Analysis of miR-9-5p, miR-124-3p, miR-21-5p, miR-138-5p, and miR-1-3p in Glioblastoma Cell Lines and Extracellular Vesicles
Abstract
1. Introduction
2. Results
2.1. Selection of a Set of miRNAs (miR-9-5p, miR-21-5p, miR-124-3p, miR-138-5p, and miR-1-3p) That Target VIM, NCL, NAP1L1, FREM2, and SPRY1 Genes
2.2. miR-9-5p, miR-21-5p, miR-124-3p, and miR-1-3p Are Involved in Cancer and Brain-Specific Processes
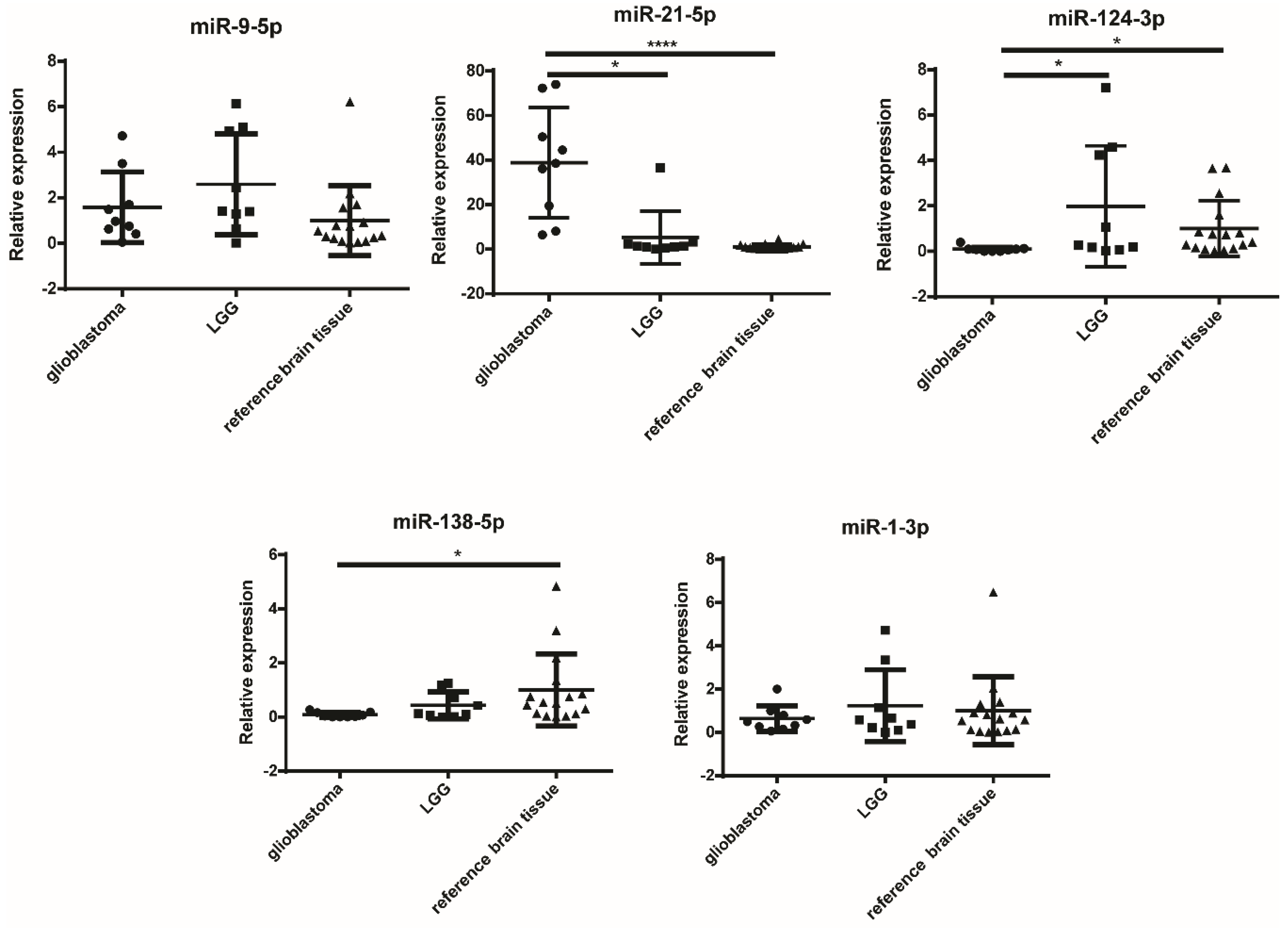
2.3. miR-124-3p and miR-138-5p Are Overexpressed in Reference Brain Tissue Compared to GBM
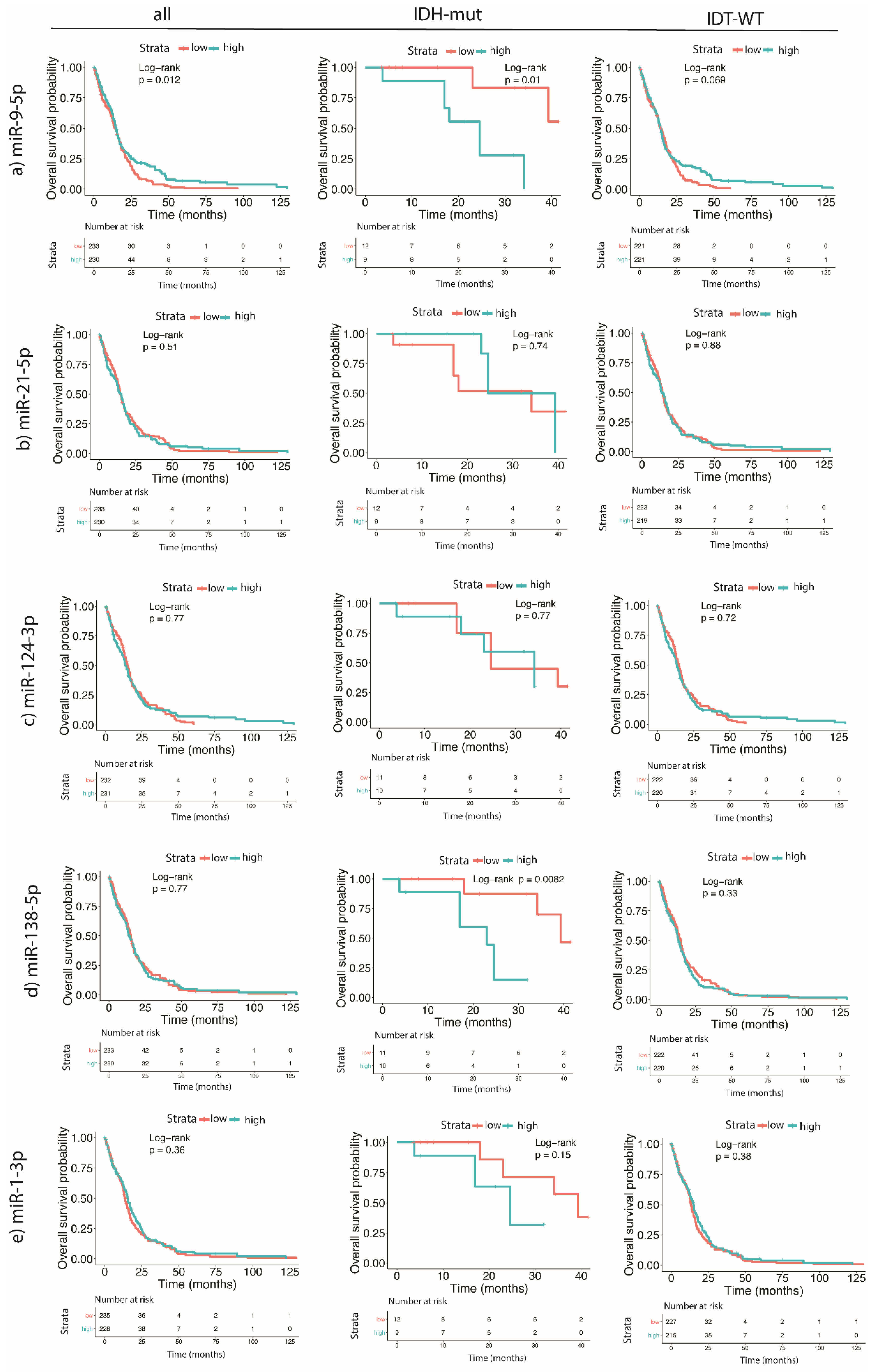
2.4. Higher Expression Levels of miR-9-5p and miR-138-5p Are Correlated to the Shorter Survival of GBM Patients Carrying IDH Mutation
2.5. Characterization of sEVs
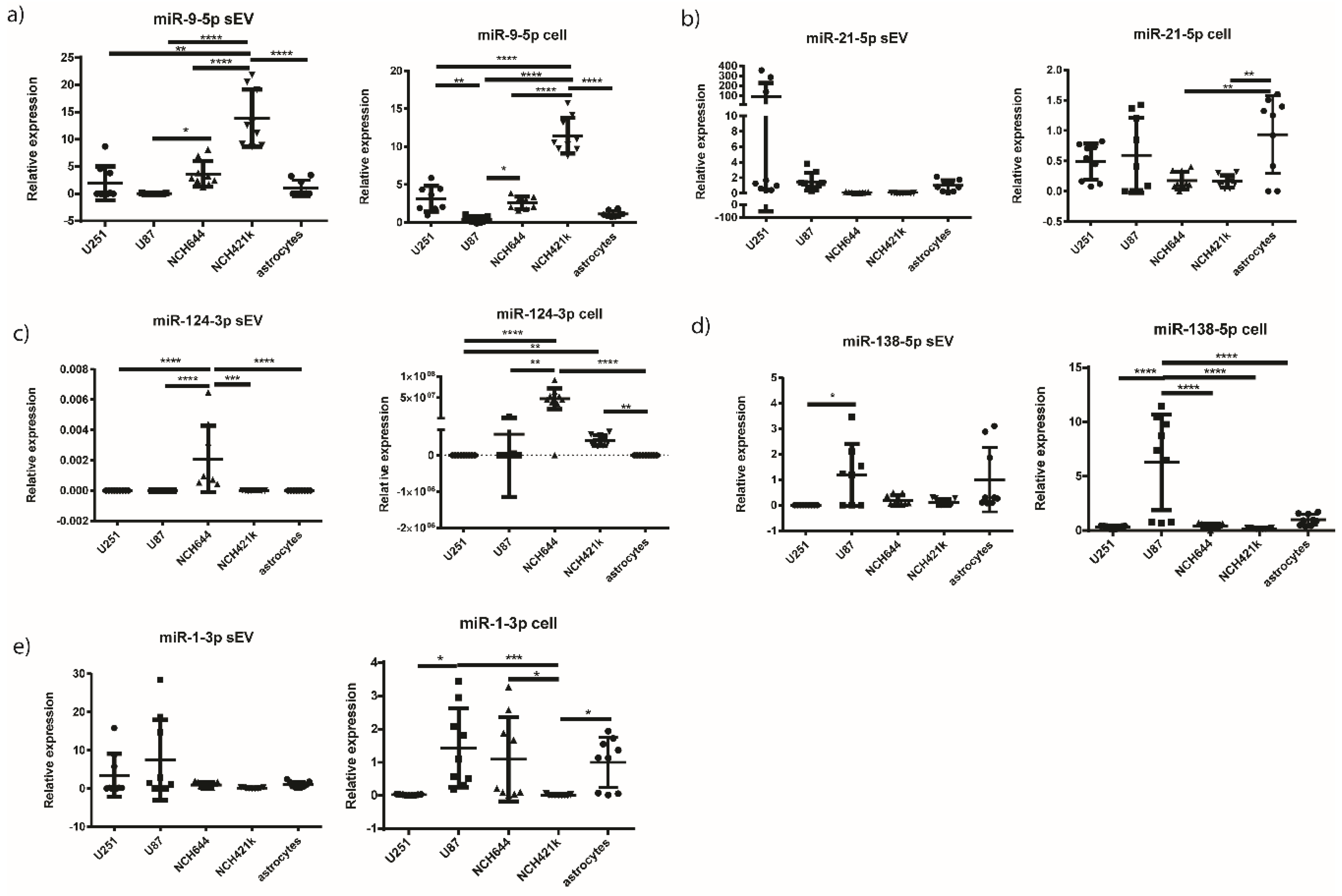
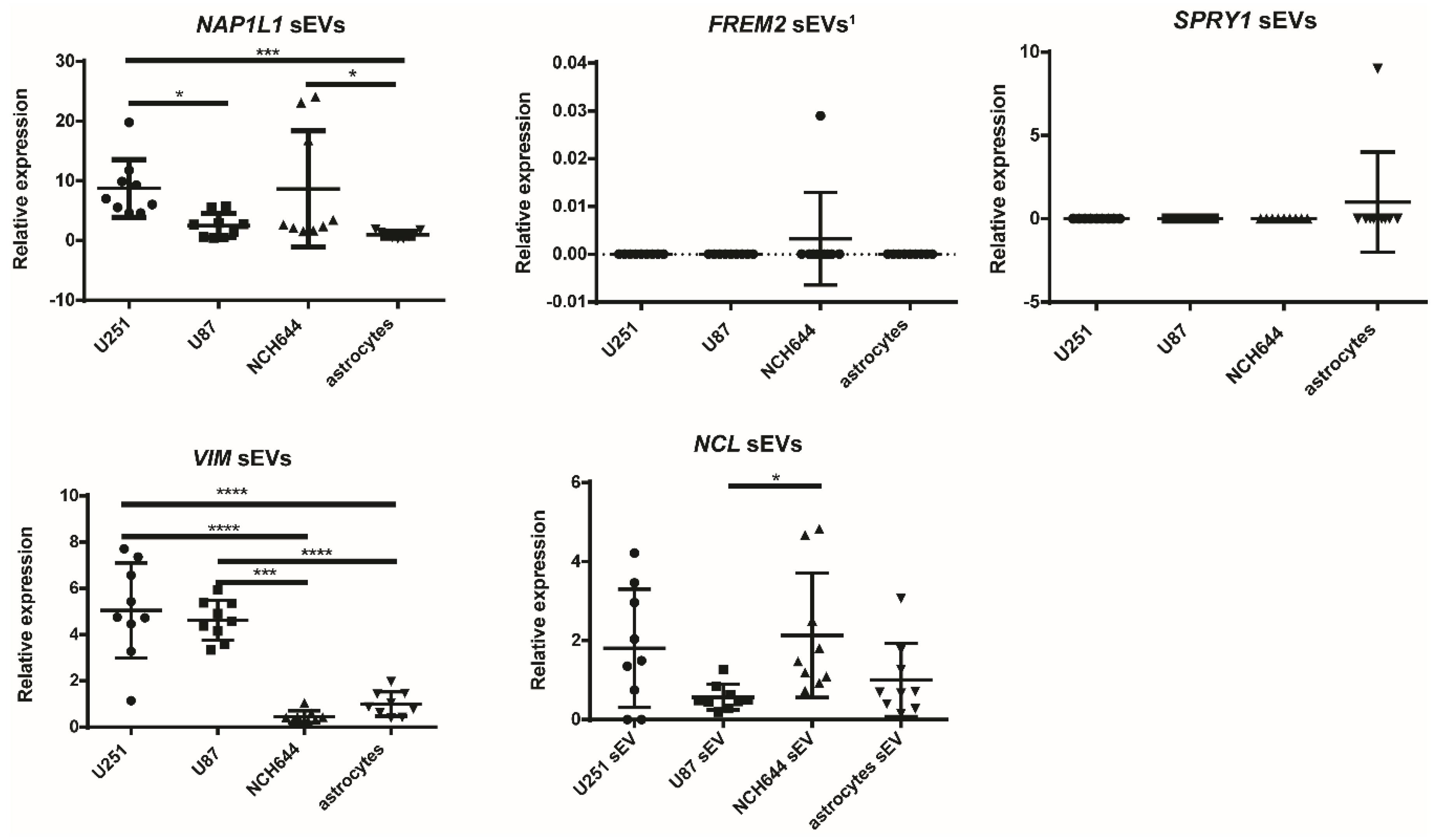
2.6. miR-9-5p and miR-124-3p are Overexpressed in the sEVs of NCH421k and NCH644 GBM Stem Cell Lines, Respectively, in Comparison to the sEVs of GBM Cell Lines and Astrocytes
2.7. Vimentin Gene Is Overexpressed in sEVs Originating from U251 and U87 GBM Cell Lines
3. Discussion
4. Materials and Methods
4.1. Selection of Candidate miRNAs
4.2. Pathway Analysis
4.3. Tissues and Ethical Approval
4.4. miRNA Quantification in Tissues
4.5. In Silico Survival Analysis
4.6. Cell Lines
CD133 Flow Cytometry
4.7. Small Extracellular Vesicles (Small EV) Isolation and Characerization
4.7.1. Protein Isolation and Western Blot
4.7.2. Nanoparticle Tracking Analysis
4.7.3. Transmission Electron Microscopy
4.8. Total RNA Isolation from Cells and sEVs
4.9. Quantification of Cellular and sEV miRNA, and sEV mRNA
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| EV | Extracellular vesicle |
| FDR | False-discovery rate |
| FREM2 | FRAS1-related extracellular matrix protein 2 |
| GBM | Glioblastoma |
| IDH | Isocitrate dehydrogenase |
| LGG | Lower-grade glioma |
| MGMT | O-6-methylguanine-DNA-methyltransferase |
| MRI | Magnetic-resonance imaging |
| NAP1L1 | Nucleosome assembly protein 1-like 1 |
| NCL | Nucleolin |
| NTA | Nanoparticle-tracking analysis |
| sEV | Small extracellular vesicle |
| SPRY1 | Protein sprouty homolog 1 |
| VIM | Vimentin |
References
- Alexander, B.M.; Cloughesy, T.F. Adult glioblastoma. J. Clin. Oncol. 2017, 35, 2402–2409. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zuo, J.; Cui, Y.; Song, C.; Wu, X.; Feng, H.; Li, J.; Li, S.; Xu, Q.; Wei, W.; et al. Characterizing the molecular mechanisms of acquired temozolomide resistance in the u251 glioblastoma cell line by protein microarray. Oncol. Rep. 2018, 39, 2333–2341. [Google Scholar] [CrossRef] [PubMed]
- Crocetti, E.; Trama, A.; Stiller, C.; Caldarella, A.; Soffietti, R.; Jaal, J.; Weber, D.C.; Ricardi, U.; Slowinski, J.; Brandes, A.; et al. Epidemiology of glial and non-glial brain tumours in europe. Eur. J. Cancer 2012, 48, 1532–1542. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Rev. Dis. Primers 2015, 1, 15017. [Google Scholar] [CrossRef]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 who classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef]
- Jovcevska, I.; Zottel, A.; Samec, N.; Mlakar, J.; Sorokin, M.; Nikitin, D.; Buzdin, A.A.; Komel, R. High frem2 gene and protein expression are associated with favorable prognosis of idh-wt glioblastomas. Cancers 2019, 11, 1060. [Google Scholar] [CrossRef]
- Miranda, A.; Blanco-Prieto, M.; Sousa, J.; Pais, A.; Vitorino, C. Breaching barriers in glioblastoma. Part i: Molecular pathways and novel treatment approaches. Int. J. Pharm. 2017, 531, 372–388. [Google Scholar] [CrossRef]
- Zhong, J.; Paul, A.; Kellie, S.J.; O’Neill, G.M. Mesenchymal migration as a therapeutic target in glioblastoma. J. Oncol. 2010, 2010, 430142. [Google Scholar] [CrossRef]
- Noroxe, D.S.; Poulsen, H.S.; Lassen, U. Hallmarks of glioblastoma: A systematic review. ESMO Open 2016, 1, e000144. [Google Scholar] [CrossRef]
- Taylor, O.G.; Brzozowski, J.S.; Skelding, K.A. Glioblastoma multiforme: An overview of emerging therapeutic targets. Front. Oncol. 2019, 9, 963. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.F.; Patel, T.; Wong, D.; Das, S.; Freedman, J.E.; Laurent, L.C.; Carter, B.S.; Hochberg, F.; Van Keuren-Jensen, K.; Huentelman, M.; et al. Extracellular rnas: Development as biomarkers of human disease. J. Extracell Vesicles 2015, 4, 27495. [Google Scholar] [CrossRef] [PubMed]
- Tezcan, G.; Tunca, B.; Bekar, A.; Preusser, M.; Berghoff, A.S.; Egeli, U.; Cecener, G.; Ricken, G.; Budak, F.; Taskapilioglu, M.O.; et al. Microrna expression pattern modulates temozolomide response in gbm tumors with cancer stem cells. Cell Mol. Neurobiol. 2014, 34, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Chen, Y.; Zhou, X. The roles of micrornas in epigenetic regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell. Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Quezada, C.; Torres, A.; Niechi, I.; Uribe, D.; Contreras-Duarte, S.; Toledo, F.; San Martin, R.; Gutierrez, J.; Sobrevia, L. Role of extracellular vesicles in glioma progression. Mol. Aspects Med. 2018, 60, 38–51. [Google Scholar] [CrossRef]
- Saadatpour, L.; Fadaee, E.; Fadaei, S.; Nassiri Mansour, R.; Mohammadi, M.; Mousavi, S.M.; Goodarzi, M.; Verdi, J.; Mirzaei, H. Glioblastoma: Exosome and microrna as novel diagnosis biomarkers. Cancer Gene Ther. 2016, 23, 415–418. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (misev2018): A position statement of the international society for extracellular vesicles and update of the misev2014 guidelines. J. Extracell Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Vidak, M.; Jovcevska, I.; Samec, N.; Zottel, A.; Liovic, M.; Rozman, D.; Dzeroski, S.; Juvan, P.; Komel, R. Meta-analysis and experimental validation identified frem2 and spry1 as new glioblastoma marker candidates. Int. J. Mol. Sci. 2018, 19, 1369. [Google Scholar] [CrossRef]
- Jovcevska, I.; Zupanec, N.; Urlep, Z.; Vranic, A.; Matos, B.; Stokin, C.L.; Muyldermans, S.; Myers, M.P.; Buzdin, A.A.; Petrov, I.; et al. Differentially expressed proteins in glioblastoma multiforme identified with a nanobody-based anti-proteome approach and confirmed by oncofinder as possible tumor-class predictive biomarker candidates. Oncotarget 2017, 8, 44141–44158. [Google Scholar] [CrossRef]
- Zottel, A.; Jovčevska, I.; Šamec, N.; Mlakar, J.; Šribar, J.; Križaj, I.; Skoblar Vidmar, M.; Komel, R. Anti-vimentin, anti-tufm, anti-nap1l1 and anti-dpysl2 nanobodies display cytotoxic effect and reduce glioblastoma cell migration. Ther. Adv. Med. Oncol. 2020, 12, 1758835920915302. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microrna target sites in mammalian mrnas. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. Mirdb: An online database for prediction of functional microrna targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of functional microrna targets by integrative modeling of microrna binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. Diana-microt web server v5.0: Service integration into mirna functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef]
- Reczko, M.; Maragkakis, M.; Alexiou, P.; Grosse, I.; Hatzigeorgiou, A.G. Functional microrna targets in protein coding sequences. Bioinformatics 2012, 28, 771–776. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mrnas are conserved targets of micrornas. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. Interactivenn: A web-based tool for the analysis of sets through venn diagrams. BMC Bioinf. 2015, 16, 169. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. Webgestalt 2019: Gene set analysis toolkit with revamped uis and apis. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Wang, J.; Duncan, D.; Shi, Z.; Zhang, B. Web-based gene set analysis toolkit (webgestalt): Update 2013. Nucleic Acids Res. 2013, 41, W77–W83. [Google Scholar] [CrossRef]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. Webgestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017, 45, W130–W137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Kirov, S.; Snoddy, J. Webgestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005, 33, W741–W748. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. Panther version 14: More genomes, a new panther go-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. String v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Yin, S.; Chen, F.F.; Yang, G.F. Vimentin immunohistochemical expression as a prognostic factor in gastric cancer: A meta-analysis. Pathol. Res. Pract. 2018, 214, 1376–1380. [Google Scholar] [CrossRef]
- Jain, N.; Zhu, H.; Khashab, T.; Ye, Q.; George, B.; Mathur, R.; Singh, R.K.; Berkova, Z.; Wise, J.F.; Braun, F.K.; et al. Targeting nucleolin for better survival in diffuse large b-cell lymphoma. Leukemia 2018, 32, 663–674. [Google Scholar] [CrossRef]
- Schimmack, S.; Taylor, A.; Lawrence, B.; Alaimo, D.; Schmitz-Winnenthal, H.; Buchler, M.W.; Modlin, I.M.; Kidd, M. A mechanistic role for the chromatin modulator, nap1l1, in pancreatic neuroendocrine neoplasm proliferation and metastases. Epigen. Chromatin. 2014, 7, 15. [Google Scholar] [CrossRef]
- Nagaishi, M.; Kim, Y.H.; Mittelbronn, M.; Giangaspero, F.; Paulus, W.; Brokinkel, B.; Vital, A.; Tanaka, Y.; Nakazato, Y.; Legras-Lachuer, C.; et al. Amplification of the stoml3, frem2, and lhfp genes is associated with mesenchymal differentiation in gliosarcoma. Am. J. Pathol. 2012, 180, 1816–1823. [Google Scholar] [CrossRef]
- He, Q.; Jing, H.; Liaw, L.; Gower, L.; Vary, C.; Hua, S.; Yang, X. Suppression of spry1 inhibits triple-negative breast cancer malignancy by decreasing egf/egfr mediated mesenchymal phenotype. Sci. Rep. 2016, 6, 23216. [Google Scholar] [CrossRef] [PubMed]
- Masoumi-Moghaddam, S.; Amini, A.; Wei, A.Q.; Robertson, G.; Morris, D.L. Sprouty 1 predicts prognosis in human epithelial ovarian cancer. Am. J. Cancer Res. 2015, 5, 1531–1541. [Google Scholar] [PubMed]
- Sullivan, T.B.; Robert, L.C.; Teebagy, P.A.; Morgan, S.E.; Beatty, E.W.; Cicuto, B.J.; Nowd, P.K.; Rieger-Christ, K.M.; Bryan, D.J. Spatiotemporal microrna profile in peripheral nerve regeneration: Mir-138 targets vimentin and inhibits schwann cell migration and proliferation. Neural Regen. Res. 2018, 13, 1253–1262. [Google Scholar] [PubMed]
- Liu, X.; Wang, C.; Chen, Z.; Jin, Y.; Wang, Y.; Kolokythas, A.; Dai, Y.; Zhou, X. Microrna-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem. J. 2011, 440, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Schraivogel, D.; Weinmann, L.; Beier, D.; Tabatabai, G.; Eichner, A.; Zhu, J.Y.; Anton, M.; Sixt, M.; Weller, M.; Beier, C.P.; et al. Camta1 is a novel tumour suppressor regulated by mir-9/9* in glioblastoma stem cells. EMBO J. 2011, 30, 4309–4322. [Google Scholar] [CrossRef]
- Ben-Hamo, R.; Zilberberg, A.; Cohen, H.; Efroni, S. Hsa-mir-9 controls the mobility behavior of glioblastoma cells via regulation of mapk14 signaling elements. Oncotarget 2016, 7, 23170–23181. [Google Scholar] [CrossRef]
- Masliah-Planchon, J.; Garinet, S.; Pasmant, E. Ras-mapk pathway epigenetic activation in cancer: Mirnas in action. Oncotarget 2016, 7, 38892–38907. [Google Scholar] [CrossRef]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.W.; Weiss, W.A. Epidermal growth factor receptor and egfrviii in glioblastoma: Signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef]
- Lange, A.M.; Lo, H.W. Inhibiting trk proteins in clinical cancer therapy. Cancers 2018, 10, 105. [Google Scholar] [CrossRef]
- Munoz, J.L.; Bliss, S.A.; Greco, S.J.; Ramkissoon, S.H.; Ligon, K.L.; Rameshwar, P. Delivery of functional anti-mir-9 by mesenchymal stem cell-derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Mol. Ther. Nucleic Acids 2013, 2, e126. [Google Scholar] [CrossRef]
- Sharifzad, F.; Ghavami, S.; Verdi, J.; Mardpour, S.; Mollapour Sisakht, M.; Azizi, Z.; Taghikhani, A.; Los, M.J.; Fakharian, E.; Ebrahimi, M.; et al. Glioblastoma cancer stem cell biology: Potential theranostic targets. Drug Resist. Updat 2019, 42, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Aldaz, B.; Sagardoy, A.; Nogueira, L.; Guruceaga, E.; Grande, L.; Huse, J.T.; Aznar, M.A.; Diez-Valle, R.; Tejada-Solis, S.; Alonso, M.M.; et al. Involvement of mirnas in the differentiation of human glioblastoma multiforme stem-like cells. PLoS ONE 2013, 8, e77098. [Google Scholar] [CrossRef] [PubMed]
- Godlewski, J.; Newton, H.B.; Chiocca, E.A.; Lawler, S.E. Micrornas and glioblastoma; the stem cell connection. Cell Death Differ. 2010, 17, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Ramakrishnan, V.; Kim, R.; Skog, J.; Nakano, I.; Pingle, S.; Kalinina, J.; Hua, W.; Kesari, S.; Mao, Y.; et al. Mir-21 in the extracellular vesicles (evs) of cerebrospinal fluid (csf): A platform for glioblastoma biomarker development. PLoS ONE 2013, 8, e78115. [Google Scholar] [CrossRef] [PubMed]
- Le Rhun, E.; Preusser, M.; Roth, P.; Reardon, D.A.; van den Bent, M.; Wen, P.; Reifenberger, G.; Weller, M. Molecular targeted therapy of glioblastoma. Cancer Treat. Rev. 2019, 80, 101896. [Google Scholar] [CrossRef] [PubMed]
- Tuncel, G.; Kalkan, R. Receptor tyrosine kinase-ras-pi 3 kinase-akt signaling network in glioblastoma multiforme. Med. Oncol. 2018, 35, 122. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, R.; Wu, M.; Johnson, K.; Kim, H.; Celebre, A.; Shahzad, U.; Graham, M.S.; Kessler, J.A.; Chuang, J.H.; Karamchandani, J.; et al. Bmp signaling mediates glioma stem cell quiescence and confers treatment resistance in glioblastoma. Sci. Rep. 2019, 9, 14569. [Google Scholar] [CrossRef]
- Cheng, L.C.; Pastrana, E.; Tavazoie, M.; Doetsch, F. Mir-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009, 12, 399–408. [Google Scholar] [CrossRef]
- Lang, M.F.; Yang, S.; Zhao, C.; Sun, G.; Murai, K.; Wu, X.; Wang, J.; Gao, H.; Brown, C.E.; Liu, X.; et al. Genome-wide profiling identified a set of mirnas that are differentially expressed in glioblastoma stem cells and normal neural stem cells. PLoS ONE 2012, 7, e36248. [Google Scholar] [CrossRef]
- Milanovic, M.; Fan, D.N.Y.; Belenki, D.; Dabritz, J.H.M.; Zhao, Z.; Yu, Y.; Dorr, J.R.; Dimitrova, L.; Lenze, D.; Monteiro Barbosa, I.A.; et al. Senescence-associated reprogramming promotes cancer stemness. Nature 2018, 553, 96–100. [Google Scholar] [CrossRef]
- Jiang, W.; Finniss, S.; Cazacu, S.; Xiang, C.; Brodie, Z.; Mikkelsen, T.; Poisson, L.; Shackelford, D.B.; Brodie, C. Repurposing phenformin for the targeting of glioma stem cells and the treatment of glioblastoma. Oncotarget 2016, 7, 56456–56470. [Google Scholar] [CrossRef] [PubMed]
- Ferrarese, R.; Harsh, G.R.t.; Yadav, A.K.; Bug, E.; Maticzka, D.; Reichardt, W.; Dombrowski, S.M.; Miller, T.E.; Masilamani, A.P.; Dai, F.; et al. Lineage-specific splicing of a brain-enriched alternative exon promotes glioblastoma progression. J. Clin. Investig. 2014, 124, 2861–2876. [Google Scholar] [CrossRef] [PubMed]
- Yeh, M.; Oh, C.S.; Yoo, J.Y.; Kaur, B.; Lee, T.J. Pivotal role of microrna-138 in human cancers. Am. J. Cancer Res. 2019, 9, 1118–1126. [Google Scholar] [PubMed]
- Sha, H.H.; Wang, D.D.; Chen, D.; Liu, S.W.; Wang, Z.; Yan, D.L.; Dong, S.C.; Feng, J.F. Mir-138: A promising therapeutic target for cancer. Tumour. Biol. 2017, 39, 1010428317697575. [Google Scholar] [CrossRef]
- Qiu, S.; Huang, D.; Yin, D.; Li, F.; Li, X.; Kung, H.F.; Peng, Y. Suppression of tumorigenicity by microrna-138 through inhibition of ezh2-cdk4/6-prb-e2f1 signal loop in glioblastoma multiforme. Biochim. Biophys. Acta 2013, 1832, 1697–1707. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, L.; Dong, X.; Liu, L.; Huo, L.; Chen, H. High expression of vimentin is associated with progression and a poor outcome in glioblastoma. Appl. Immunohistochem. Mol. Morphol. 2018, 26, 337–344. [Google Scholar] [CrossRef]
- Quick, Q.; Paul, M.; Skalli, O. Roles and potential clinical applications of intermediate filament proteins in brain tumors. Semin. Pediatr. Neurol. 2015, 22, 40–48. [Google Scholar] [CrossRef]
- Trendowski, M. Exploiting the cytoskeletal filaments of neoplastic cells to potentiate a novel therapeutic approach. Biochim. Biophys. Acta 2014, 1846, 599–616. [Google Scholar] [CrossRef]
- Barbagallo, D.; Condorelli, A.; Ragusa, M.; Salito, L.; Sammito, M.; Banelli, B.; Caltabiano, R.; Barbagallo, G.; Zappala, A.; Battaglia, R.; et al. Dysregulated mir-671-5p/cdr1-as/cdr1/vsnl1 axis is involved in glioblastoma multiforme. Oncotarget 2016, 7, 4746–4759. [Google Scholar] [CrossRef]
- Anaya, J. Oncolnc: Linking tcga survival data to mrnas, mirnas, and lncrnas. PeerJ Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef]
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a shared vision for cancer genomic data. N. Engl. J. Med. 2016, 375, 1109–1112. [Google Scholar] [CrossRef]
- Stok, U.; Blokar, E.; Lenassi, M.; Holcar, M.; Frank-Bertoncelj, M.; Erman, A.; Resnik, N.; Sodin-Semrl, S.; Cucnik, S.; Pirkmajer, K.P.; et al. Characterization of plasma-derived small extracellular vesicles indicates ongoing endothelial and platelet activation in patients with thrombotic antiphospholipid syndrome. Cells 2020, 9, 1211. [Google Scholar] [CrossRef]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 3. [Google Scholar] [CrossRef] [PubMed]
- Manda, S.V.; Kataria, Y.; Tatireddy, B.R.; Ramakrishnan, B.; Ratnam, B.G.; Lath, R.; Ranjan, A.; Ray, A. Exosomes as a biomarker platform for detecting epidermal growth factor receptor-positive high-grade gliomas. J. Neurosurg. 2018, 128, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Rohn, G.; Koch, A.; Krischek, B.; Stavrinou, P.; Goldbrunner, R.; Timmer, M. Actb and sdha are suitable endogenous reference genes for gene expression studies in human astrocytomas using quantitative rt-pcr. Technol. Cancer Res. Treat. 2018, 17, 1533033818802318. [Google Scholar] [CrossRef]
- Blazquez, C.; Salazar, M.; Carracedo, A.; Lorente, M.; Egia, A.; Gonzalez-Feria, L.; Haro, A.; Velasco, G.; Guzman, M. Cannabinoids inhibit glioma cell invasion by down-regulating matrix metalloproteinase-2 expression. Cancer Res. 2008, 68, 1945–1952. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative rt-pcr data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
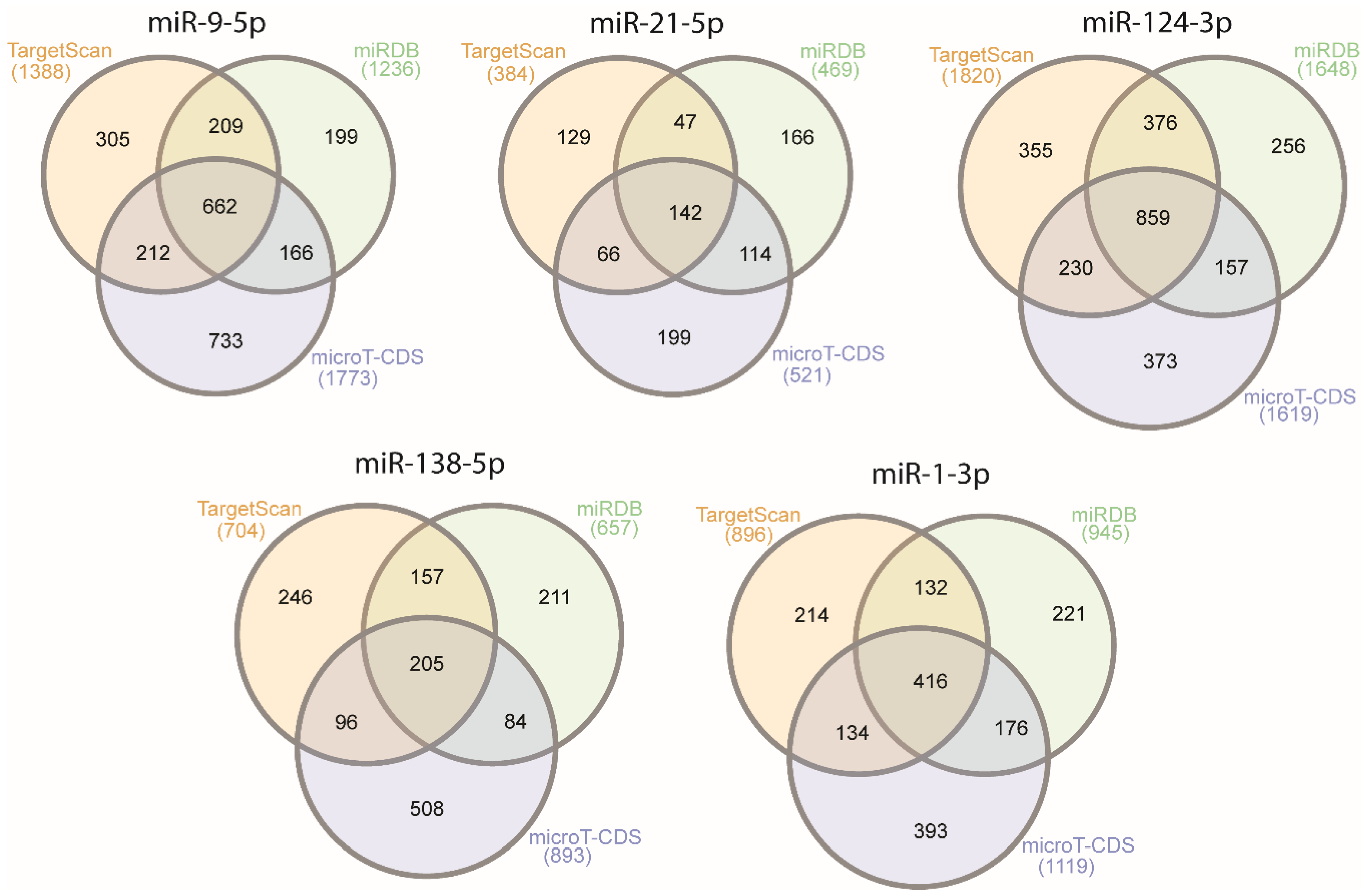

| miRNA | Gene | TargetScan (Aggregate Pct) | miRDB (Target Score) | microT-CDS (miTG Score) |
|---|---|---|---|---|
| hsa-miR-9-5p | FREM2 | 0.96 | 90 | 0.99 |
| NAP1L1 | 0.91 | 74 | 0.92 | |
| hsa-miR-21-5p | SPRY1 | 0.70 | 95 | 0.96 |
| hsa-miR-124-3p | VIM | 0.86 | 91 | 0.74 |
| NAP1L1 | 0.92 | 61 | 0.82 | |
| SPRY1 | 0.87 | X | 0.72 | |
| hsa-miR-138-5p | VIM | 0.70 | 94 | X |
| NCL | X | X | 0.80 | |
| hsa-miR-1-3p | NCL | 0.98 | 90 | 1 |
| Description | FDR | Description | FDR | ||||
|---|---|---|---|---|---|---|---|
| miR-9-5p | KEGG | Focal adhesion | 0.021 | miR-124-3p | KEGG | Apelin signaling pathway | 0.012 |
| Prostate cancer | 0.021 | Cellular senescence | 0.012 | ||||
| ErbB signaling pathway | 0.021 | Axon guidance | 0.012 | ||||
| Cellular senescence | 0.021 | Pathways in cancer | 0.012 | ||||
| Regulation of actin cytoskeleton | 0.021 | Panther | x | ||||
| MicroRNAs in cancer | 0.021 | Reactome | Receptor-type tyrosine-protein phosphatases | 0.031 | |||
| Neurotrophin signaling pathway | 0.021 | NOTCH2 Activation and Transmission of Signal to the Nucleus | 0.031 | ||||
| Hepatocellular carcinoma | 0.021 | Semaphorin interactions | 0.031 | ||||
| Panther | Ras Pathway | 0.009 | Signaling by Receptor Tyrosine Kinases | 0.031 | |||
| Reactome | X | Signaling by MET | 0.031 | ||||
| MiR-21-5p | KEGG | Proteoglycans in cancer | 0.019 | MiR-138-5p | KEGG | X | |
| Ras signaling pathway | 0.032 | Panther | X | ||||
| MicroRNAs in cancer | 0.038 | Reactome | X | ||||
| Neurotrophin signaling pathway | 0.043 | MiR-1-3p | KEGG | MicroRNAs in cancer | 0.000 | ||
| EGFR tyrosine kinase inhibitor resistance | 0.043 | Adherens junction | 0.010 | ||||
| Panther | X | Bacterial invasion of epithelial cells | 0.010 | ||||
| Reactome | Signaling by NTRK2 (TRKB) | 2.62 × 10−3 | Axon guidance | 0.029 | |||
| Signaling by NTRKs | 2.62 × 10−3 | Proteoglycans in cancer | 0.029 | ||||
| Signaling by BMP | 2.69 × 10−3 | EGFR tyrosine kinase inhibitor resistance | 0.035 | ||||
| Signaling by Receptor Tyrosine Kinases | 4.94 × 10−3 | Choline metabolism in cancer | 0.035 | ||||
| MET activates PI3K/AKT signaling | 4.94 × 10−3 | Regulation of actin cytoskeleton | 0.035 | ||||
| PI-3K cascade:FGFR3 | 5.14 × 10−3 | Hepatitis B | 0.035 | ||||
| PI-3K cascade:FGFR4 | 5.97 × 10−3 | Cocaine addiction | 0.042 | ||||
| Signaling by FGFR3 | 5.97 × 10−3 | Viral carcinogenesis | 0.042 | ||||
| Signaling by FGFR4 | 5.97 × 10−3 | Renal cell carcinoma | 0.042 | ||||
| Signaling by EGFR | 5.97 × 10−3 | Hippo signaling pathway | 0.042 | ||||
| Signaling by FGFR3 in disease | 5.97 × 10−3 | Spliceosome | 0.043 | ||||
| Signaling by FGFR3 point mutants in cancer | 5.97 × 10−3 | Human papillomavirus infection | 0.044 | ||||
| PI-3K cascade:FGFR2 | 6.63 × 10−3 | Thyroid cancer | 0.044 | ||||
| Downstream signaling of activated FGFR3 | 8.67 × 10−3 | Cellular senescence | 0.044 | ||||
| Signaling by FGFR3 fusions in cancer | 9.29 × 10−3 | Pathogenic Escherichia coli infection | 0.044 | ||||
| Signaling by FGFR1 | 9.29 × 10−3 | Panther | X | ||||
| Downstream signaling of activated FGFR4 | 9.77 × 10−3 | Reactome | Signaling by Receptor Tyrosine Kinases | 0.000 | |||
| Signaling by FGFR4 in disease | 0.010 | Clathrin-mediated endocytosis | 0.010 | ||||
| GAB1 signalosome | 0.010 | RHO GTPases Activate WASPs and WAVEs | 0.010 | ||||
| PIP3 activates AKT signaling | 0.010 | Signaling by VEGF | 0.013 | ||||
| Downstream signaling of activated FGFR2 | 0.012 | RNA Polymerase II Transcription | 0.028 | ||||
| Signaling by FGFR in disease | 0.020 | VEGFA-VEGFR2 Pathway | 0.028 | ||||
| Intracellular signaling by second messengers | 0.022 | Gene expression (Transcription) | 0.035 | ||||
| MAPK family signaling cascades | 0.022 | Signaling by RAS mutants | 0.035 | ||||
| Signaling by TGF-beta family members | 0.022 | Diseases of signal transduction | 0.035 | ||||
| Signaling by FGFR1 in disease | 0.025 | EPH-Ephrin signaling | 0.041 | ||||
| Interleukin-4 and Interleukin-13 signaling | 0.028 | Generic Transcription Pathway | 0.041 | ||||
| Constitutive Signaling by Aberrant PI3K in Cancer | 0.028 | Platelet activation, signaling and aggregation | 0.042 | ||||
| Signaling by FGFR2 | 0.031 | RHO GTPase Effectors | 0.042 | ||||
| Signaling by FGFR2 in disease | 0.034 | EPHB-mediated forward signaling | 0.042 | ||||
| Diseases of signal transduction | 0.034 | Hemostasis | 0.045 | ||||
| Generic Transcription Pathway | 0.035 | Regulation of actin dynamics for phagocytic cup formation | 0.048 | ||||
| PI3K Cascade | 0.035 | Membrane Trafficking | 0.049 | ||||
| Signaling by NODAL | 0.038 | ||||||
| Gene expression (Transcription) | 0.038 | ||||||
| MET activates PTPN11 | 0.039 | ||||||
| PI-3K cascade:FGFR1 | 0.040 | ||||||
| IRS-mediated signalling | 0.042 | ||||||
| miRNA | Tissues | Significance | p-Value |
|---|---|---|---|
| miR-9-5p | ns | ||
| miR-21-5p | GBM vs. LGG | * | 0.0146 |
| GBM vs. reference brain tissue | **** | <0.0001 | |
| miR-124-3p | GBM vs. reference brain tissue | * | 0.0178 |
| GBM vs. LGG | * | 0.0278 | |
| miR-138-5p | GBM vs. reference brain tissue | * | 0.0143 |
| miR-1-3p | ns |
| miRNA | sEV | Cell | ||||
|---|---|---|---|---|---|---|
| miR-9-5p | U251 vs. NCH421k | ** | 0.0048 | U251 vs. U87 | ** | 0.0028 |
| U87 vs. NCH644 | * | 0.0362 | U251 vs. NCH421k | **** | <0.0001 | |
| U87 vs. NCH421k | **** | <0.0001 | U87 vs. NCH644 | * | 0.0284 | |
| NCH644 vs. NCH421k | **** | <0.0001 | U87 vs. NCH421k | **** | <0.0001 | |
| NCH421k vs. astrocytes | **** | <0.0001 | NCH644 vs. NCH421k | **** | <0.0001 | |
| NCH421k vs. astrocytes | **** | <0.0001 | ||||
| miR-21-5p | ns | NCH644 vs. astrocytes | ** | 0.0052 | ||
| NCH421k vs. astrocytes | ** | 0.0042 | ||||
| miR-124-3p | U251 vs. NCH644 | **** | <0.0001 | U251 vs. NCH644 | **** | <0.0001 |
| U87 vs. NCH644 | **** | <0.0001 | U251 vs. NCH421k | ** | 0.0092 | |
| NCH644 vs. NCH421k | *** | 0.0002 | U87 vs. NCH644 | ** | 0.0028 | |
| NCH644 vs. astrocytes | **** | <0.0001 | NCH644 vs. astrocytes | **** | <0.0001 | |
| NCH421k vs. astrocytes | ** | 0.0075 | ||||
| miR-138-5p | U251 vs. U87 | * | 0.0301 | U251 vs. U87 | **** | <0.0001 |
| U87 vs. NCH644 | **** | <0.0001 | ||||
| U87 vs. NCH421k | **** | <0.0001 | ||||
| U87 vs. astrocytes | **** | <0.0001 | ||||
| miR-1-3p | ns | U87 vs. NCH421k | *** | 0.0007 | ||
| U251 vs. U87 | * | 0.013 | ||||
| NCH644 vs. NCH421k | * | 0.0242 | ||||
| NCH421k vs. astrocytes | * | 0.0140 | ||||
| Gene | Cells | Significance | p-Value |
|---|---|---|---|
| NAP1L1 | U251 vs. U87 | * | 0.0466 |
| U251 vs. astrocytes | *** | 0.0002 | |
| NCH644 vs. astrocytes | * | 0.0300 | |
| VIM | U251 vs. NCH644 | **** | <0.0001 |
| U251 vs. astrocytes | **** | <0.0001 | |
| U87 vs. NCH644 | *** | 0.0003 | |
| U87 vs. astrocytes | **** | <0.0001 | |
| NCL | NCH644 vs. U87 | * | 0.0418 |
| miRNA or mRNA | sEVs | Cell Lines | Tissue |
|---|---|---|---|
| miR-9-5p miRNA | ↑ NCH421k | ↑ NCH421k | ↑ associated with the poor survival of IDH mutant GBM patients ↓ associated with the better survival of GBM patients (regardless of IDH mutation) |
| miR-21-5p miRNA | / | ↓ NCH421k, NCH644 | ↑ GBM vs. LGG, reference tissue |
| miR-124-3p miRNA | ↑ NCH644 | ↑ NCH421k, NCH644 | ↓ GBM vs. LGG, reference tissue |
| miR-138-5p miRNA | / | / | ↓ GBM vs. reference tissue ↑ associated with the poor survival of IDH mutant GBM patients |
| VIM mRNA | ↑ U251, U87 | / |
| miRNA | Assay |
|---|---|
| miR-9-5p | 478214_mir |
| miR-21-5p | 477975_mir |
| miR-124-3p | 480901_mir |
| miR-138-5p | 477905_mir |
| miR-1-3p | 477820_mir |
| miR-99a-5p | 478519_mir |
| SPRY1 | Hs00398096_m1 |
| vimentin | Hs00958111_m1 |
| FREM2 | Hs01388268_m1 |
| NAP1L1 | Hs01590181_g1 |
| nucleolin | Hs01066668_m1 |
| GAPDH | Hs99999905_m1 |
| 18S | Hs99999901_s1 |
| Beta-actin | Hs99999903_m1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zottel, A.; Šamec, N.; Kump, A.; Raspor Dall’Olio, L.R.; Pužar Dominkuš, P.; Romih, R.; Hudoklin, S.; Mlakar, J.; Nikitin, D.; Sorokin, M.; et al. Analysis of miR-9-5p, miR-124-3p, miR-21-5p, miR-138-5p, and miR-1-3p in Glioblastoma Cell Lines and Extracellular Vesicles. Int. J. Mol. Sci. 2020, 21, 8491. https://doi.org/10.3390/ijms21228491
Zottel A, Šamec N, Kump A, Raspor Dall’Olio LR, Pužar Dominkuš P, Romih R, Hudoklin S, Mlakar J, Nikitin D, Sorokin M, et al. Analysis of miR-9-5p, miR-124-3p, miR-21-5p, miR-138-5p, and miR-1-3p in Glioblastoma Cell Lines and Extracellular Vesicles. International Journal of Molecular Sciences. 2020; 21(22):8491. https://doi.org/10.3390/ijms21228491
Chicago/Turabian StyleZottel, Alja, Neja Šamec, Ana Kump, Lucija Raspor Raspor Dall’Olio, Pia Pužar Dominkuš, Rok Romih, Samo Hudoklin, Jernej Mlakar, Daniil Nikitin, Maxim Sorokin, and et al. 2020. "Analysis of miR-9-5p, miR-124-3p, miR-21-5p, miR-138-5p, and miR-1-3p in Glioblastoma Cell Lines and Extracellular Vesicles" International Journal of Molecular Sciences 21, no. 22: 8491. https://doi.org/10.3390/ijms21228491
APA StyleZottel, A., Šamec, N., Kump, A., Raspor Dall’Olio, L. R., Pužar Dominkuš, P., Romih, R., Hudoklin, S., Mlakar, J., Nikitin, D., Sorokin, M., Buzdin, A., Jovčevska, I., & Komel, R. (2020). Analysis of miR-9-5p, miR-124-3p, miR-21-5p, miR-138-5p, and miR-1-3p in Glioblastoma Cell Lines and Extracellular Vesicles. International Journal of Molecular Sciences, 21(22), 8491. https://doi.org/10.3390/ijms21228491









