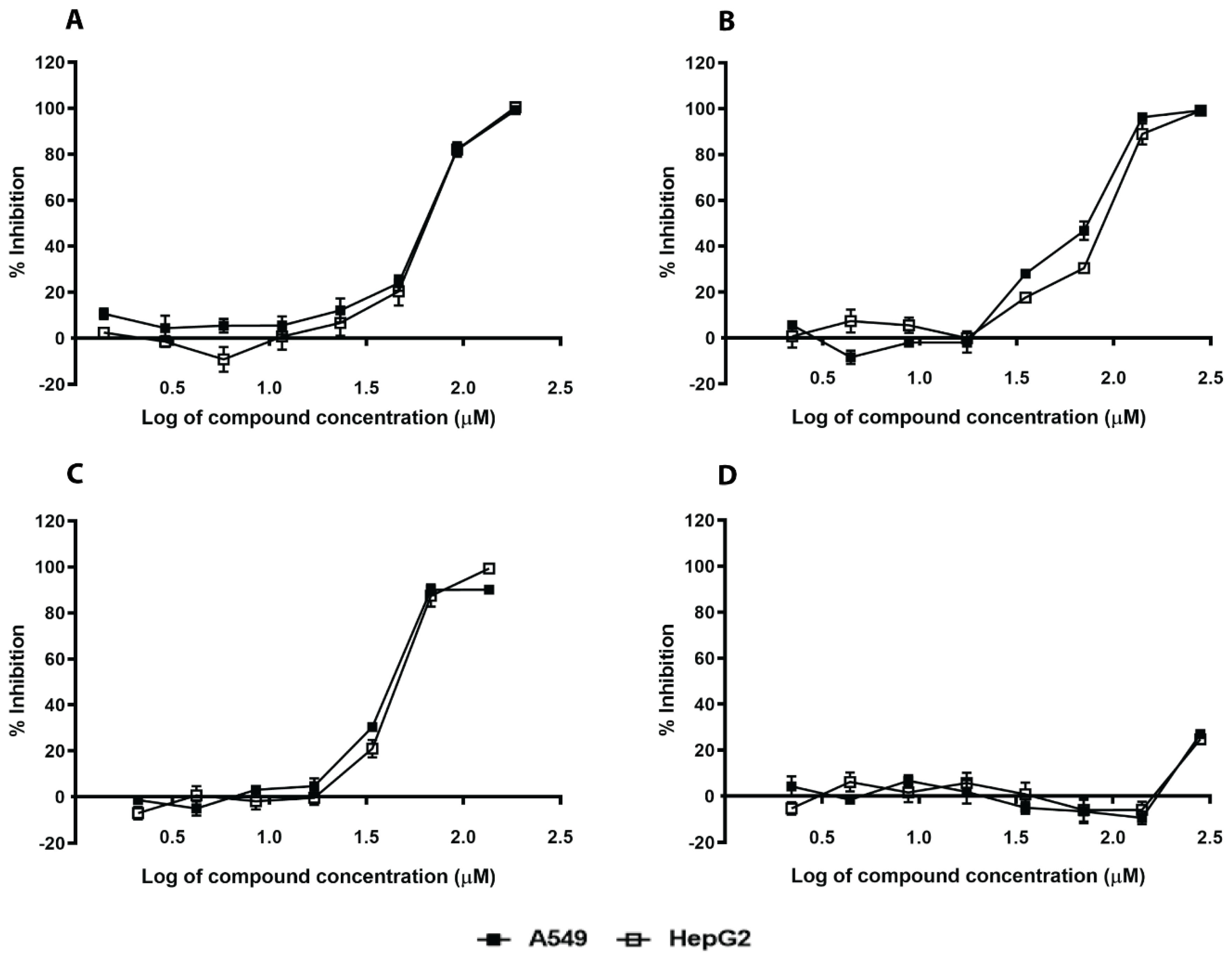
2.3. In Silico Hits Affect S. aureus Growth and Have Moderate Mammalian Toxicity
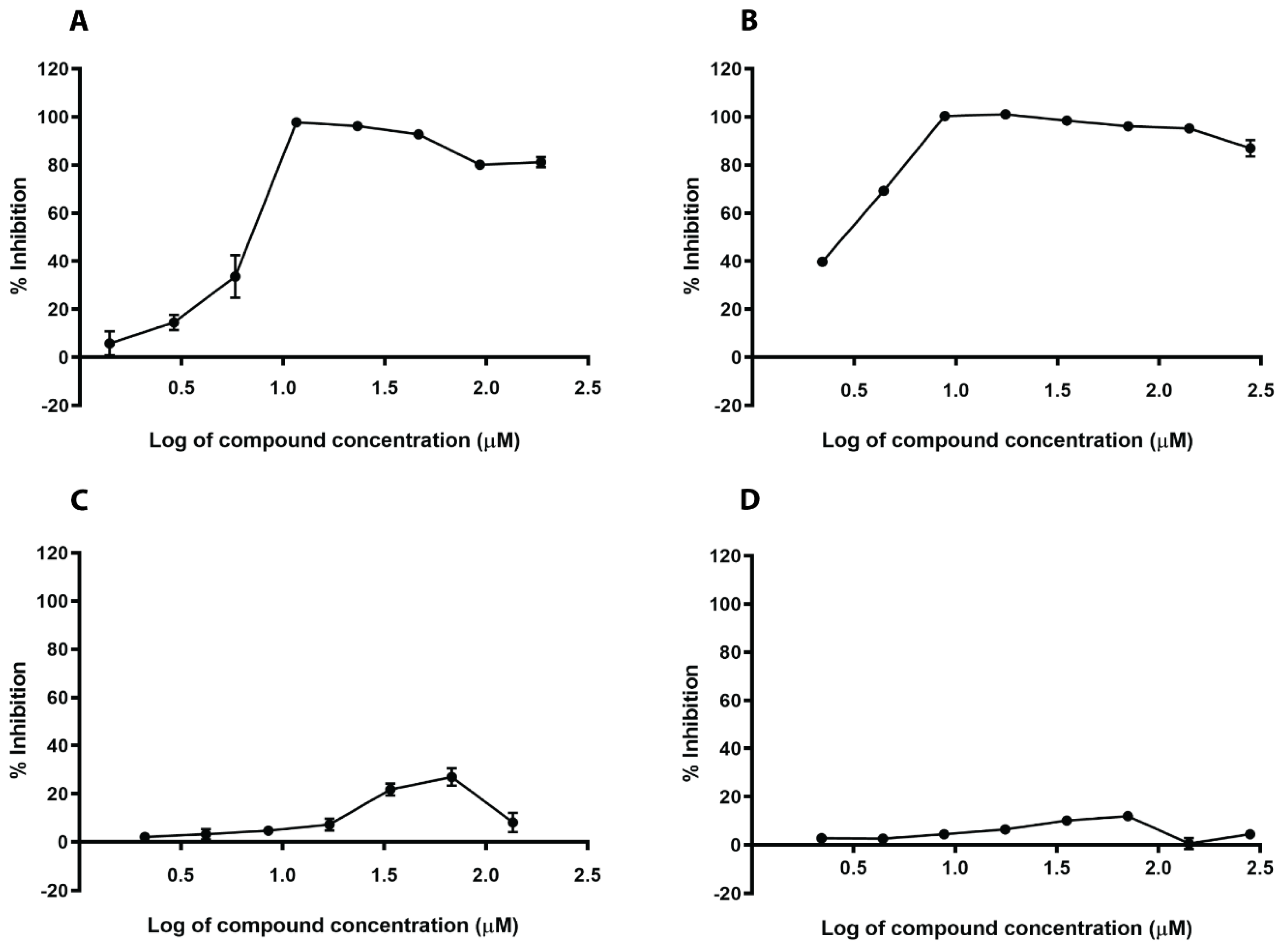
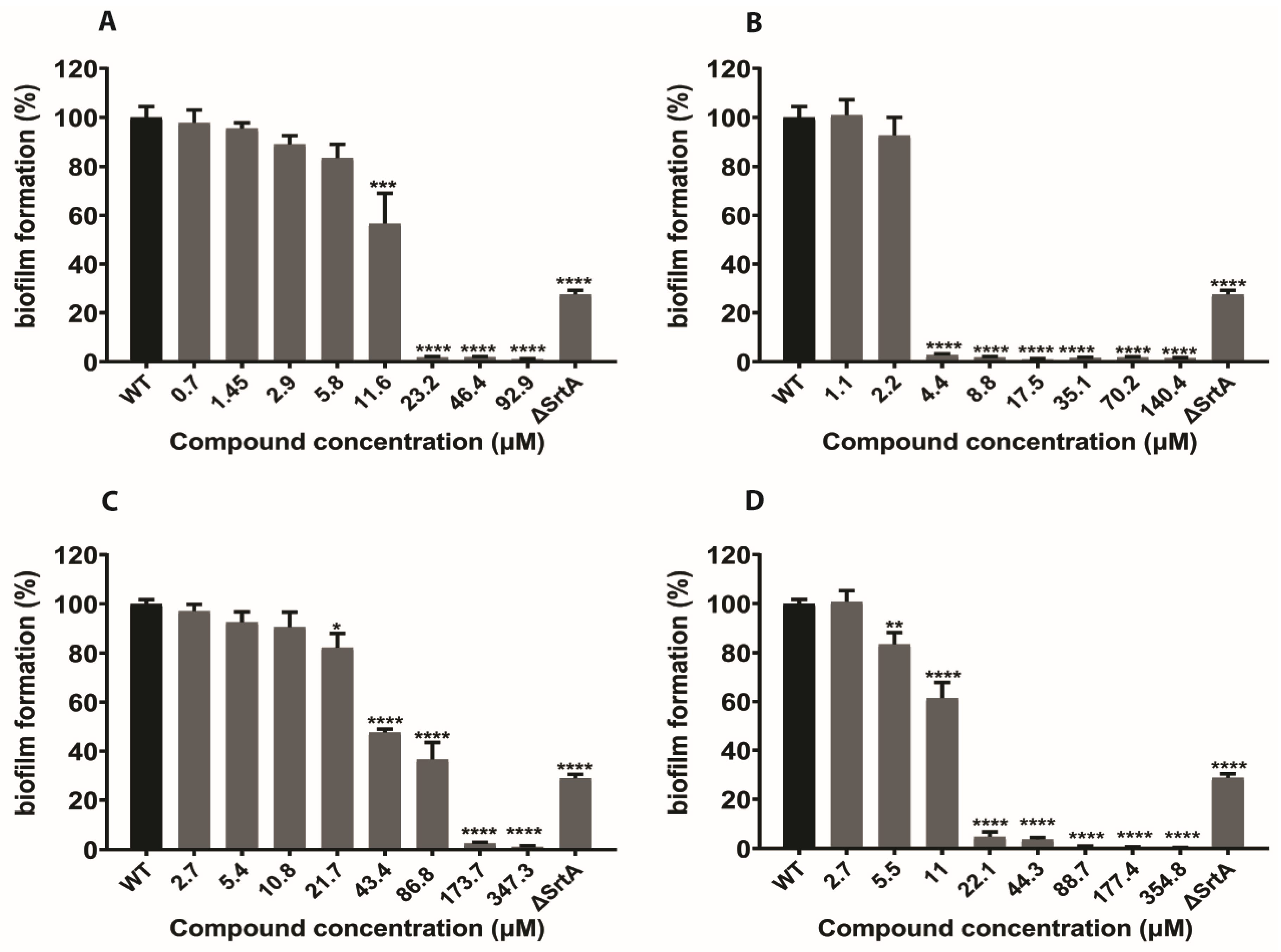
2.4. In Silico Hits Reduce Biofilm Formation
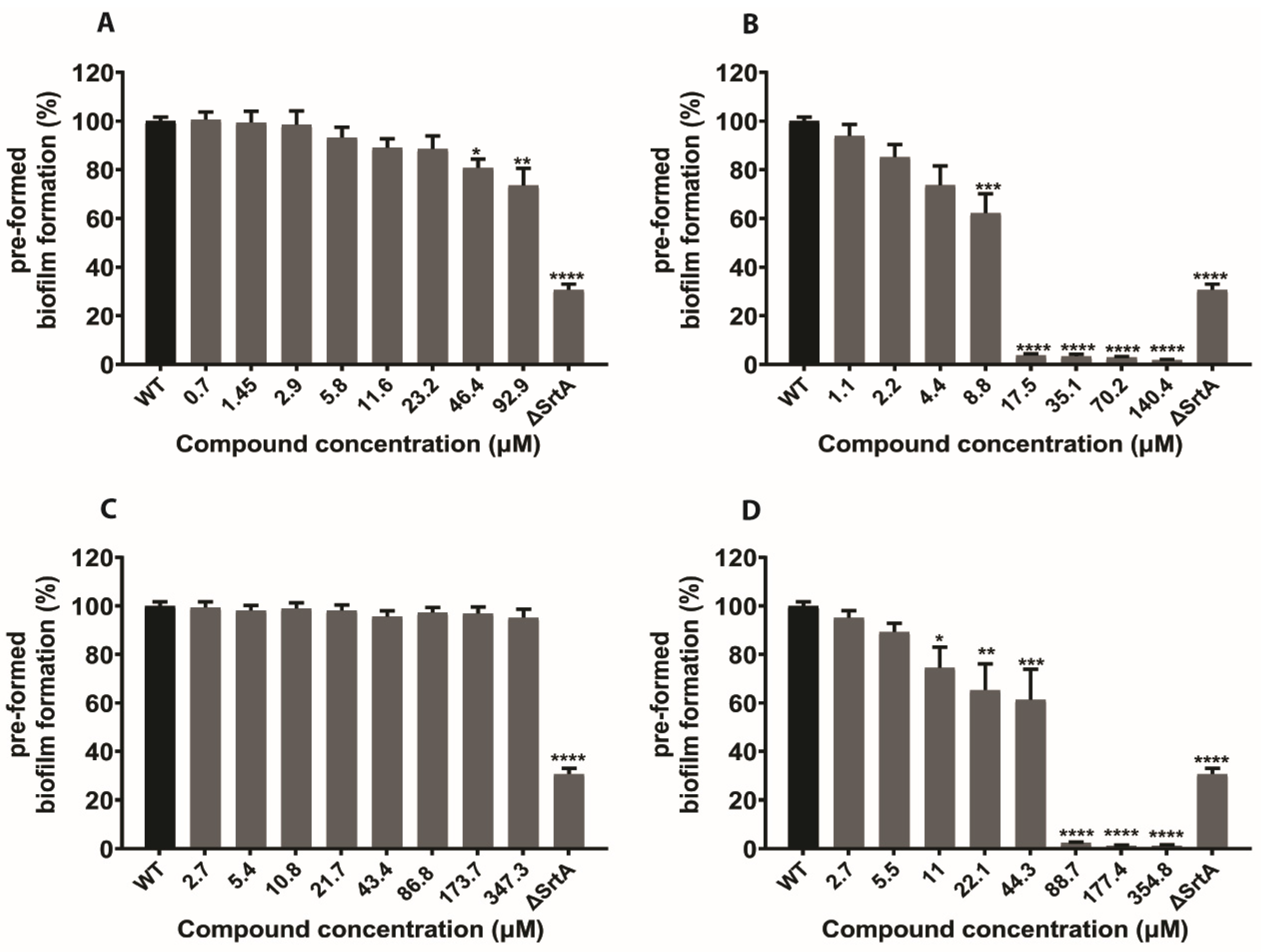
2.5. In Silico Hits Reduce the Adherence of S. aureus to Fibrinogen
3. Discussion
4. Materials and Methods
4.1. Reagents, Bacterial Strains and Cell Lines
4.2. Chemical Compounds
4.3. Virtual Screening
4.4. SrtA Enzyme Inhibition Assay
4.5. S. aureus Whole Cell Assay
4.6. Biofilm Inhibitory Assay
4.7. Assessment of Biofilms Metabolic Activity
4.8. Fibrinogen Binding Assay
4.9. Mammalian Cell Cytotoxicity Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BSA | bovine serum albumin |
| CFU | colony-forming unit |
| CV | Crystal violet |
| DMSO | dimethyl sulfoxide |
| FRET | fluorescence resonance energy transfer |
| p-HMB | p-hydroxymercuribenzoic acid |
| IC50 | half maximal inhibitory concentration |
| MDR | multidrug-resistant |
| MIC | minimum inhibitory concentration |
| PBS | Phosphate buffer saline |
| SrtA | sortase A |
| TSB | Tryptic soya broth |
References
- Piddock, L.J.V. Reflecting on the final report of the O’Neill Review on Antimicrobial Resistance. Lancet Infect. Dis. 2016, 16, 767–768. [Google Scholar] [CrossRef]
- Adeyi, O.; Baris, E.; Jonas, O.; Irwin, A.; Berthe, F.; Le Gall, F.; Marquez, P.; Nikolic, I.; Plante, C.; Schneidman, M. Drug-Resistant Infections: A Threat to Our Economic Future; World Bank Group: Washington, DC, USA, 2017. [Google Scholar]
- Spaulding, C.N.; Klein, R.D.; Schreiber, H.L.; Janetka, J.W.; Hultgren, S.J. Precision antimicrobial therapeutics: The path of least resistance? NPJ Biofilms Microbiomes 2018, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Parrino, B.; Schillaci, D.; Carnevale, I.; Giovannetti, E.; Diana, P.; Cirrincione, G.; Cascioferro, S. Synthetic small molecules as anti-biofilm agents in the struggle against antibiotic resistance. Eur. J. Med. Chem. 2019, 161, 154–178. [Google Scholar] [CrossRef]
- Thappeta, K.R.V.; Vikhe, Y.S.; Yong, A.M.H.; Chan-Park, M.B.; Kline, K.A. Combined Efficacy of an Antimicrobial Cationic Peptide Polymer with Conventional Antibiotics to Combat Multidrug-Resistant Pathogens. ACS Infect. Dis. 2020, 6, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Parrino, B.; Carbone, D.; Cirrincione, G.; Diana, P.; Cascioferro, S. Inhibitors of antibiotic resistance mechanisms: Clinical applications and future perspectives. Future Med. Chem. 2020, 12, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Ericson, J.E.; Popoola, V.O.; Smith, P.B.; Benjamin, D.K.; Fowler, V.G.; Benjamin, D.K., Jr.; Clark, R.H.; Milstone, A.M. Burden of Invasive Staphylococcus Aureus Infections in Hospitalized Infants. JAMA Pediatr. 2015, 169, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Defres, S.; Marwick, C.; Nathwani, D. MRSA as a cause of lung infection including airway infection, community-acquired pneumonia and hospital-acquired pneumonia. Eur. Respir. J. 2009, 34, 1470–1476. [Google Scholar] [CrossRef]
- Ippolito, G.; Leone, S.; Lauria, F.N.; Nicastri, E.; Wenzel, R.P. Methicillin-resistant Staphylococcus aureus: The superbug. Int. J. Infect. Dis. 2010, 14, S7–S11. [Google Scholar] [CrossRef]
- Boyle-Vavra, S.; Daum, R.S. Community-acquired methicillin-resistant Staphylococcus aureus: The role of Panton–Valentine leukocidin. Lab. Investig. 2007, 87, 3–9. [Google Scholar] [CrossRef]
- Zetola, N.; Francis, J.S.; Nuermberger, E.L.; Bishai, W.R. Community-acquired meticillin-resistant Staphylococcus aureus: An emerging threat. Lancet Infect. Dis. 2005, 5, 275–286. [Google Scholar] [CrossRef]
- Cascioferro, S.; Carbone, D.; Parrino, B.; Pecoraro, C.; Giovannetti, E.; Cirrincione, G.; Diana, P. Therapeutic strategies to counteract antibiotic resistance in MRSA biofilm-associated infections. ChemMedChem 2020. [Google Scholar] [CrossRef] [PubMed]
- Pallen, M.J.; Lam, A.C.; Antonio, M.; Dunbar, K. An embarrassment of sortases—A richness of substrates? Trends Microbiol. 2001, 9, 97–101. [Google Scholar] [CrossRef]
- Maresso, A.W.; Schneewind, O. Sortase as a Target of Anti-Infective Therapy. Pharmacol. Rev. 2008, 60, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Schneewind, O.; Missiakas, D.M. Protein secretion and surface display in Gram-positive bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 1123–1139. [Google Scholar] [CrossRef] [PubMed]
- Spirig, T.; Weiner, E.M.; Clubb, R.T. Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 2011, 82, 1044–1059. [Google Scholar] [CrossRef]
- Clancy, K.W.; Melvin, J.A.; McCafferty, D.G. Sortase transpeptidases: Insights into mechanism, substrate specificity, and inhibition. Biopolymers 2010, 94, 385–396. [Google Scholar] [CrossRef]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef]
- Jonsson, I.-M.; Mazmanian, S.K.; Schneewind, O.; Verdrengh, M.; Bremell, T.; Tarkowski, A. On the Role of Staphylococcus aureus Sortase and Sortase-Catalyzed Surface Protein Anchoring in Murine Septic Arthritis. J. Infect. Dis. 2002, 185, 1417–1424. [Google Scholar] [CrossRef]
- Bradshaw, W.J.; Davies, A.H.; Chambers, C.J.; Roberts, A.K.; Shone, C.C.; Acharya, K.R. Molecular features of the sortase enzyme family. FEBS J. 2015, 282, 2097–2114. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, G.; Jensen, E.R.; Lenoy, E.; Schneewind, O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. USA 2000, 97, 5510–5515. [Google Scholar] [CrossRef]
- Oh, K.-B.; Oh, M.-N.; Kim, J.-G.; Shin, D.-S.; Shin, J. Inhibition of sortase-mediated Staphylococcus aureus adhesion to fibronectin via fibronectin-binding protein by sortase inhibitors. Appl. Microbiol. Biotechnol. 2006, 70, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 207–228. [Google Scholar] [PubMed]
- Suree, N.; Liew, C.K.; Villareal, V.A.; Thieu, W.; Fadeev, E.A.; Clemens, J.J.; Jung, M.E.; Clubb, R.T. The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized. J. Biol. Chem. 2009, 284, 24465–24477. [Google Scholar] [CrossRef] [PubMed]
- Cascioferro, S.; Totsika, M.; Schillaci, D. Sortase A: An ideal target for anti-virulence drug development. Microb. Pathog. 2014, 77, 105–112. [Google Scholar] [CrossRef]
- Suree, N.; Jung, M.E.; Clubb, R.T. Recent Advances Towards New Anti-Infective Agents that Inhibit Cell Surface Protein Anchoring in Staphylococcus aureus and Other Gram-Positive Pathogens. Mini-Rev. Med. Chem. 2007, 7, 991–1000. [Google Scholar] [CrossRef]
- Mühlen, S.; Dersch, P. Anti-virulence Strategies to Target Bacterial Infections. In How to Overcome the Antibiotic Crisis: Facts, Challenges, Technologies and Future Perspectives; Stadler, M., Dersch, P., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 147–183. ISBN 978-3-319-49284-1. [Google Scholar]
- Silva, L.N.; Zimmer, K.R.; Macedo, A.J.; Trentin, D.S. Plant Natural Products Targeting Bacterial Virulence Factors. Chem. Rev. 2016, 116, 9162–9236. [Google Scholar] [CrossRef]
- Ng, S.B.; Kanagasundaram, Y.; Fan, H.; Arumugam, P.; Eisenhaber, B.; Eisenhaber, F. The 160K Natural Organism Library, a unique resource for natural products research. Nat. Biotechnol. 2018, 36, 570–573. [Google Scholar] [CrossRef]
- Irwin, J.J.; Shoichet, B.K. Docking Screens for Novel Ligands Conferring New Biology. J. Med. Chem. 2016, 59, 4103–4120. [Google Scholar] [CrossRef]
- Zong, Y.; Bice, T.W.; Ton-That, H.; Schneewind, O.; Narayana, S.V.L. Crystal structures of Staphylococcus aureus sortase A and its substrate complex. J. Biol. Chem. 2004, 279, 31383–31389. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System; Version 1.4.1; DeLano Scientific: San Carlos, CA, USA, 2011. [Google Scholar]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef]
- Cerca, N.; Martins, S.; Cerca, F.; Jefferson, K.K.; Pier, G.B.; Oliveira, R.; Azeredo, J. Comparative assessment of antibiotic susceptibility of coagulase-negative staphylococci in biofilm versus planktonic culture as assessed by bacterial enumeration or rapid XTT colorimetry. J. Antimicrob. Chemother. 2005, 56, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Roche, F.M.; Massey, R.; Peacock, S.J.; Day, N.P.; Visai, L.; Speziale, P.; Lam, A.; Pallen, M.; Foster, T.J. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 2003, 149, 643–654. [Google Scholar] [CrossRef] [PubMed]
- DeDent, A.; Bae, T.; Missiakas, D.M.; Schneewind, O. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J. 2008, 27, 2656–2668. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. MRSA virulence and spread. Cell. Microbiol. 2012, 14, 1513–1521. [Google Scholar] [CrossRef] [PubMed]
- Welsh, K.J.; Abbott, A.N.; Lewis, E.M.; Gardiner, J.M.; Kruzel, M.C.; Lewis, C.T.; Mohr, J.F.; Wanger, A.; Armitige, L.Y. Clinical characteristics, outcomes, and microbiologic features associated with methicillin-resistant Staphylococcus aureus bacteremia in pediatric patients treated with vancomycin. J. Clin. Microbiol. 2010, 48, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef]
- Rollinger, J.M.; Stuppner, H.; Langer, T. Virtual screening for the discovery of bioactive natural products. In Natural Compounds as Drugs Volume I; Petersen, F., Amstutz, R., Eds.; Birkhäuser Basel: Basel, Switzerland, 2008; pp. 211–249. ISBN 978-3-7643-8117-2. [Google Scholar]
- Pereira, F.; Aires-de-Sousa, J. Computational Methodologies in the Exploration of Marine Natural Product Leads. Mar. Drugs 2018, 16, 236. [Google Scholar] [CrossRef]
- Parker, J.C.; McPherson, R.K.; Andrews, K.M.; Levy, C.B.; Dubins, J.S.; Chin, J.E.; Perry, P.V.; Hulin, B.; Perry, D.A.; Inagaki, T.; et al. Effects of skyrin, a receptor-selective glucagon antagonist, in rat and human hepatocytes. Diabetes 2000, 49, 2079. [Google Scholar] [CrossRef][Green Version]
- Jahn, L.; Schafhauser, T.; Wibberg, D.; Rückert, C.; Winkler, A.; Kulik, A.; Weber, T.; Flor, L.; van Pée, K.-H.; Kalinowski, J.; et al. Linking secondary metabolites to biosynthesis genes in the fungal endophyte Cyanodermella asteris: The anti-cancer bisanthraquinone skyrin. J. Biotechnol. 2017, 257, 233–239. [Google Scholar] [CrossRef]
- Cascioferro, S.; Raimondi, M.V.; Cusimano, M.G.; Raffa, D.; Maggio, B.; Daidone, G.; Schillaci, D. Pharmaceutical Potential of Synthetic and Natural Pyrrolomycins. Molecules 2015, 20, 21658–21671. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, M.V.; Listro, R.; Cusimano, M.G.; La Franca, M.; Faddetta, T.; Gallo, G.; Schillaci, D.; Collina, S.; Leonchiks, A.; Barone, G. Pyrrolomycins as antimicrobial agents. Microwave-assisted organic synthesis and insights into their antimicrobial mechanism of action. Bioorg. Med. Chem. 2019, 27, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Schillaci, D.; Petruso, S.; Raimondi, M.V.; Cusimano, M.G.; Cascioferro, S.; Scalisi, M.; La Giglia, M.A.; Vitale, M. Pyrrolomycins as potential anti-staphylococcal biofilms agents. Biofouling 2010, 26, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Nitulescu, G.; Nicorescu, I.M.; Olaru, O.T.; Ungurianu, A.; Mihai, D.P.; Zanfirescu, A.; Nitulescu, G.M.; Margina, D. Molecular Docking and Screening Studies of New Natural Sortase A Inhibitors. Int. J. Mol. Sci. 2017, 18, 2217. [Google Scholar] [CrossRef] [PubMed]
- Oniga, S.D.; Araniciu, C.; Palage, M.D.; Popa, M.; Chifiriuc, M.-C.; Marc, G.; Pirnau, A.; Stoica, C.I.; Lagoudis, I.; Dragoumis, T.; et al. New 2-Phenylthiazoles as Potential Sortase A Inhibitors: Synthesis, Biological Evaluation and Molecular Docking. Molecules 2017, 22, 1827. [Google Scholar] [CrossRef]
- Kahlon, A.K.; Negi, A.S.; Kumari, R.; Srivastava, K.K.; Kumar, S.; Darokar, M.P.; Sharma, A. Identification of 1-chloro-2-formyl indenes and tetralenes as novel antistaphylococcal agents exhibiting sortase A inhibition. Appl. Microbiol. Biotechnol. 2014, 98, 2041–2051. [Google Scholar] [CrossRef]
- Suree, N.; Yi, S.W.; Thieu, W.; Marohn, M.; Damoiseaux, R.; Chan, A.; Jung, M.E.; Clubb, R.T. Discovery and Structure-Activity Relationship Analysis of Staphylococcus aureus Sortase A Inhibitors. Bioorg. Med. Chem. 2009, 17, 7174–7185. [Google Scholar] [CrossRef]
- Irwin, J.J.; Shoichet, B.K.; Mysinger, M.M.; Huang, N.; Colizzi, F.; Wassam, P.; Cao, Y. Automated docking screens: A feasibility study. J. Med. Chem. 2009, 52, 5712–5720. [Google Scholar] [CrossRef]
- Krivov, G.G.; Shapovalov, M.V.; Dunbrack, R.L., Jr. Improved prediction of protein side-chain conformations with SCWRL4. Proteins 2009, 77, 778–795. [Google Scholar] [CrossRef]
- Mysinger, M.M.; Shoichet, B.K. Rapid Context-Dependent Ligand Desolvation in Molecular Docking. J. Chem. Inf. Model. 2010, 50, 1561–1573. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, 2437. [Google Scholar] [CrossRef] [PubMed]
- Torres, N.S.; Abercrombie, J.J.; Srinivasan, A.; Lopez-Ribot, J.L.; Ramasubramanian, A.K.; Leung, K.P. Screening a commercial library of pharmacologically active small molecules against Staphylococcus aureus biofilms. Antimicrob. Agents Chemother. 2016, 60, 5663–5672. [Google Scholar] [CrossRef] [PubMed]
- Elgalai, I.; Foster, H.A. Comparison of adhesion of wound isolates of Staphylococcus aureus to immobilized proteins. J. Appl. Microbiol. 2003, 94, 413–420. [Google Scholar] [CrossRef] [PubMed]
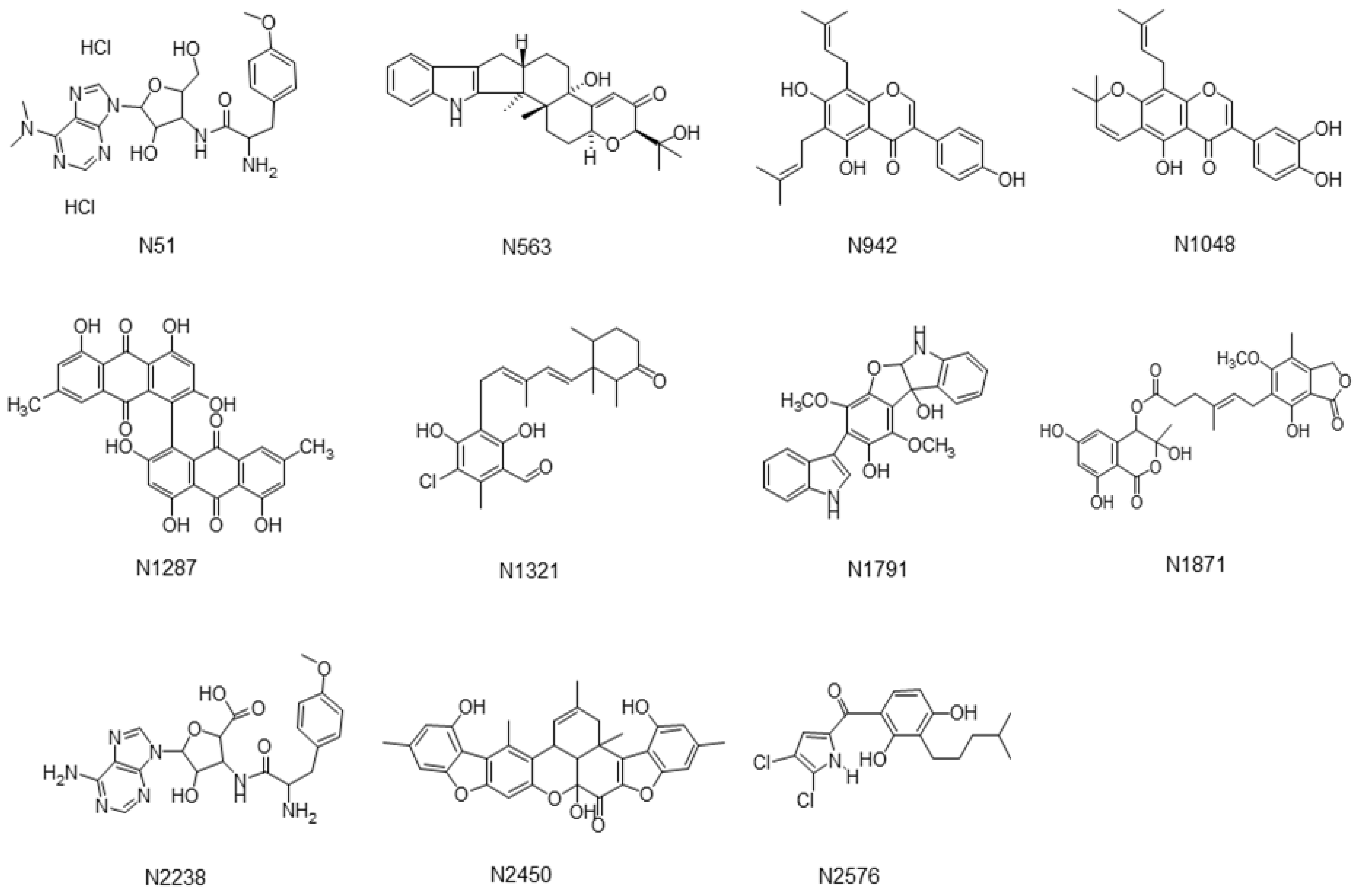

| Compounds | Docking Scores/Ranks from 3 Different Sortase A Structures | |||||
|---|---|---|---|---|---|---|
| 1T2P | 1T2W | 1T2W_C184 | ||||
| Score | Rank | Score | Rank | Score | Rank | |
| N51 | −44.39 | 41 | −39.14 | 97 | −36.86 | 139 |
| N563 | −35.85 | 410 | −37.95 | 141 | −36.83 | 143 |
| N942 | −40.61 | 142 | −41.59 | 33 | −42.19 | 11 |
| N1048 | −36.9 | 330 | −38.10 | 135 | −39.76 | 4 |
| N1287 | −41.87 | 88 | −36.01 | 322 | N/A | N/A |
| N1321 | −38.63 | 233 | −37.60 | 158 | −38.15 | 85 |
| N1791 | −40.29 | 164 | −35.15 | 372 | −36.33 | 176 |
| N1871 | −46.58 | 12 | −40.03 | 67 | −40.31 | 35 |
| N2238 | −45.30 | 25 | −45.93 | 8 | −50.42 | 2 |
| N2450 | −38.36 | 251 | −39.76 | 76 | −42.81 | 9 |
| N2576 | −34.61 | 464 | −33.88 | 435 | −36.65 | 156 |
| Curcumin | −47.53 | 10 | −35.85 | 339 | −31.24 | 491 |
| Chlorogenic acid | −28.19 | >500 | −32.24 | >500 | −28.19 | >500 |
| Compound ID | Compound Name | Sortase A IC50 (µM) (DABCYL-LPETG-EDANS Substrate; 4 h) | Sortase A IC50 (µM) (5-FAM/QXL Substrate; 10 min) |
|---|---|---|---|
| N51 | Puromycin hydrochloride | >200 | >200 |
| N563 | Paxilline | >200 | >200 |
| N942 | 6,8-diprenylgenistein | >200 | >200 |
| N1048 | Auriculasin | >200 | >200 |
| N1287 | Skyrin | 24 ± 2.1 | 31 ± 0.3 |
| N1321 | Ascochlorin | >200 | >200 |
| N1791 | Demethyl asterriquinone | >200 | >200 |
| N1871 | Antibiotic F 13459 | >200 | >200 |
| N2238 | Chryscandin | >200 | >200 |
| N2450 | Asticolorin B | >200 | >200 |
| N2576 | (4,5-dichloro-1H-pyrrol-2-yl)-[2,4-dihydroxy-3-(4-methyl-pentyl)-phenyl]-methanone | 47 ± 4.2 | 32 ± 6.3 |
| Curcumin | 7.0 ± 1.8 | 107 ± 16 | |
| Chlorogenic acid | 88 ± 5.6 | 144 ± 16 | |
| p-hydroxymercuribenzoic acid (p-HMB) | 0.20 ± 0.07 | 0.18 ± 0.08 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thappeta, K.R.V.; Zhao, L.N.; Nge, C.E.; Crasta, S.; Leong, C.Y.; Ng, V.; Kanagasundaram, Y.; Fan, H.; Ng, S.B. In-Silico Identified New Natural Sortase A Inhibitors Disrupt S. aureus Biofilm Formation. Int. J. Mol. Sci. 2020, 21, 8601. https://doi.org/10.3390/ijms21228601
Thappeta KRV, Zhao LN, Nge CE, Crasta S, Leong CY, Ng V, Kanagasundaram Y, Fan H, Ng SB. In-Silico Identified New Natural Sortase A Inhibitors Disrupt S. aureus Biofilm Formation. International Journal of Molecular Sciences. 2020; 21(22):8601. https://doi.org/10.3390/ijms21228601
Chicago/Turabian StyleThappeta, Kishore Reddy Venkata, Li Na Zhao, Choy Eng Nge, Sharon Crasta, Chung Yan Leong, Veronica Ng, Yoganathan Kanagasundaram, Hao Fan, and Siew Bee Ng. 2020. "In-Silico Identified New Natural Sortase A Inhibitors Disrupt S. aureus Biofilm Formation" International Journal of Molecular Sciences 21, no. 22: 8601. https://doi.org/10.3390/ijms21228601
APA StyleThappeta, K. R. V., Zhao, L. N., Nge, C. E., Crasta, S., Leong, C. Y., Ng, V., Kanagasundaram, Y., Fan, H., & Ng, S. B. (2020). In-Silico Identified New Natural Sortase A Inhibitors Disrupt S. aureus Biofilm Formation. International Journal of Molecular Sciences, 21(22), 8601. https://doi.org/10.3390/ijms21228601





