Genome-Wide Identification and Expression Analysis of Udp-Glucuronosyltransferases in the Whitefly Bemisia Tabaci (Gennadius) (HemipterA: Aleyrodidae)
Abstract
1. Introduction
2. Results
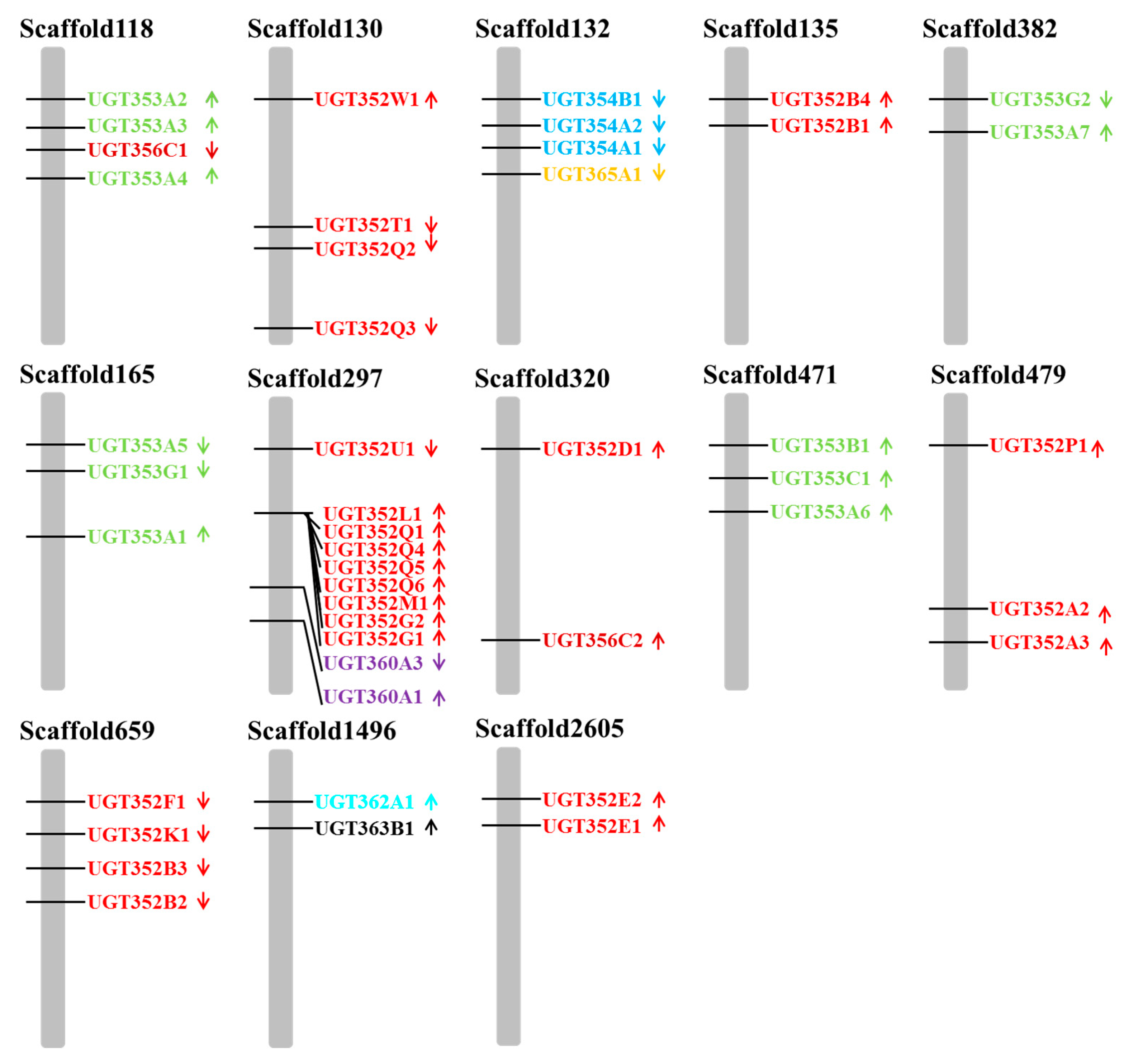
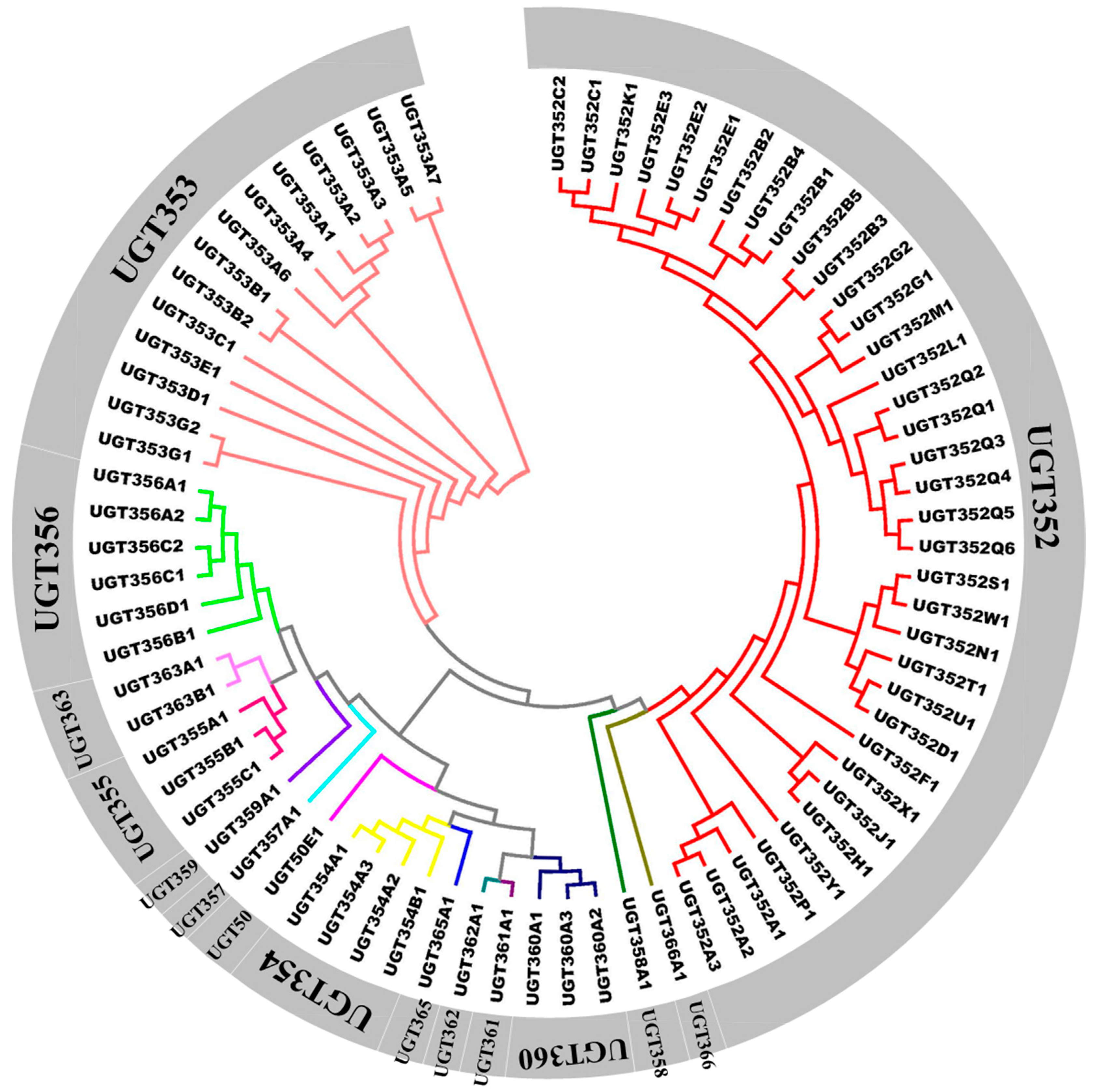
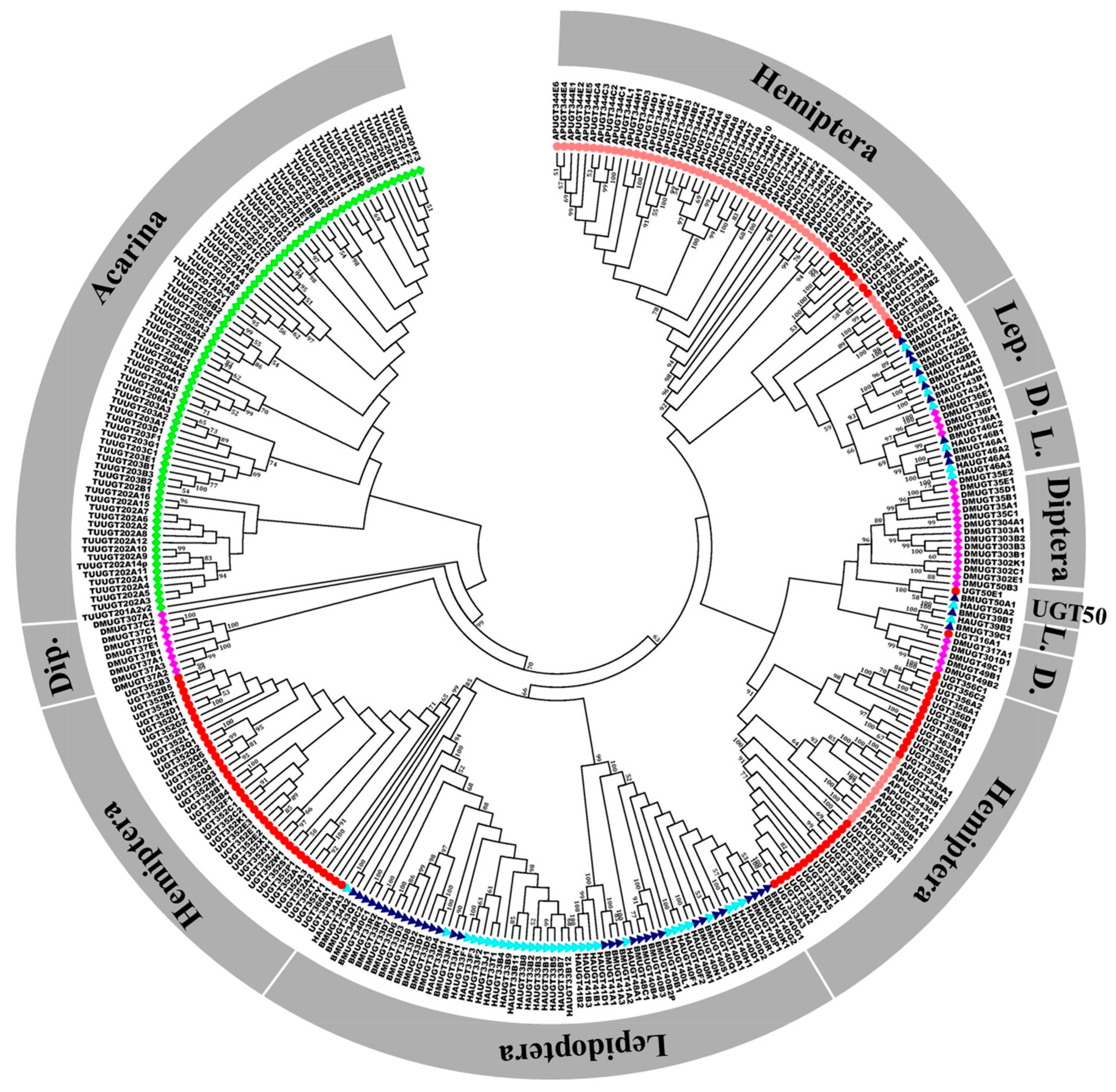
2.1. Identification and Phylogenetic Analysis of B. tabaci MEAM1 UGT Genes
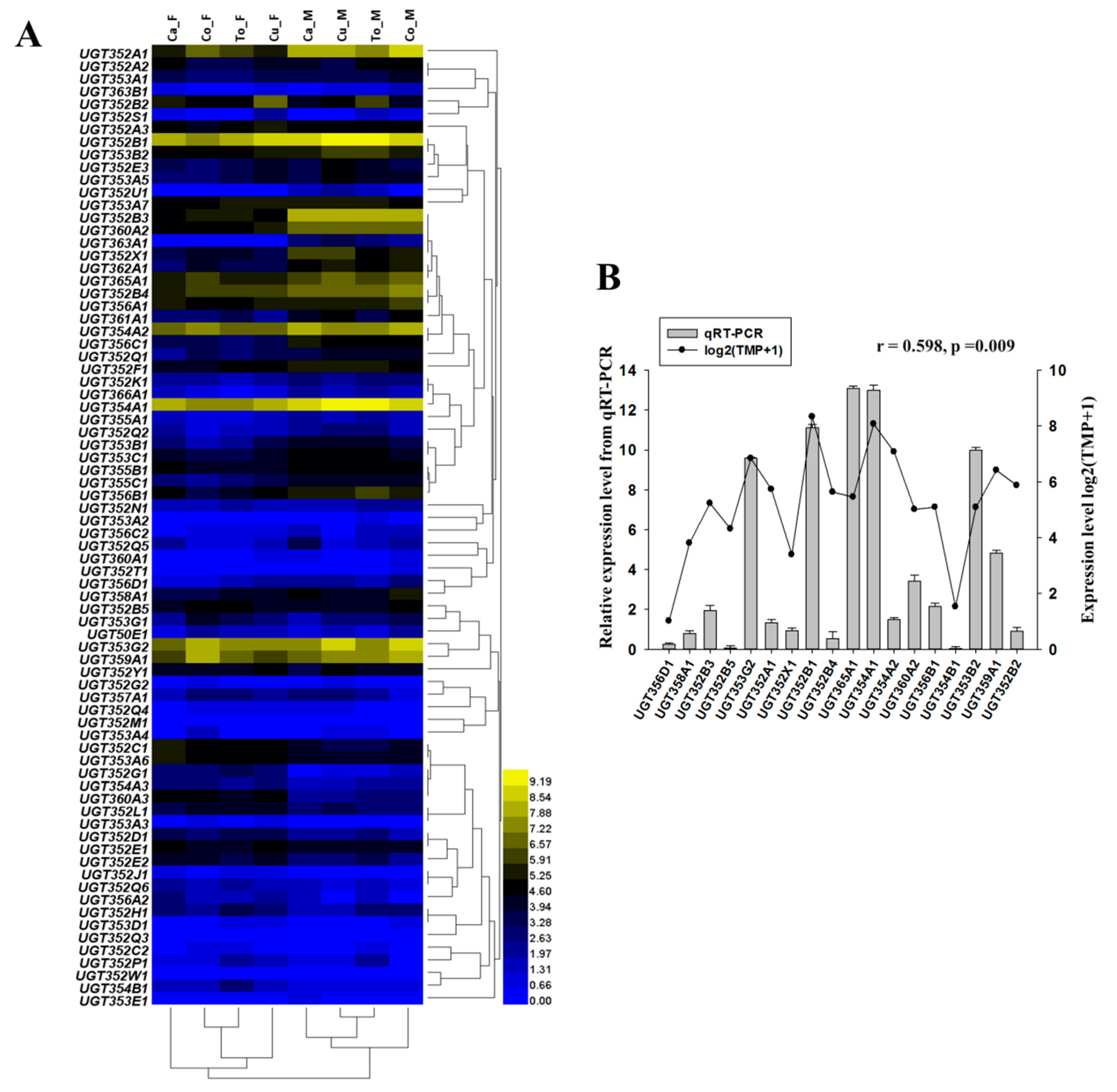
2.2. Expression of the UGT Genes in Different Host Plants and Developmental Stages
2.3. mRNA Expressions of Selected UGT Genes by RT-qPCR
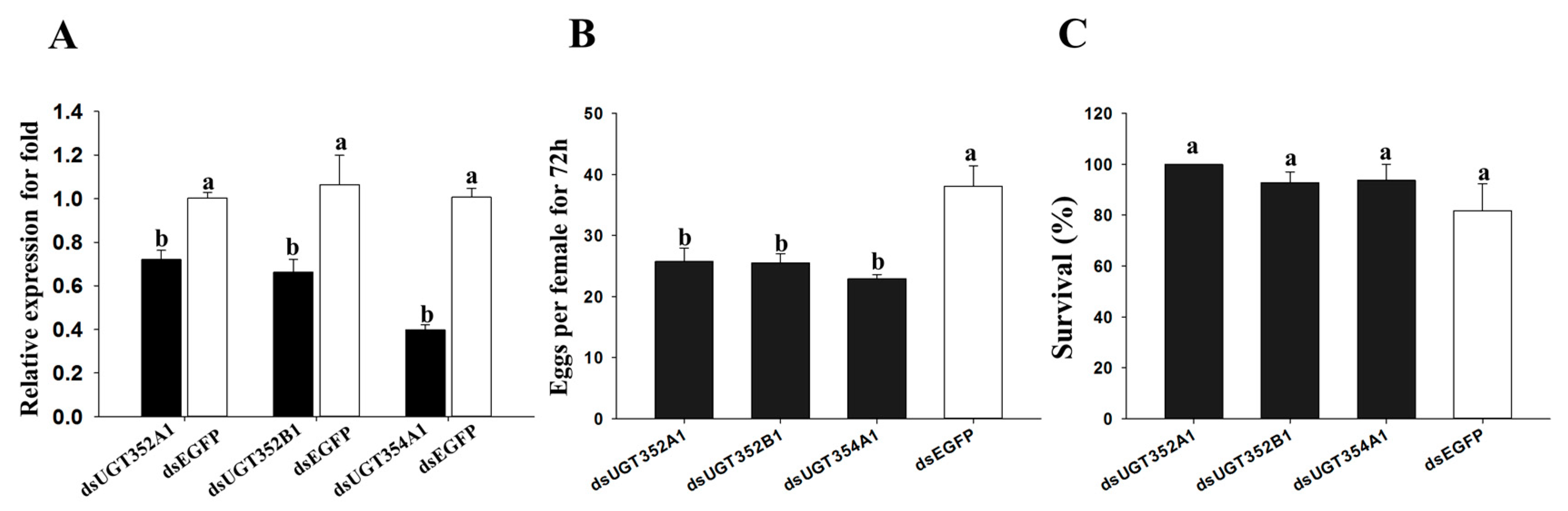
2.4. Effect of RNAi of UGTs on B. tabaci MEAM1 Adaptability
3. Discussion
4. Materials and Methods
4.1. Insect Strain
4.2. Identification of UGTs in B. tabaci and Other Insects
4.3. Phylogenetic Analysis of B. tabaci MEAM1 UGT Genes
4.4. Expression profiling of UGT Genes
4.5. RNA Interference
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| UDP | Uridine diphosphate |
| UGTs | Uridine diphosphate glucuronosyltransferases |
| EF-1α | The translation elongation factor 1 alpha gene |
| MEAM1 | Middle East-Asia Minor 1 |
| TPM | Transcripts Per Million |
References
- Liu, P.; Ma, H.; Zhu, Q.S.; Chen, B.C.; Gao, J.; Lin, X.Q. Research progress of insect adaptability to their host plants. Biol. Disaster Sci. 2016, 39, 250–254. [Google Scholar]
- Sousa, V.C.; Zélé, F.; Rodrigues, L.R.; Godinho, D.P.; de la Masselière, M.C.; Magalhães, S. Rapid host-plant adaptation in the herbivorous spider mite Tetranychus urticae occurs at low cost. Curr. Opin. Insect Sci. 2019, 36, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Grbic, M.; Van Leeuwen, T.; Clark, R.M.; Rombauts, S.; Rouze, P.; Grbic, V.; Osborne, E.J.; Dermauw, W.; Ngoc, P.C.T.; Ortego, F.; et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 2011, 479, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Santamaría, M.E.; Hernández-Crespo, P.; Ortego, F.; Grbic, V.; Grbic, M.; Diaz, I.; Martinez, M. Cysteine peptidases and their inhibitors in Tetranychus urticae: A comparative genomic approach. BMC Genom. 2012, 13, 307. [Google Scholar] [CrossRef]
- Dermauw, W.; Osborne, E.J.; Clark, R.M.; Grbic, M.; Tirry, L.; Van Leeuwen, T. A burst of ABC genes in the genome of the polyphagous spider mite Tetranychus urticae. BMC Genom. 2013, 14, 317. [Google Scholar] [CrossRef]
- Dermauw, W.; Wybouw, N.; Rombauts, S.; Menten, B.; Vontas, J.; Grbic, M.; Clark, R.M.; Feyereisen, R.; Van Leeuwen, T. A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proc. Natl. Acad. Sci. USA 2013, 110, E113–E122. [Google Scholar] [CrossRef]
- Wybouw, N.; Balabanidou, V.; Ballhorn, D.J.; Dermauw, W.; Grbić, M.; Vontas, J.; Van Leeuwen, T. A horizontally transferred cyanase gene in the spider mite Tetranychus urticae is involved in cyanate metabolism and is differentially expressed upon host plant change. Insect Biochem. Mol. Biol. 2012, 42, 881–889. [Google Scholar] [CrossRef]
- Wybouw, N.; Dermauw, W.; Tirry, L.; Stevens, C.; Grbic, M.; Feyereisen, R.; Van Leeuwen, T. A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning. eLife 2014, 3, e02365. [Google Scholar] [CrossRef]
- Wybouw, N.; Van Leeuwen, T.; Dermauw, W. A massive incorporation of microbial genes into the genome of Tetranychus urticae, a polyphagous arthropod herbivore. Insect Mol. Biol. 2018, 27, 333–351. [Google Scholar] [CrossRef]
- Schlachter, C.R.; Daneshian, L.; Amaya, J.; Klapper, V.; Wybouw, N.; Borowski, T.; Van Leeuwen, T.; Grbic, V.; Grbic, M.; Makris, T.M.; et al. Structural and functional characterization of an intradiol ring-cleavage dioxygenase from the polyphagous spider mite herbivore Tetranychus urticae Koch. Insect Biochem. Mol. Biol. 2019, 107, 19–30. [Google Scholar] [CrossRef]
- Snoeck, S.; Pavlidi, N.; Dermauw, W.; Van Leeuwen, T. Substrate specificity and promiscuity of UDP-glycosyltransferases in the polyphagous arthropod Tetranychus urticae. Insect Biochem. Mol. Biol. 2019, 109, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Wybouw, N.; Zhurov, V.; Martel, C.; Bruinsma, K.A.; Hendrickx, F.; Grbić, V.; Van Leeuwen, T. Adaptation of a polyphagous herbivore to a novel host plant extensively shapes the transcriptome of herbivore and host. Mol. Ecol. 2015, 24, 4647–4663. [Google Scholar] [CrossRef] [PubMed]
- Zhurov, V.; Navarro, M.; Bruinsma, K.A.; Arbona, V.; Estrella Santamaria, M.; Cazaux, M.; Wybouw, N.; Osborne, E.J.; Ens, C.; Rioja, C.; et al. Reciprocal responses in the interaction between Arabidopsis and the cell-content-feeding chelicerate herbivore spider mite. Plant. Physiol. 2014, 164, 384–399. [Google Scholar] [CrossRef] [PubMed]
- Snoeck, S.; Wybouw, N.; Van Leeuwen, T.; Dermauw, W. Transcriptomic plasticity in the arthropod generalist Tetranychus urticae upon long-term acclimation to different host plants. G3 Genes Genomes Genet. 2018, 8, 3865–3879. [Google Scholar] [CrossRef]
- Ahn, S.J.; Vogel, H.; Heckel, D.G. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 2012, 42, 133–147. [Google Scholar] [CrossRef]
- Krempl, C.; Sporer, T.; Reichelt, M.; Ahn, S.J.; Heidel-Fischer, H.; Vogel, H.; Heckel, D.G.; Jouben, N. Potential detoxification of gossypol by UDP-glycosyltransferases in the two Heliothine moth species Helicoverpa armigera and Heliothis virescens. Insect Biochem. Mol. Biol. 2016, 71, 49–57. [Google Scholar] [CrossRef]
- Govind, G.; Mittapalli, O.; Griebel, T.; Allmann, S.; Böcker, S.; Baldwin, I.T. Unbiased transcriptional comparisons of generalist and specialist herbivores feeding on progressively defenseless Nicotiana attenuata plants. PLoS ONE 2010, 5, e8735. [Google Scholar] [CrossRef]
- Celorio-Mancera, M.P.; Heckel, D.G.; Vogel, H. Transcriptional analysis of physiological pathways in a generalist herbivore: Responses to different host plants and plant structures by the cotton bollworm, Helicoverpa armigera. Entomol. Exp. Appl. 2012, 144, 123–133. [Google Scholar] [CrossRef]
- De la Paz, C.-M.M.; Wheat, C.W.; Vogel, H.; Söderlind, L.; Janz, N.; Nylin, S. Mechanisms of macroevolution: Polyphagous plasticity in butterfly larvae revealed by RNA-Seq. Mol. Ecol. 2013, 22, 4884–4895. [Google Scholar] [CrossRef]
- Ragland, G.J.; Almskaar, K.; Vertacnik, K.L.; Gough, H.M.; Feder, J.L.; Hahn, D.A.; Schwarz, D. Differences in performance and transcriptome-wide gene expression associated with Rhagoletis (Diptera: Tephritidae) larvae feeding in alternate host fruit environments. Mol. Ecol. 2015, 24, 2759–2776. [Google Scholar] [CrossRef]
- Xu, H.X.; Hong, Y.; Zhang, M.Z.; Wang, Y.L.; Liu, S.S.; Wang, X.W. Transcriptional responses of invasive and indigenous whiteflies to different host plants reveal their disparate capacity of adaptation. Sci. Rep. 2015, 5, 10774. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.J.; Dermauw, W.; Wybouw, N.; Heckel, D.G.; Leeuwen, T.V. Bacterial origin of a diverse family of UDP-glycosyltransferase genes in the Tetranychus urticae genome. Insect Biochem. Mol. Biol. 2014, 50, 43–57. [Google Scholar] [CrossRef] [PubMed]
- McKenna, D.D.; Scully, E.D.; Pauchet, Y.; Hoover, K.; Kirsch, R.; Geib, S.M.; Mitchell, R.F.; Waterhouse, R.M.; Ahn, S.J.; Arsala, D.; et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle-plant interface. Genome Biol. 2016, 17, 227. [Google Scholar] [CrossRef]
- Brown, J.K.; Frohlich, D.R.; Rosell, R.C. The sweetpotato or silverleaf whiteflies: Biotypes of Bemisia tabaci or a species complex? Ann. Rev. Entomol. 1995, 40, 511–534. [Google Scholar] [CrossRef]
- De Barro, P.J.; Liu, S.S.; Boykin, L.M.; Dinsdale, A. Bemisia tabaci: A statement of species status. Ann. Rev. Entomol. 2011, 56, 1–19. [Google Scholar] [CrossRef]
- Delatte, H.; Duyck, P.; Triboire, A.; David, P.; Becker, N.; Bonato, O.; Rrynaud, B. Differential invasion success among biotypes: Case of Bemisia tabaci. Biol. Invasion 2009, 11, 1059–1070. [Google Scholar] [CrossRef]
- Dinsdale, A.; Cook, L.; Riginos, C.; Buckley, Y.M.; De Barro, P. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann. Entomol Soc. Am. 2010, 103, 196–208. [Google Scholar] [CrossRef]
- Pan, H.; Chu, D.; Yan, W.Q.; Su, Q.; Liu, B.M.; Wang, S.L.; Wu, Q.J.; Xie, W.; Jiao, X.G.; Li, R.M.; et al. Rapid spread of Tomato yellow leaf curl virus in China is aided differentially by two invasive whiteflies. PLoS ONE 2012, 7, e34817. [Google Scholar] [CrossRef]
- Gilbertson, R.L.; Batuman, O.; Webster, C.G.; Adkins, S. Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Ann. Rev. Virol. 2015, 2, 67–93. [Google Scholar] [CrossRef]
- Zang, L.S.; Chen, W.Q.; Liu, S.S. Comparison of performance on different host plants between the B biotype and a non-B biotype of Bemisia tabaci from Zhejiang, China. Entomol. Exp. Appl. 2006, 121, 221–227. [Google Scholar] [CrossRef]
- Jiao, X.G.; Xie, W.; Guo, L.T.; Liu, B.M.; Wang, S.L.; Wu, Q.J.; Zhang, Y.J. Differing effects of cabbage and pepper on B and Q putative species of Bemisia tabaci. J. Pest. Sci. 2014, 87, 629–637. [Google Scholar] [CrossRef]
- Jiao, X.G.; Xie, W.; Zeng, Y.; Wan, C.; Liu, B.M.; Wang, S.L.; Wu, Q.J.; Zhang, Y.J. Lack of correlation between host choice and feeding efficiency for the B and Q putative species of Bemisia tabaci on four pepper genotypes. J. Pest. Sci. 2018, 91, 133–143. [Google Scholar] [CrossRef]
- Douglas, A.E. Phloem-sap feeding by animals: Problems and solutions. J. Exp. Bot. 2006, 57, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.Y.; Guo, L.T.; Wang, S.L.; Xie, W.; Jiao, X.G.; Wu, Q.J.; Zhang, Y.J. The ability to manipulate plant glucosinolates and nutrients explains the better performance of Bemisia tabaci Middle East-Asia Minor 1 than Mediterranean on cabbage plants. Ecol. Evol. 2017, 7, 6141–6150. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.T.; Su, Q.; Yang, Z.Z.; Xie, W.; Wang, S.L.; Wu, Q.J.; Cui, H.Y.; Zhang, Y.J. Amino Acid Utilization May Explain Why Bemisia tabaci Q and B Differ in Their Performance on Plants Infected by the Tomato yellow leaf curl virus. Front. Physiol. 2019, 10, 489. [Google Scholar] [CrossRef]
- Elbaz, M.; Halon, E.; Malka, O.; Malitsky, S.; Blum, E.; Aharoni, A.; Morin, S. Asymmetric adaptation to indolic and aliphatic glucosinolates in the B and Q sibling species of Bemisia tabaci (Hemiptera: Aleyrodidae). Mol. Ecol. 2012, 21, 4533–4546. [Google Scholar] [CrossRef] [PubMed]
- Markovich, O.; Kafle, D.; Elbaz, M.; Malitsky, S.; Aharoni, A.; Schwarzkopf, A.; Gershenzon, J.; Morin, S. Arabidopsis thaliana plants with different levels of aliphatic- and indolyl-glucosinolates affect host selection and performance of Bemisia tabaci. J. Chem. Ecol. 2013, 39, 1361–1372. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, X.; Zhao, H.P.; Xue, M.; Wang, D. Phenolic compounds induced by Bemisia tabaci and Trialeurodes vaporariorum in Nicotiana tabacum L. and their relationship with the salicylic acid signaling pathway. Arthropod Plant. Interact. 2017, 11, 659–667. [Google Scholar] [CrossRef]
- Yang, J.; Xie, W.; Liu, B.; Wang, S.; Wu, Q.; He, Y.; Zhang, Y.; Jiao, X. Phenolics, rather than glucosinolates, mediate host choice of Bemisia tabaci MEAM1 and MED on five cabbage genotypes. J. Appl. Entomol. 2020, 144, 287–296. [Google Scholar] [CrossRef]
- Mackenzie, P.I.; Owens, I.S.; Burchell, B.; Bock, K.W.; Bairoch, A.; Belanger, A.; FournelGigleux, S.; Green, M.; Hum, D.W.; Iyanagi, T.; et al. The UDP glycosyltransferase gene superfamily: Recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 1997, 7, 255–269. [Google Scholar] [CrossRef]
- Chen, W.B.; Hasegawa, D.K.; Kaur, N.; Kliot, A.; Pinheiro, P.V.; Luan, J.B.; Stensmyr, M.C.; Zheng, Y.; Liu, W.L.; Sun, H.H.; et al. The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 2016, 14, 110. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.W.; Tang, C.Y.; Ma, K.S.; Xia, J.; Song, D.L.; Gao, X.W. Overexpression of UDP-glycosyltransferase potentially involved in insecticide resistance in Aphis gossypii Glover collected from Bt cotton fields in China. Pest. Manag. Sci. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kaplanoglu, E.; Chapman, P.; Scott, I.M.; Donly, C. Overexpression of a cytochrome P450 and a UDP-glycosyltransferase is associated with imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata. Sci. Rep. 2017, 7, 1762. [Google Scholar] [CrossRef]
- Li, X.X.; Zhu, B.; Gao, X.W.; Liang, P. Over-expression of UDP-glycosyltransferase gene UGT2B17 is involved in chlorantraniliprole resistance in Plutella xylostella (L.). Pest. Manag. Sci. 2017, 73, 1402–1409. [Google Scholar] [CrossRef]
- Wang, M.Y.; Liu, X.Y.; Shi, L.; Liu, J.L.; Shen, G.M.; Zhang, P.; Lu, W.C.; He, L. Functional analysis of UGT201D3 associated with abamectin resistance in Tetranychus cinnabarinus (Boisduval). Insect Sci. 2018, 27, 276–291. [Google Scholar] [CrossRef]
- Tian, F.J.; Wang, Z.B.; Li, C.F.; Liu, J.L.; Zeng, X.N. UDP-Glycosyltransferases are involved in imidacloprid resistance in the Asian citrus psyllid, Diaphorina citri (Hemiptera: Lividae). Pestic. Biochem. Physiol. 2019, 154, 23–31. [Google Scholar] [CrossRef]
- Zhou, Y.; Fu, W.B.; Si, F.L.; Yan, Z.T.; Zhang, Y.J.; He, Q.Y.; Chen, B. UDP-glycosyltransferase genes and their association and mutations associated with pyrethroid resistance in Anopheles sinensis (Diptera: Culicidae). Malar. J. 2019, 18, 62. [Google Scholar] [CrossRef]
- Su, Q.; Li, S.X.; Shi, C.H.; Zhang, J.M.; Zhang, G.H.; Jin, Z.Y.; Li, C.R.; Wang, W.K.; Zhang, Y.J. Implication of heat-shock protein 70 and UDP-glucuronosyltransferase in thiamethoxam-induced whitefly Bemisia tabaci thermotolerance. J. Pest. Sci. 2018, 91, 469–478. [Google Scholar] [CrossRef]
- Hu, B.; Zhang, S.H.; Ren, M.M.; Tian, X.R.; Wei, Q.; Mburu, D.K.; Su, J.Y. The expression of Spodoptera exigua P450 and UGT genes: Tissue specificity and response to insecticides. Insect Sci. 2019, 26, 199–216. [Google Scholar] [CrossRef]
- Pan, Y.; Xu, P.; Zeng, X.; Liu, X.; Shang, Q. Characterization of UDP-Glucuronosyltransferases and the Potential Contribution to Nicotine Tolerance in Myzus Persicae. Int. J. Mol. Sci. 2019, 20, 3637. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Wan, F.H.; Zhang, Y.J.; Brown, J.K. Change in the biotype composition of Bemisia tabaci in Shandong Province of China from 2005 to 2008. Environ. Entomol. 2010, 39, 1028–1036. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Wu, Q.; Wang, S.; Jiao, X.; Guo, L.; Zhou, X.; Zhang, Y. Transcriptome analysis of host-associated differentiation in Bemisia tabaci (Hemiptera: Aleyrodidae). Front. Physiol. 2014, 5, 487. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. CD-HIT: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Luque, T.; Okano, K.; O’Reilly, D.R. Characterization of a novel silkworm (Bombyx mori) phenol UDP-glucosyltransferase. Eur J. Biochem. 2002, 269, 819–825. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Xia, J.X.; Pan, H.P.; Cheng, G.; Xie, W.; Guo, Z.J.; Zheng, H.X.; Yang, X.; Yang, F.S.; Wu, Q.J.; et al. Genome-wide characterization and expression profiling of sugar transporter family in the whitefly, Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Front. Physiol. 2017, 8, 322. [Google Scholar] [CrossRef]
- Li, R.; Xie, W.; Wang, S.; Wu, Q.; Yang, N.; Yang, X.; Pan, H.; Zhou, X.; Bai, L.; Xu, B.; et al. Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS ONE 2013, 8, e53006. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yang, X.; He, C.; Xie, W.; Liu, Y.; Xia, J.; Yang, Z.; Guo, L.; Wen, Y.; Wang, S.; Wu, Q.; et al. Glutathione S-transferases are involved in thiamethoxam resistance in the field whitefly Bemisia tabaci Q (Hemiptera: Aleyrodidae). Pestic. Biochem. Physiol. 2016, 134, 73–78. [Google Scholar] [CrossRef]

| Species | Number a | Food Type | Diet Breadth |
|---|---|---|---|
| Bemisia tabaci | 76 | Phytophagous | Polyphagous |
| Tetranychus urticae | 81 | Phytophagous | Polyphagous |
| Acyrthosiphon pisum | 72 | Phytophagous | Polyphagous |
| Locusta migratoria | 68 | Phytophagous | Polyphagous |
| Helicoverpa armigera | 42 | Phytophagous | Polyphagous |
| Diaphorina citri | 37 | Phytophagous | Oligophagous |
| Rhodnius prolixus | 16 | Phytophagous | Oligophagous |
| Danaus plexippus | 35 | Phytophagous | Oligophagous |
| Apis mellifera | 11 | Phytophagous | Oligophagous |
| Nilaparvata lugens | 20 | Phytophagous | Monophagous |
| Bombyx mori | 38 | Phytophagous | Monophagous |
| Nasonia vitripennis | 22 | Carnivorous | Monophagous |
| Camponotus floridanus | 21 | Omnivorous | Polyphagous |
| Tribolium castaneum | 27 | Omnivorous | Polyphagous |
| Anopheles gambiae | 24 | Sanguivorous | Oligophagous |
| Pediculus humanus | 4 | Sanguivorous | Oligophagous |
| Drosophila melanogaster | 35 | Saprophagous | Polyphagous |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, L.; Xie, W.; Yang, Z.; Xu, J.; Zhang, Y. Genome-Wide Identification and Expression Analysis of Udp-Glucuronosyltransferases in the Whitefly Bemisia Tabaci (Gennadius) (HemipterA: Aleyrodidae). Int. J. Mol. Sci. 2020, 21, 8492. https://doi.org/10.3390/ijms21228492
Guo L, Xie W, Yang Z, Xu J, Zhang Y. Genome-Wide Identification and Expression Analysis of Udp-Glucuronosyltransferases in the Whitefly Bemisia Tabaci (Gennadius) (HemipterA: Aleyrodidae). International Journal of Molecular Sciences. 2020; 21(22):8492. https://doi.org/10.3390/ijms21228492
Chicago/Turabian StyleGuo, Litao, Wen Xie, Zezhong Yang, Jianping Xu, and Youjun Zhang. 2020. "Genome-Wide Identification and Expression Analysis of Udp-Glucuronosyltransferases in the Whitefly Bemisia Tabaci (Gennadius) (HemipterA: Aleyrodidae)" International Journal of Molecular Sciences 21, no. 22: 8492. https://doi.org/10.3390/ijms21228492
APA StyleGuo, L., Xie, W., Yang, Z., Xu, J., & Zhang, Y. (2020). Genome-Wide Identification and Expression Analysis of Udp-Glucuronosyltransferases in the Whitefly Bemisia Tabaci (Gennadius) (HemipterA: Aleyrodidae). International Journal of Molecular Sciences, 21(22), 8492. https://doi.org/10.3390/ijms21228492








