Lost in the Crowd: How Does Human 8-Oxoguanine DNA Glycosylase 1 (OGG1) Find 8-Oxoguanine in the Genome?
Abstract
1. 8-Oxoguanine: Biological Relevance and Repair
1.1. Biological Prominence and Consequences of 8-Oxoguanine
1.2. 8-oxoG Repair in Mammalian Cells
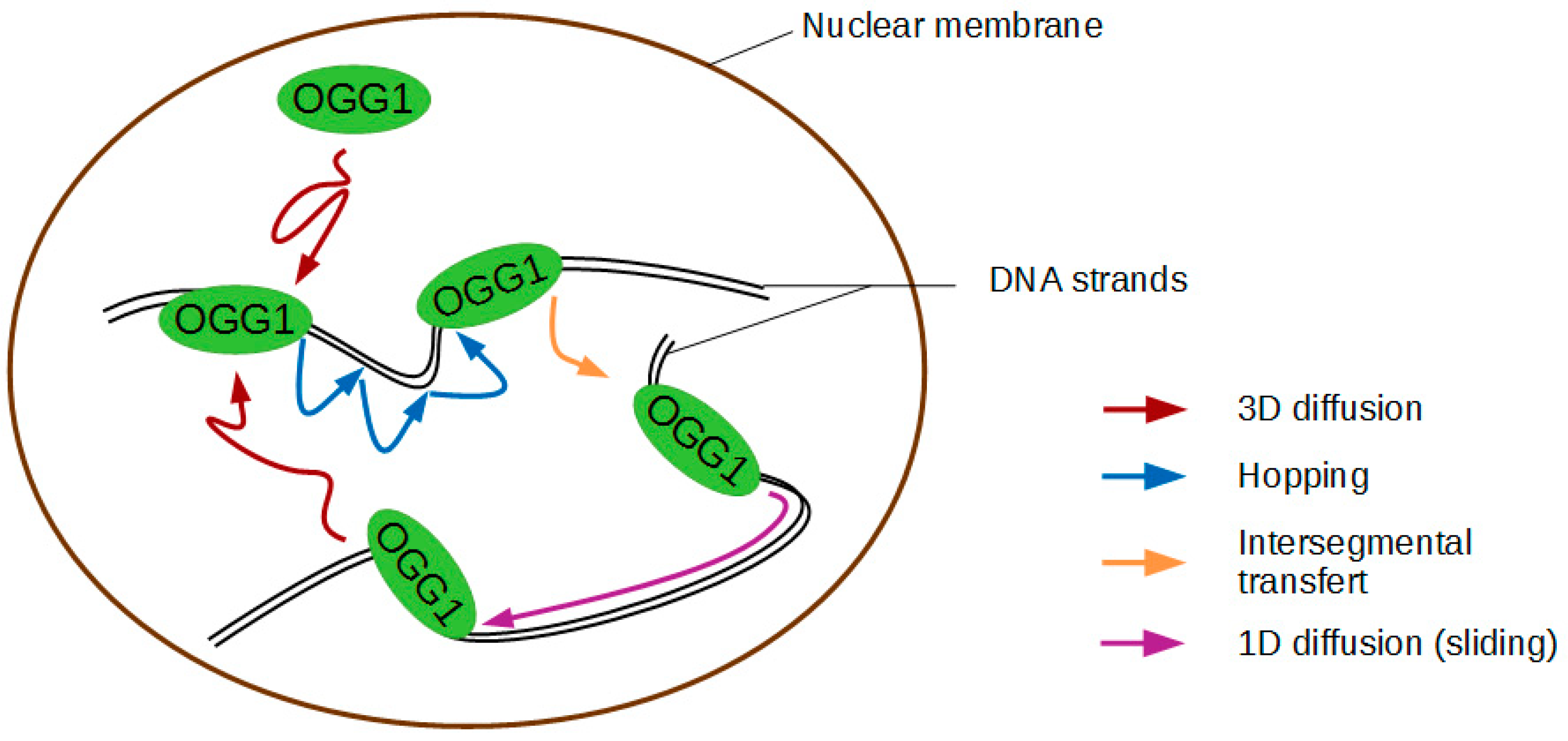
2. How Does OGG1 Find 8-oxoG Amongst a Million-Fold Excess of Gs?
2.1. 8-oxoG Recognition by OGG1 on Naked DNA
2.2. Impact of the Nucleosome on 8-oxoG Excision by OGG1
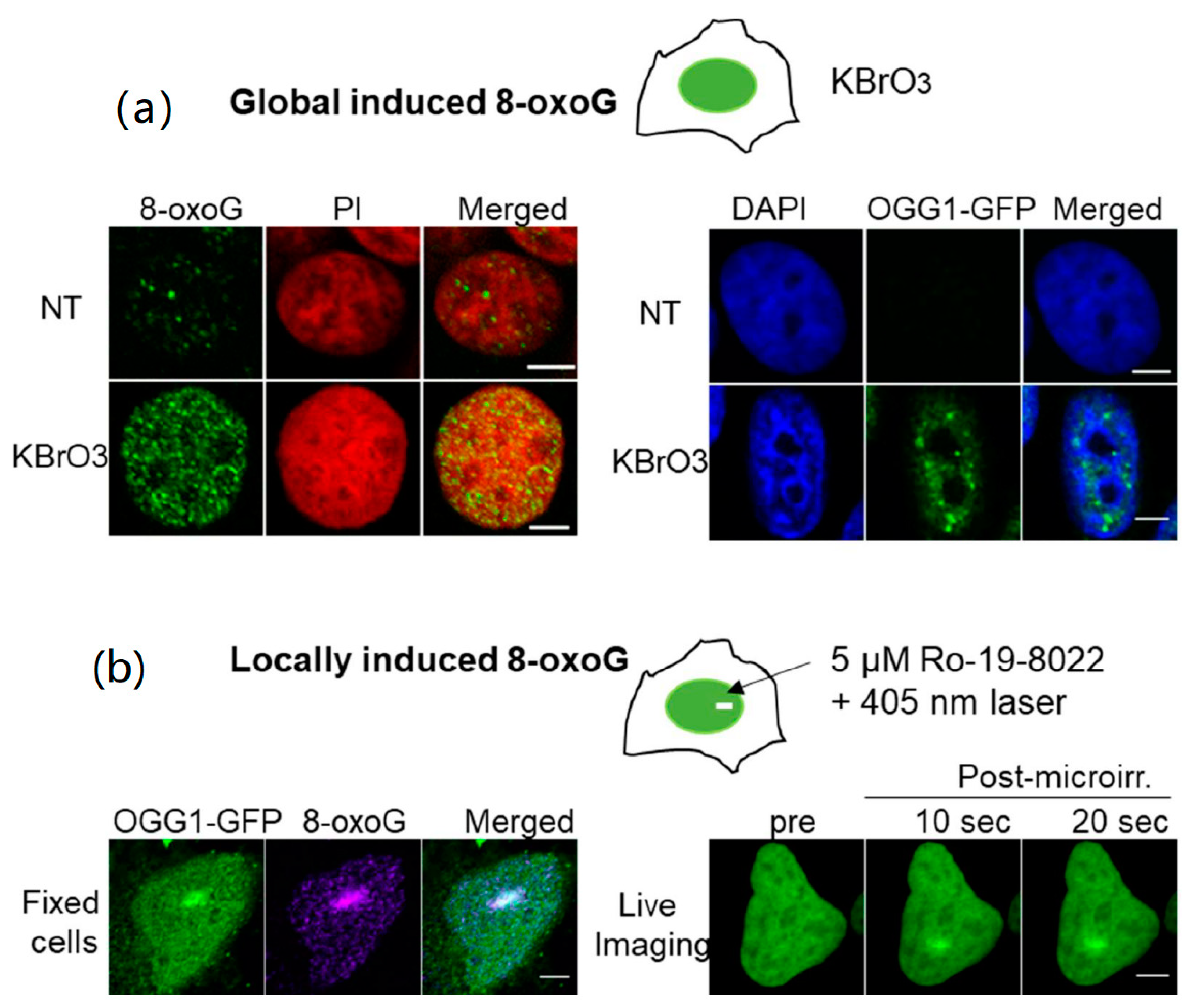
2.3. Mechanisms Triggering the Recruitment of OGG1 to 8-oxoG Lesions in the Nuclear Context
2.3.1. Efficient Recruitment of OGG1 to Chromatin Is Promoted by Several Cofactors
2.3.2. Potential Role of Post-Translational Modifications in OGG1 Nuclear Dynamics
2.4. Preferential Recruitment of OGG1 to Open Chromatin Regions: A Link with Transcription?
3. Open Questions Regarding the Influence of the Nuclear Environment on OGG1 Functions
3.1. Does Macromolecular Crowding Facilitate or Impair OGG1 Dynamics within the Nucleus?
3.2. How Much Time Does It Take to Find an 8-oxoG Lesion within the Complex Architecture of the Nucleus?
3.3. Does Facilitated Diffusion of OGG1 Indeed Occur in the Cell Nucleus?
Funding
Conflicts of Interest
References
- Marnett, L.J. Oxyradicals and DNA damage. Carcinogenesis 2000, 21, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J. Oxidative damage to DNA: Formation, measurement and biochemical features. Mutat. Res. Mol. Mech. Mutagen. 2003, 531, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Gedik, C.M.; Collins, A. Establishing the background level of base oxidation in human lymphocyte DNA: Results of an interlaboratory validation study. FASEB J. 2005, 19, 82–84. [Google Scholar] [CrossRef] [PubMed]
- Ohno, M.; Miura, T.; Furuichi, M.; Tominaga, Y.; Tsuchimoto, D.; Sakumi, K.; Nakabeppu, Y. A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome. Genome Res. 2006, 16, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Kasai, H.; Tanooka, H.; Nishimura, S. Formation of 8-hydroxyguanine residues in DNA by X-irradiation. Gan 1984, 75, 1037–1039. [Google Scholar]
- Neeley, W.L.; Essigmann, J.M. Mechanisms of Formation, Genotoxicity, and Mutation of Guanine Oxidation Products. Chem. Res. Toxicol. 2006, 19, 491–505. [Google Scholar] [CrossRef]
- Hall, D.B.; Holmlin, R.E.; Barton, J.K. Oxidative DNA damage through long-range electron transfer. Nature 1996, 382, 731–735. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Sampath, H.; McCullough, A.K.; Lloyd, R.S. Regulation of DNA glycosylases and their role in limiting disease. Free Radic. Res. 2012, 46, 460–478. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Z.; Oka, S.; Tsuchimoto, D.; Abolhassani, N.; Nomaru, H.; Sakumi, K.; Yamada, H.; Nakabeppu, Y. 8-Oxoguanine causes neurodegeneration during MUTYH-mediated DNA base excision repair. J Clin. Investig. 2012, 122, 4344–4361. [Google Scholar] [CrossRef]
- Siddiqui, A.; Rivera-Sánchez, S.; Castro, M.D.R.; Acevedo-Torres, K.; Rane, A.; Torres-Ramos, C.A.; Nicholls, D.G.; Andersen, J.K.; Ayala-Torres, S. Mitochondrial DNA damage Is associated with reduced mitochondrial bioenergetics in Huntington’s disease. Free Radic. Biol. Med. 2012, 53, 1478–1488. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Pena, S. Role of oxidative DNA damage in mitochondrial dysfunction and Huntington’s disease pathogenesis. Free Radic. Biol. Med. 2013, 62, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Bradley-Whitman, M.A.; Timmons, M.D.; Beckett, T.L.; Murphy, M.P.; Lynn, B.C.; Lovell, M.A. Nucleic acid oxidation: An early feature of Alzheimer’s disease. J. Neurochem. 2014, 128, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Golden, T.-R.; Melov, S. Mitochondrial DNA mutations, oxidative stress, and aging. Mech. Ageing Dev. 2001, 122, 1577–1589. [Google Scholar] [CrossRef]
- Boiteux, S.; Coste, F.; Castaing, B. Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: Properties and biological roles of the Fpg and OGG1 DNA N-glycosylases. Free Radic. Biol. Med. 2017, 107, 179–201. [Google Scholar] [CrossRef]
- Plum, G.E.; Grollman, A.P.; Johnson, F.; Breslauer, K.J. Influence of the Oxidatively Damaged Adduct 8-Oxodeoxyguanosine on the Conformation, Energetics, and Thermodynamic Stability of a DNA Duplex. Biochemistry 1995, 34, 16148–16160. [Google Scholar] [CrossRef]
- Singh, S.K.; Szulik, M.W.; Ganguly, M.; Khutsishvili, I.; Stone, M.P.; Marky, L.A.; Gold, B. Characterization of DNA with an 8-oxoguanine modification. Nucleic Acids Res. 2011, 39, 6789–6801. [Google Scholar] [CrossRef]
- Lipscomb, L.A.; Peek, M.E.; Morningstar, M.L.; Verghis, S.M.; Miller, E.M.; Rich, A.; Essigmann, J.M.; Williams, L.D. X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl. Acad. Sci. USA 1995, 92, 719–723. [Google Scholar] [CrossRef]
- Oda, Y.; Uesugi, S.; Ikehara, M.; Nishimura, S.; Kawase, Y.; Ishikawa, H.; Inoue, H.; Ohtsuka, E. NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res. 1991, 19, 1407–1412. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gannett, P.M.; Sura, T.P. Base pairing of 8-oxoguanosine and 8-oxo-2’-deoxyguanosine with 2’-deoxyadenosine, 2’-deoxycytosine, 2’-deoxyguanosine, and thymidine. Chem. Res. Toxicol. 1993, 6, 690–700. [Google Scholar] [CrossRef]
- Yudkina, A.V.; Shilkin, E.S.; Endutkin, A.V.; Makarova, A.V.; Zharkov, D.O. Reading and Misreading 8-oxoguanine, a Paradigmatic Ambiguous Nucleobase. Crystals 2019, 9, 269. [Google Scholar] [CrossRef]
- Kitsera, N.; Stathis, D.; Luehnsdorf, B.; Mueller, H.; Carell, T.; Epe, B.; Khobta, A. 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res. 2011, 39, 5926–5934. [Google Scholar] [CrossRef] [PubMed]
- Kouchakdjian, M.; Bodepudi, V.; Shibutani, S.; Eisenberg, M.; Johnson, F.; Grollman, A.P.; Patel, D.J. NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H-dG(syn).cntdot.dA(anti) alignment at lesion site. Biochemistry 1991, 30, 1403–1412. [Google Scholar] [CrossRef]
- Shibutani, S.; Takeshita, M.; Grollman, A.P. Insertion of specific bases during DNA synthesis past the oxidation- damaged base 8-oxodG. Nature 1991, 349, 431–434. [Google Scholar] [CrossRef]
- Hsu, G.W.; Ober, M.; Carell, T.; Beese, L.S. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature 2004, 431, 217–221. [Google Scholar] [CrossRef]
- Grollman, A.P.; Moriya, M. Mutagenesis by 8-oxoguanine: An enemy within. Trends Genet 1993, 9, 246–249. [Google Scholar] [CrossRef]
- Saxowsky, T.T.; Meadows, K.L.; Klungland, A.; Doetsch, P.W. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 18877–18882. [Google Scholar] [CrossRef]
- Lindahl, T.; Karran, P.; Wood, R.D. DNA excision repair pathways. Curr. Opin. Genet. Dev. 1997, 7, 158–169. [Google Scholar] [CrossRef]
- Lindahl, T.; Wood, R.D. Quality Control by DNA Repair. Science 1999, 286, 1897–1905. [Google Scholar] [CrossRef]
- Krokan, H.E.; Bjoras, M. Base Excision Repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef]
- van der Kemp, P.A.; Thomas, D.; Barbey, R.; de Oliveira, R.; Boiteux, S. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. USA 1996, 93, 5197–5202. [Google Scholar] [CrossRef]
- Nash, H.M.; Bruner, S.D.; Schärer, O.D.; Kawate, T.; Addona, T.A.; Spooner, E.; Lane, W.S.; Verdine, G.L. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 1996, 6, 968–980. [Google Scholar] [CrossRef]
- Aburatani, H.; Hippo, Y.; Ishida, T.; Takashima, R.; Matsuba, C.; Kodama, T.; Takao, M.; Yasui, A.; Yamamoto, K.; Asano, M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997, 57, 2151–2156. [Google Scholar]
- Arai, K.; Morishita, K.; Shinmura, K.; Kohno, T.; Kim, S.R.; Nohmi, T.; Taniwaki, M.; Ohwada, S.; Yokota, J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene 1997, 14, 2857–2861. [Google Scholar] [CrossRef]
- Bjoras, M. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 1997, 16, 6314–6322. [Google Scholar] [CrossRef]
- Lu, R.; Nash, H.M.; Verdine, G.L. A mammalian DNA repair enzyme that excises oxidatively damaged guanines maps to a locus frequently lost in lung cancer. Curr. Biol. 1997, 7, 397–407. [Google Scholar] [CrossRef]
- Radicella, J.P.; Dherin, C.; Desmaze, C.; Fox, M.S.; Boiteux, S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1997, 94. [Google Scholar] [CrossRef]
- Roldán-Arjona, T.; Wei, Y.F.; Carter, K.C.; Klungland, A.; Anselmino, C.; Wang, R.P.; Augustus, M.; Lindahl, T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl. Acad. Sci. USA 1997, 94, 8016–8020. [Google Scholar] [CrossRef]
- Rosenquist, T.A.; Zharkov, D.O.; Grollman, A.P. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. USA 1997, 94, 7429–7434. [Google Scholar] [CrossRef]
- Dalhus, B.; Forsbring, M.; Helle, I.H.; Vik, E.S.; Forstrøm, R.J.; Backe, P.H.; Alseth, I.; Bjørås, M. Separation-of-Function Mutants Unravel the Dual-Reaction Mode of Human 8-Oxoguanine DNA Glycosylase. Structure 2011, 19, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.E.; Hickson, I.D.; Boiteux, S.; Radicella, J.P. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: Bypass of the AP lyase activity step. Nucleic Acids Res. 2001, 29, 1285–1292. [Google Scholar] [CrossRef]
- Hill, J.W. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: Potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001, 29, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Reardon, J.T.; Bessho, T.; Kung, H.C.; Bolton, P.H.; Sancar, A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: Possible explanation for neurodegeneration in Xeroderma pigmentosum patients. Proc. Natl. Acad. Sci. USA 1997, 94, 9463–9468. [Google Scholar] [CrossRef]
- Bergeron, F.; Auvre, F.; Radicella, J.P.; Ravanat, J.L. HO* radicals induce an unexpected high proportion of tandem base lesions refractory to repair by DNA glycosylases. Proc. Natl. Acad. Sci. USA 2010, 107, 5528–5533. [Google Scholar] [CrossRef]
- David, S.S.; O’Shea, V.L.; Kundu, S. Base-excision repair of oxidative DNA damage. Nature 2007, 447, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Bruner, S.D.; Norman, D.P.G.; Verdine, G.L. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 2000, 403, 859–866. [Google Scholar] [CrossRef]
- Cheng, X.; Kelso, C.; Hornak, V.; de los Santos, C.; Grollman, A.P.; Simmerling, C. Dynamic Behavior of DNA Base Pairs Containing 8-Oxoguanine. J. Am. Chem. Soc. 2005, 127, 13906–13918. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Haushalter, K.A.; Lieber, C.M.; Verdine, G.L. Direct visualization of a DNA glycosylase searching for damage. Chem. Biol. 2002, 9, 345–350. [Google Scholar] [CrossRef]
- Banerjee, A.; Yang, W.; Karplus, M.; Verdine, G.L. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature 2005, 434, 612–618. [Google Scholar] [CrossRef]
- Blainey, P.C.; Van Oijen, A.M.; Banerjee, A.; Verdine, G.L.; Xie, X.S. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. USA 2006, 103, 5752–5757. [Google Scholar] [CrossRef]
- Crenshaw, C.M.; Nam, K.; Oo, K.; Kutchukian, P.S.; Bowman, B.R.; Karplus, M.; Verdine, G.L. Enforced Presentation of an Extrahelical Guanine to the Lesion Recognition Pocket of Human 8-Oxoguanine Glycosylase, hOGG1. J. Biol. Chem. 2012, 287, 24916–24928. [Google Scholar] [CrossRef]
- Li, H.; Endutkin, A.V.; Bergonzo, C.; Fu, L.; Grollman, A.; Zharkov, D.O.; Simmerling, C. DNA Deformation-Coupled Recognition of 8-Oxoguanine: Conformational Kinetic Gating in Human DNA Glycosylase. J. Am. Chem. Soc. 2017, 139, 2682–2692. [Google Scholar] [CrossRef]
- Kuznetsov, N.A.; Koval, V.V.; Nevinsky, G.A.; Douglas, K.T.; Zharkov, D.O.; Fedorova, O.S. Kinetic Conformational Analysis of Human 8-Oxoguanine-DNA Glycosylase. J. Biol. Chem. 2007, 282, 1029–1038. [Google Scholar] [CrossRef]
- Tyugashev, T.E.; Vorobjev, Y.N.; Kuznetsova, A.A.; Lukina, M.V.; Kuznetsov, N.A.; Fedorova, O.S. Roles of Active-Site Amino Acid Residues in Specific Recognition of DNA Lesions by Human 8-Oxoguanine-DNA Glycosylase (OGG1). J. Phys. Chem. 2019, 123, 4878–4887. [Google Scholar] [CrossRef]
- Berg, O.G.; Winter, R.B.; Von Hippel, P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry 1981, 20, 6929–6948. [Google Scholar] [CrossRef]
- Riggs, A.D.; Bourgeois, S.; Cohn, M. The lac represser-operator interaction. J. Mol. Biol. 1970, 53, 401–417. [Google Scholar] [CrossRef]
- Blainey, P.C.; Luo, G.; Kou, S.C.; Mangel, W.F.; Verdine, G.L.; Bagchi, B.; Xie, X.S. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009, 16, 1224–1229. [Google Scholar] [CrossRef]
- Ahmadi, A.; Rosnes, I.; Blicher, P.; Diekmann, R.; Schüttpelz, M.; Glette, K.; Tørresen, J.; Bjørås, M.; Dalhus, B.; Rowe, A.D. Breaking the speed limit with multimode fast scanning of DNA by Endonuclease V. Nat. Commun. 2018, 9, 5381. [Google Scholar] [CrossRef]
- Lee, A.J.; Majumdar, C.; Kathe, S.D.; Van Ostrand, R.P.; Vickery, H.R.; Averill, A.M.; Nelson, S.R.; Manlove, A.H.; McCord, M.A.; David, S.S. Detection of OG: A Lesion Mispairs by MutY Relies on a Single His Residue and the 2-Amino Group of 8-Oxoguanine. J. Am. Chem. Soc. 2020, 142, 13283–13287. [Google Scholar] [CrossRef]
- Dunn, A.R.; Kad, N.M.; Nelson, S.R.; Warshaw, D.M.; Wallace, S.S. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011, 39, 7487–7498. [Google Scholar] [CrossRef]
- Rowland, M.M.; Schonhoft, J.D.; McKibbin, P.L.; David, S.S.; Stivers, J.T. Microscopic mechanism of DNA damage searching by hOGG1. Nucleic Acids Res. 2014, 42, 9205–9303. [Google Scholar] [CrossRef]
- Lee, A.J.; Wallace, S.S. Hide and seek: How do DNA glycosylases locate oxidatively damaged DNA bases amidst a sea of undamaged bases? Free Radic. Biol. Med. 2017, 107, 170–178. [Google Scholar] [CrossRef]
- Smerdon, M.J. DNA repair and the role of chromatin structure. Curr. Opin. Cell Biol. 1991, 3, 422–428. [Google Scholar] [CrossRef]
- Green, C.M.; Almouzni, G. When repair meets chromatin. EMBO Rep. 2002, 3, 28–33. [Google Scholar] [CrossRef]
- Nilsen, H.; Lindahl, T.; Verreault, A. DNA base excision repair of uracil residues in reconstituted nucleosome core particles. EMBO J. 2002, 21, 5943–5952. [Google Scholar] [CrossRef] [PubMed]
- Beard, B.C.; Wilson, S.H.; Smerdon, M.J. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proc. Natl. Acad. Sci. USA 2003, 100, 7465–7470. [Google Scholar] [CrossRef] [PubMed]
- Bilotti, K.; Kennedy, E.E.; Li, C.; Delaney, S. Human OGG1 activity in nucleosomes is facilitated by transient unwrapping of DNA and is influenced by the local histone environment. DNA Repair 2017, 59, 1–8. [Google Scholar] [CrossRef]
- Menoni, H.; Gasparutto, D.; Hamiche, A.; Cadet, J.; Dimitrov, S.; Bouvet, P.; Angelov, D. ATP-Dependent Chromatin Remodeling Is Required for Base Excision Repair in Conventional but Not in Variant H2A.Bbd Nucleosomes. Mol. Cell. Biol. 2007, 27, 5949–5956. [Google Scholar] [CrossRef]
- Menoni, H.; Hoeijmakers, J.H.J.; Vermeulen, W. Nucleotide excision repair-initiating proteins bind to oxidative DNA lesions in vivo. J. Cell Biol. 2012, 199, 1037–1046. [Google Scholar] [CrossRef]
- Amouroux, R.; Campalans, A.; Epe, B.; Radicella, J.P. Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic Acids Res. 2010, 38. [Google Scholar] [CrossRef]
- Campalans, A.; Kortulewski, T.; Amouroux, R.; Menoni, H.; Vermeulen, W.; Radicella, J.P. Distinct spatiotemporal patterns and PARP dependence of XRCC1 recruitment to single-strand break and base excision repair. Nucleic Acids Res. 2013, 41, 3115–3129. [Google Scholar] [CrossRef]
- Lebraud, E.; Pinna, G.; Siberchicot, C.; Depagne, J.; Busso, D.; Fantini, D.; Irbah, L.; Robeska, E.; Kratassiouk, G.; Ravanat, J.-L.; et al. Chromatin recruitment of OGG1 requires cohesin and mediator and is essential for efficient 8-oxoG removal. Nucleic Acids Res. 2020, 48, 9082–9097. [Google Scholar] [CrossRef]
- Visnes, T.; Cázares-Körner, A.; Hao, W.; Wallner, O.; Masuyer, G.; Loseva, O.; Mortusewicz, O.; Wiita, E.; Sarno, A.; Manoilov, A.; et al. Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation. Science 2018, 362, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Nakajima, S.; Wei, L.; Sun, L.; Hsieh, C.L.; Sobol, R.W.; Bruchez, M.; Van Houten, B.; Yasui, A.; Levine, A.S. Novel method for site-specific induction of oxidative DNA damage reveals differences in recruitment of repair proteins to heterochromatin and euchromatin. Nucleic Acids Res. 2014, 42, 2330–2345. [Google Scholar] [CrossRef]
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.; Lebeaupin, T.; Juhász, S.; Chapuis, C.; D’Augustin, O.; Dutertre, S.; Burkovics, P.; Biertümpfel, C.; Timinszky, G.; Huet, S. Poly(ADP-ribose)-dependent chromatin unfolding facilitates the association of DNA-binding proteins with DNA at sites of damage. Nucleic Acids Res. 2019, 47, 11250–11267. [Google Scholar] [CrossRef]
- Kumar, N.; Raja, S.; Van Houten, B. The involvement of nucleotide excision repair proteins in the removal of oxidative DNA damage. Nucleic Acids Res. 2020, gkaa777. [Google Scholar] [CrossRef]
- Jang, S.; Kumar, N.; Beckwitt, E.C.; Kong, M.; Fouquerel, E.; Rapić-Otrin, V.; Prasad, R.; Watkins, S.C.; Khuu, C.; Majumdar, C.; et al. Damage sensor role of UV-DDB during base excision repair. Nat. Struct. Mol. Biol. 2019, 26, 695–703. [Google Scholar] [CrossRef] [PubMed]
- van der Kemp, P.A.; Charbonnier, J.B.; Audebert, M.; Boiteux, S. Catalytic and DNA-binding properties of the human Ogg1 DNA N-glycosylase/AP lyase: Biochemical exploration of H270, Q315 and F319, three amino acids of the 8-oxoguanine-binding pocket. Nucleic Acids Res. 2004, 32, 570–578. [Google Scholar] [CrossRef]
- Kumar, N.; Moreno, N.C.; Feltes, B.C.; Menck, C.F.; Van Houten, B.; Kumar, N.; Moreno, N.C.; Feltes, B.C.; Menck, C.F.; Houten, B. Cooperation and interplay between base and nucleotide excision repair pathways: From DNA lesions to proteins. Genet. Mol. Biol. 2020, 43. [Google Scholar] [CrossRef]
- D’Errico, M.; Parlanti, E.; Teson, M.; De Jesus, B.M.B.; Degan, P.; Calcagnile, A.; Jaruga, P.; Bjørås, M.; Crescenzi, M.; Pedrini, A.M.; et al. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 2006, 25, 4305–4315. [Google Scholar] [CrossRef] [PubMed]
- Menoni, H.; Wienholz, F.; Theil, A.F.; Janssens, R.C.; Lans, H.; Campalans, A.; Radicella, J.P.; Marteijn, J.A.; Vermeulen, W. The transcription-coupled DNA repair-initiating protein CSB promotes XRCC1 recruitment to oxidative DNA damage. Nucleic Acids Res. 2018, 46, 7747–7756. [Google Scholar] [CrossRef] [PubMed]
- Fortini, P.; Parlanti, E.; Sidorkina, O.M.; Laval, J.; Dogliotti, E. The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J. Biol. Chem. 1999, 274, 15230–15236. [Google Scholar] [CrossRef]
- Paquet, N.; Adams, M.N.; Leong, V.; Ashton, N.W.; Touma, C.; Gamsjaeger, R.; Cubeddu, L.; Beard, S.; Burgess, J.T.; Bolderson, E.; et al. HSSB1 (NABP2/OBFC2B) is required for the repair of 8-oxo-guanine by the hOGG1-mediated base excision repair pathway. Nucleic Acids Res. 2015, 43, 8817–8829. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huang, S.C.; Glenn St Hilaire, B.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–320.e24. [Google Scholar] [CrossRef]
- Soutourina, J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 2018, 19, 262–274. [Google Scholar] [CrossRef]
- Kim, J.S.; Krasieva, T.B.; LaMorte, V.; Malcolm, A.; Taylor, R.; Yokomori, K. Specific recruitment of human cohesin to laser-induced DNA damage. J. Biol. Chem. 2002, 277, 45149–45153. [Google Scholar] [CrossRef]
- Eyboulet, F.; Cibot, C.; Eychenne, T.; Neil, H.; Alibert, O.; Werner, M.; Soutourina, J. Mediator links transcription and DNA repair by facilitating Rad2/XPG recruitment. Genes Dev. 2013, 27, 2549–2562. [Google Scholar] [CrossRef]
- Ström, L.; Lindroos, H.B.; Shirahige, K.; Sjögren, C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 2004, 16, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Dorsett, D.; Ström, L. The ancient and evolving roles of cohesin in DNA repair and gene expression. Curr. Biol. 2012, 22, 240–250. [Google Scholar] [CrossRef]
- Bhakat, K.K.; Mokkapati, S.K.; Boldogh, I.; Hazra, T.K.; Mitra, S. Acetylation of Human 8-Oxoguanine-DNA Glycosylase by p300 and Its Role in 8-Oxoguanine Repair In Vivo. Mol. Cell. Biol. 2006, 26, 1654–1665. [Google Scholar] [CrossRef] [PubMed]
- Tini, M.; Benecke, A.; Um, S.J.; Torchia, J.; Evans, R.M.; Chambon, P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell 2002, 9, 256–277. [Google Scholar] [CrossRef]
- Hu, J.; Imam, S.Z.; Hashiguchi, K.; de Souza-Pinto, N.C.; Bohr, V.A. Phosphorylation of human oxoguanine DNA glycosylase (α-OGG1) modulates its function. Nucleic Acids Res. 2005, 33, 3271–3282. [Google Scholar] [CrossRef]
- Dantzer, F.; Luna, L.; Bjørås, M.; Seeberg, E. Human OGG1 undergoes serine phosphorylation and associates with the nuclear matrix and mitotic chromatin in vivo. Nucleic Acids Res. 2002, 30, 2349–2357. [Google Scholar] [CrossRef]
- Verschure, P.J.; van der Kraan, I.; Manders, E.M.M.; Hoogstraten, D.; Houtsmuller, A.B.; van Driel, R. Condensed chromatin domains in the mammalian nucleus are accessible to large macromolecules. EMBO Rep. 2003, 4, 861–866. [Google Scholar] [CrossRef]
- Kagey, M.H.; Newman, J.J.; Bilodeau, S.; Zhan, Y.; Orlando, D.A.; van Berkum, N.L.; Ebmeier, C.C.; Goossens, J.; Rahl, P.B.; Levine, S.S.; et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010, 467, 430–435. [Google Scholar] [CrossRef]
- Yoshihara, M.; Jiang, L.; Akatsuka, S.; Suyama, M.; Toyokuni, S. Genome-wide profiling of 8-oxoguanine reveals its association with spatial positioning in nucleus. DNA Res. 2014, 21, 603–612. [Google Scholar] [CrossRef]
- Amente, S.; Di Palo, G.; Scala, G.; Castrignanò, T.; Gorini, F.; Cocozza, S.; Moresano, A.; Pucci, P.; Ma, B.; Stepanov, I.; et al. Genome-wide mapping of 8-oxo-7,8-dihydro-2′-deoxyguanosine reveals accumulation of oxidatively-generated damage at DNA replication origins within transcribed long genes of mammalian cells. Nucleic Acids Res. 2019, 47, 221–236. [Google Scholar] [CrossRef]
- Ding, Y.; Fleming, A.M.; Burrows, C.J. Sequencing the Mouse Genome for the Oxidatively Modified Base 8-Oxo-7,8-dihydroguanine by OG-Seq. J. Am. Chem. Soc. 2017, 139, 2569–2572. [Google Scholar] [CrossRef]
- Pastukh, V.; Roberts, J.T.; Clark, D.W.; Bardwell, G.C.; Patel, M.; Al-Mehdi, A.B.; Borchert, G.M.; Gillespie, M.N. An oxidative DNA “damage” and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 309, L1367–L1375. [Google Scholar] [CrossRef]
- Sampath, H.; Lloyd, R.S. Roles of OGG1 in transcriptional regulation and maintenance of metabolic homeostasis. DNA Repair 2019, 81. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. Interplay of Guanine Oxidation and G-Quadruplex Folding in Gene Promoters. J. Am. Chem. Soc. 2020, 142, 1115–1136. [Google Scholar] [CrossRef]
- Fleming, A.M.; Ding, Y.; Burrows, C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. USA 2017, 114, 2601–2609. [Google Scholar] [CrossRef]
- Ba, X. Boldogh, lstvan 8-Oxoguanine DNA glycosylase 1: Beyond repair of the oxidatively modified base lesions. Redox Biol. 2018, 14, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Beaudouin, J.; Mora-Bermúdez, F.; Klee, T.; Daigle, N.; Ellenberg, J. Dissecting the Contribution of Diffusion and Interactions to the Mobility of Nuclear Proteins. Biophys. J. 2006, 90, 1878–1894. [Google Scholar] [CrossRef]
- Ou, H.D.; Phan, S.; Deerinck, T.J.; Thor, A.; Ellisman, M.H.; O’Shea, C.C. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 2017, 357, eaag0025. [Google Scholar] [CrossRef]
- Lebeaupin, T.; Smith, R.; Huet, S.; Timinszky, G. Poly(ADP-ribose)-dependent chromatin remodeling in DNA repair. Methods Mol. Biol. 2017, 1608, 165–183. [Google Scholar] [CrossRef]
- Joyner, R.P.; Tang, J.H.; Helenius, J.; Dultz, E.; Brune, C.; Holt, L.J.; Huet, S.; Müller, D.J.; Weis, K. A glucose-starvation response regulates the diffusion of macromolecules. Elife 2016, 5. [Google Scholar] [CrossRef]
- Bancaud, A.; Huet, S.; Daigle, N.; Mozziconacci, J.; Beaudouin, J.; Ellenberg, J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 2009, 28, 3785–3798. [Google Scholar] [CrossRef] [PubMed]
- Cravens, S.L.; Stivers, J.T. Comparative Effects of Ions, Molecular Crowding, and Bulk DNA on the Damage Search Mechanisms of hOGG1 and hUNG. Biochemistry 2016, 55, 5230–5242. [Google Scholar] [CrossRef]
- Dey, P.; Bhattacherjee, A. Mechanism of Facilitated Diffusion of DNA Repair Proteins in Crowded Environment: Case Study with Human Uracil DNA Glycosylase. J. Phys. Chem. B 2019, 123, 10354–10364. [Google Scholar] [CrossRef]
- Winter, R.B.; Berg, O.G.; Von Hippel, P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor-operator interaction: Kinetic measurements and conclusions. Biochemistry 1981, 20, 6961–6977. [Google Scholar] [CrossRef]
- Coppey, M.; Bénichou, O.; Voituriez, R.; Moreau, M. Kinetics of Target Site Localization of a Protein on DNA: A Stochastic Approach. Biophys. J. 2004, 87, 1640–1649. [Google Scholar] [CrossRef]
- Chupeau, M.; Bénichou, O.; Voituriez, R. Cover times of random searches. Nat. Phys. 2015, 11, 844–847. [Google Scholar] [CrossRef]
- Uphoff, S.; Reyes-Lamothe, R.; Garza de Leon, F.; Sherratt, D.J.; Kapanidis, A.N. Single-molecule DNA repair in live bacteria. Proc. Natl. Acad. Sci. USA 2013, 110, 8063–8068. [Google Scholar] [CrossRef]
- Kashida, H.; Kurihara, A.; Kawai, H.; Asanuma, H. Orientation-dependent FRET system reveals differences in structures and flexibilities of nicked and gapped DNA duplexes. Nucleic Acids Res. 2017, 45, e105. [Google Scholar] [CrossRef]
- Bénichou, O.; Chevalier, C.; Klafter, J.; Meyer, B.; Voituriez, R. Geometry-controlled kinetics. Nat. Chem. 2010, 2, 472–477. [Google Scholar] [CrossRef]
- Izeddin, I.; Récamier, V.; Bosanac, L.; Cissé, I.I.; Boudarene, L.; Dugast-Darzacq, C.; Proux, F.; Bénichou, O.; Voituriez, R.; Bensaude, O.; et al. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. Elife 2014, 3, e02230. [Google Scholar] [CrossRef] [PubMed]
- Keizer, V.I.P.; Coppola, S.; Houtsmuller, A.B.; Geverts, B.; van Royen, M.E.; Schmidt, T.; Schaaf, M.J.M. Repetitive switching between DNA-binding modes enables target finding by the glucocorticoid receptor. J. Cell Sci. 2019, 132, jcs217455. [Google Scholar] [CrossRef]
- Condamin, S.; Tejedor, V.; Voituriez, R.; Benichou, O.; Klafter, J. Probing microscopic origins of confined subdiffusion by first-passage observables. Proc. Natl. Acad. Sci. USA 2008, 105, 5675–5680. [Google Scholar] [CrossRef]
- Amitai, A. Chromatin Configuration Affects the Dynamics and Distribution of a Transiently Interacting Protein. Biophys. J. 2018, 114, 766–771. [Google Scholar] [CrossRef]
- Knight, S.C.; Xie, L.; Deng, W.; Guglielmi, B.; Witkowsky, L.B.; Bosanac, L.; Zhang, E.T.; El Beheiry, M.; Masson, J.-B.; Dahan, M.; et al. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science 2015, 350, 823–826. [Google Scholar] [CrossRef]
- von Hippel, P.H.; Berg, O.G. Facilitated target location in biological systems. J. Biol. Chem. 1989, 264, 675–678. [Google Scholar]
- Mirny, L.; Slutsky, M.; Wunderlich, Z.; Tafvizi, A.; Leith, J.; Kosmrlj, A. How a protein searches for its site on DNA: The mechanism of facilitated diffusion. J. Phys. A Math. Theor. 2009, 42, 434013. [Google Scholar] [CrossRef]
- Leven, I.; Levy, Y. Quantifying the two-state facilitated diffusion model of protein–DNA interactions. Nucleic Acids Res. 2019, 47, 5530–5538. [Google Scholar] [CrossRef] [PubMed]
- Esadze, A.; Stivers, J.T. Facilitated Diffusion Mechanisms in DNA Base Excision Repair and Transcriptional Activation. Chem. Rev. 2018, 118, 11298–11323. [Google Scholar] [CrossRef]
- Howard, M.J.; Wilson, S.H. DNA scanning by base excision repair enzymes and implications for pathway coordination. DNA Repair 2018, 71, 101–107. [Google Scholar] [CrossRef]
- Vestergaard, C.L.; Blainey, P.C.; Flyvbjerg, H. Single-particle trajectories reveal two-state diffusion-kinetics of hOGG1 proteins on DNA. Nucleic Acids Res. 2018, 46, 2446–2458. [Google Scholar] [CrossRef]
- Shvets, A.A.; Kolomeisky, A.B. Crowding on DNA in Protein Search for Targets. J. Phys. Chem. Lett. 2016, 7, 2502–2506. [Google Scholar] [CrossRef]
- Hammar, P.; Leroy, P.; Mahmutovic, A.; Marklund, E.G.; Berg, O.G.; Elf, J. The lac Repressor Displays Facilitated Diffusion in Living Cells. Science 2012, 336, 1595–1598. [Google Scholar] [CrossRef]
- Esadze, A.; Rodriguez, G.; Weiser, B.P.; Cole, P.A.; Stivers, J.T. Measurement of nanoscale DNA translocation by uracil DNA glycosylase in human cells. Nucleic Acids Res. 2017, 45, 12413–12424. [Google Scholar] [CrossRef] [PubMed]
- Stracy, M.; Jaciuk, M.; Uphoff, S.; Kapanidis, A.N.; Nowotny, M.; Sherratt, D.J.; Zawadzki, P. Single-molecule imaging of UvrA and UvrB recruitment to DNA lesions in living Escherichia coli. Nat. Commun. 2016, 7, 12568. [Google Scholar] [CrossRef]
- Kapanidis, A.N.; Uphoff, S.; Stracy, M. Understanding Protein Mobility in Bacteria by Tracking Single Molecules. J. Mol. Biol. 2018, 430, 4443–4455. [Google Scholar] [CrossRef]
- Woringer, M.; Darzacq, X. Protein motion in the nucleus: From anomalous diffusion to weak interactions. Biochem. Soc. Trans. 2018, 46, 945–956. [Google Scholar] [CrossRef]
- White, M.D.; Angiolini, J.F.; Alvarez, Y.D.; Kaur, G.; Zhao, Z.W.; Mocskos, E.; Bruno, L.; Bissiere, S.; Levi, V.; Plachta, N. Long-Lived Binding of Sox2 to DNA Predicts Cell Fate in the Four-Cell Mouse Embryo. Cell 2016, 165, 75–87. [Google Scholar] [CrossRef]
- Jones, D.L.; Leroy, P.; Unoson, C.; Fange, D.; Ćurić, V.; Lawson, M.J.; Elf, J. Kinetics of dCas9 target search in Escherichia coli. Science 2017, 357, 1420–1424. [Google Scholar] [CrossRef] [PubMed]
- Normanno, D.; Boudarène, L.; Dugast-Darzacq, C.; Chen, J.; Richter, C.; Proux, F.; Bénichou, O.; Voituriez, R.; Darzacq, X.; Dahan, M. Probing the target search of DNA-binding proteins in mammalian cells using TetR as model searcher. Nat. Commun. 2015, 6, 7357. [Google Scholar] [CrossRef]
- Hansen, A.S.; Amitai, A.; Cattoglio, C.; Tjian, R.; Darzacq, X. Guided nuclear exploration increases CTCF target search efficiency. Nat. Chem. Biol. 2020, 16, 257–266. [Google Scholar] [CrossRef]
- Doucleff, M.; Clore, G.M. Global jumping and domain-specific intersegment transfer between DNA cognate sites of the multidomain transcription factor Oct-1. Proc. Natl. Acad. Sci. USA 2008, 105, 13871–13876. [Google Scholar] [CrossRef]
- Hedglin, M.; Zhang, Y.; O’Brien, P.J. Isolating Contributions from Intersegmental Transfer to DNA Searching by Alkyladenine DNA Glycosylase. J. Biol. Chem. 2013, 288, 24550–24559. [Google Scholar] [CrossRef]
- Sheinman, M.; Kafri, Y. The effects of intersegmental transfers on target location by proteins. Phys. Biol. 2009, 6, 016003. [Google Scholar] [CrossRef][Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Augustin, O.; Huet, S.; Campalans, A.; Radicella, J.P. Lost in the Crowd: How Does Human 8-Oxoguanine DNA Glycosylase 1 (OGG1) Find 8-Oxoguanine in the Genome? Int. J. Mol. Sci. 2020, 21, 8360. https://doi.org/10.3390/ijms21218360
D’Augustin O, Huet S, Campalans A, Radicella JP. Lost in the Crowd: How Does Human 8-Oxoguanine DNA Glycosylase 1 (OGG1) Find 8-Oxoguanine in the Genome? International Journal of Molecular Sciences. 2020; 21(21):8360. https://doi.org/10.3390/ijms21218360
Chicago/Turabian StyleD’Augustin, Ostiane, Sébastien Huet, Anna Campalans, and Juan Pablo Radicella. 2020. "Lost in the Crowd: How Does Human 8-Oxoguanine DNA Glycosylase 1 (OGG1) Find 8-Oxoguanine in the Genome?" International Journal of Molecular Sciences 21, no. 21: 8360. https://doi.org/10.3390/ijms21218360
APA StyleD’Augustin, O., Huet, S., Campalans, A., & Radicella, J. P. (2020). Lost in the Crowd: How Does Human 8-Oxoguanine DNA Glycosylase 1 (OGG1) Find 8-Oxoguanine in the Genome? International Journal of Molecular Sciences, 21(21), 8360. https://doi.org/10.3390/ijms21218360




