A Genomic Approach to Investigating Ocular Surface Microorganisms: Monitoring Core Microbiota on Eyelid Margin with a Dot hybridization Assay
Abstract
1. Introduction
2. Results
2.1. A DHA Model for Postulated COSM and Pre-Test Analysis
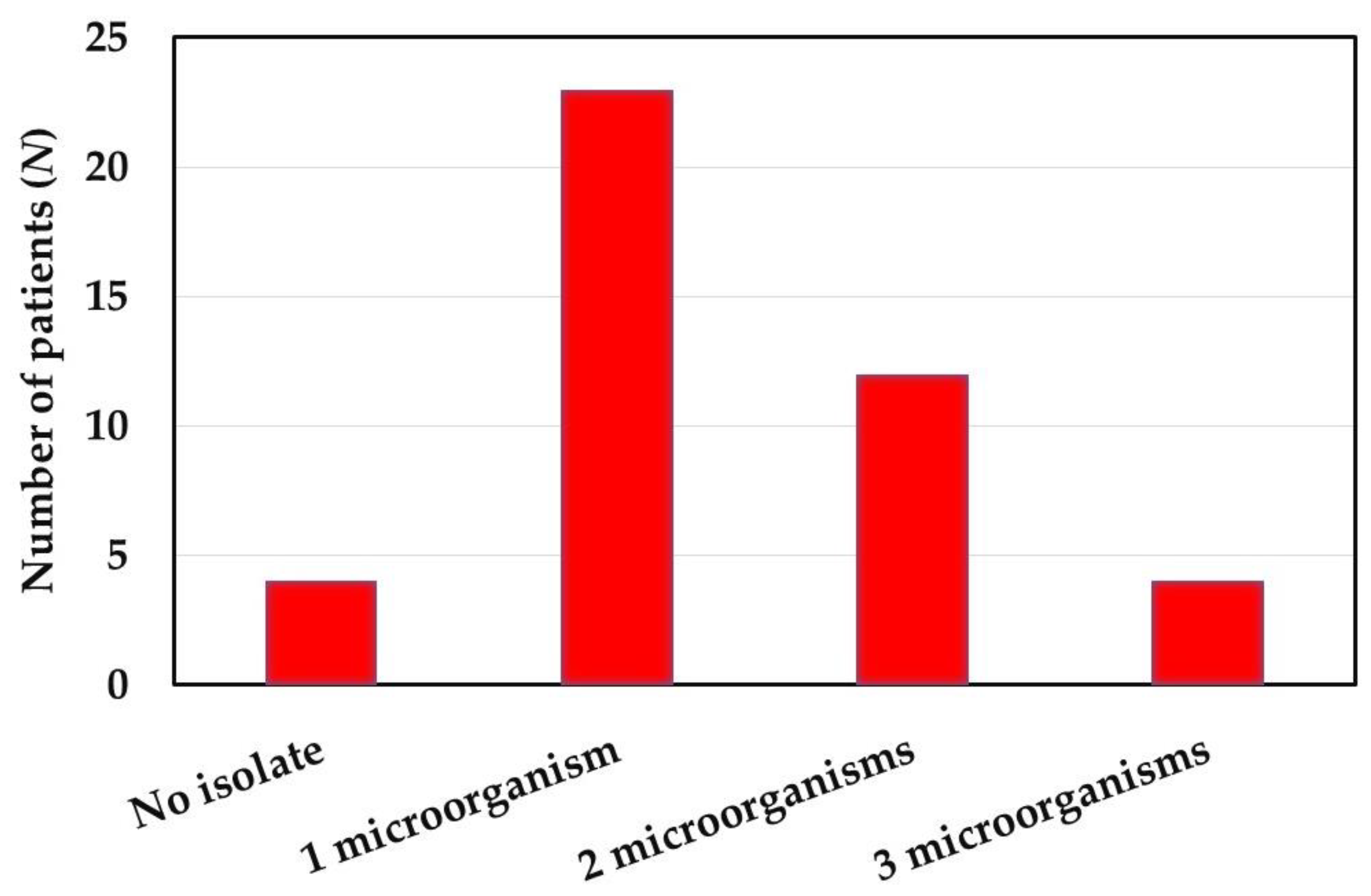
2.2. Clinical Subjects
2.3. Blood Culutre with Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MOLDI-TOF MS) for Determination of Ocular Surface Microbiota
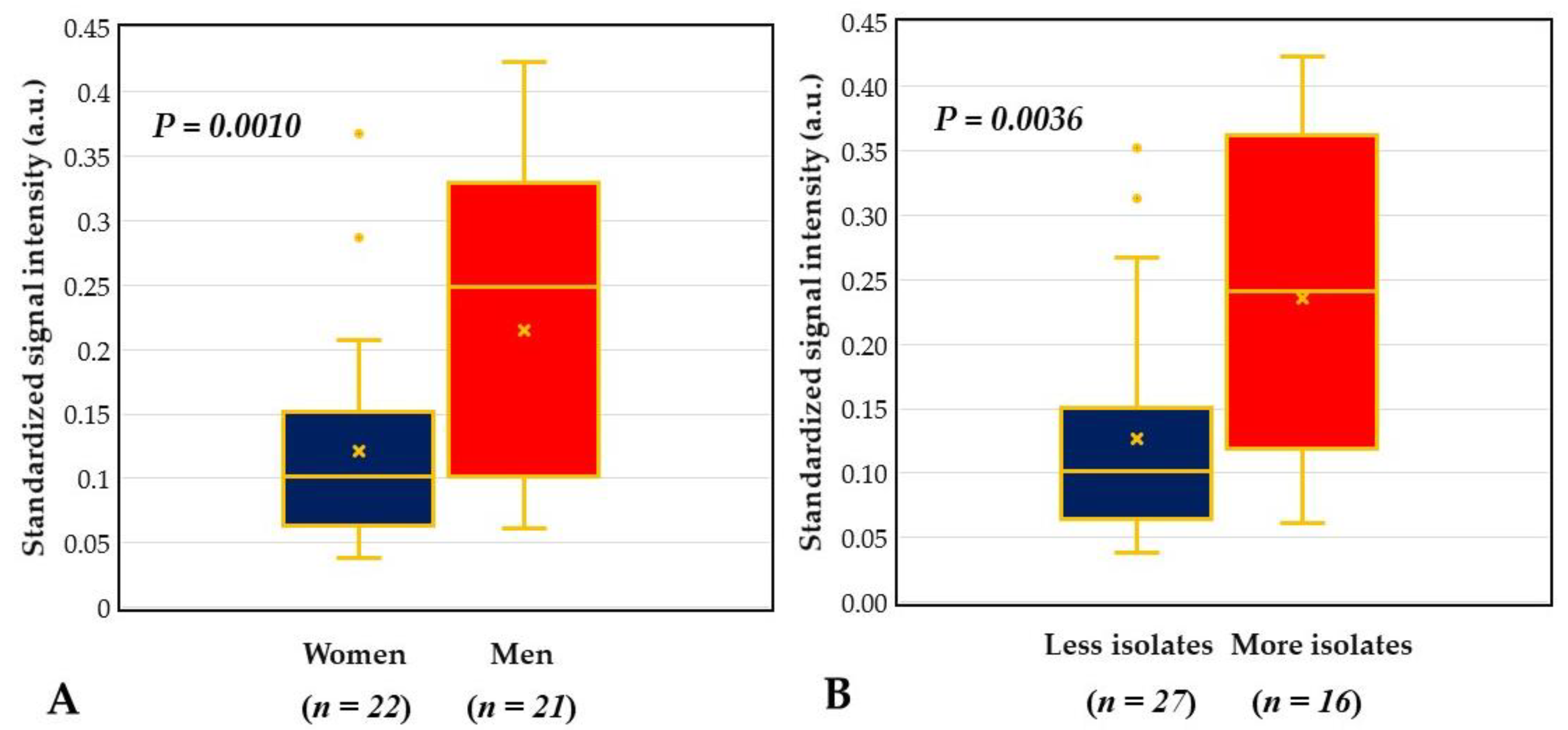
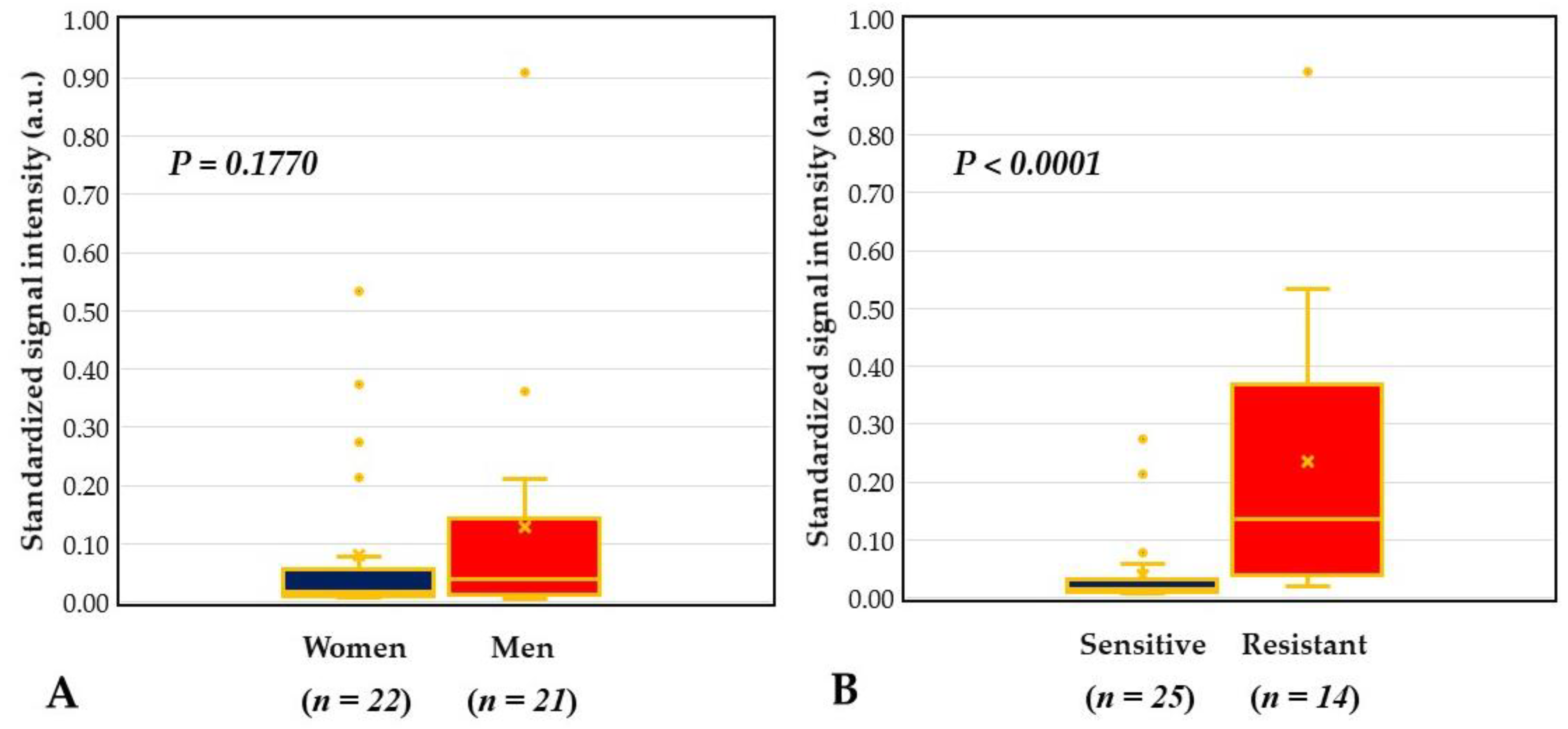
2.4. DHA Model for Evaluation of Core Ocular Surface Microbiota
2.5. Integrative Analysis for COSM from DHA and Blood Culture with MALDI-TOF MS
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Sample Collection
4.3. Identification of Cultured Microbiota by the MOLDI-TOF Mass Spectrometry
4.3.1. Sample Preparation for MALDI-TOF MS Analysis
4.3.2. Identification of Ocular Surface Microobiota by MALDI Biotyper
4.4. DNA Extraction and PCR Amplification
4.5. The Dot Hybridization Assay for Assessing COSM
4.5.1. Immobilization of Oligonucleotide Probes on a Nylon Membrane
4.5.2. Detection of Microbial DNA with the COSM DHA
4.5.3. Quantification of the Signals for Each Dot in the DHA
4.6. Data Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| a.u. | Arbitrary Unit |
| CGMH | Chang Gung Memorial Hospital |
| CoNS | Coagulase Negative Staphylococci |
| COSM | Core Ocular Surface Microbiota |
| DHA | Dot Hybridization Assay |
| MOLDI-TOF | Matrix-Assisted Laser Desorption Ionization-Time Of Flight |
| MS | Mass Spectrometry |
References
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Shore, S.A.; Cho, Y. Obesity and Asthma: Microbiome-Metabolome Interactions. Am. J. Respir. Cell Mol. Biol. 2016, 54, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R. Rheumatoid arthritis: Microbiome reflects status of RA and response to therapy. Nat. Rev. Rheumatol. 2015, 11, 502. [Google Scholar] [CrossRef]
- Devaraj, S.; Hemarajata, P.; Versalovic, J. The Human Gut Microbiome and Body Metabolism: Implications for Obesity and Diabetes. Clin. Chem. 2013, 59, 617–628. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef]
- Zegans, M.E.; Van Gelder, R.N. Considerations in Understanding the Ocular Surface Microbiome. Am. J. Ophthalmol. 2014, 158, 420–422. [Google Scholar] [CrossRef]
- Kugadas, A.; Gadjeva, M. Impact of Microbiome on Ocular Health. Ocul. Surf. 2016, 14, 342–349. [Google Scholar] [CrossRef]
- Petrillo, F.; Pignataro, D.; Lavano, M.A.; Santella, B.; Folliero, V.; Zannella, C.; Astarita, C.; Gagliano, C.; Franci, G.; Avitabile, T.; et al. Current Evidence on the Ocular Surface Microbiota and Related Diseases. Microorganism 2020, 8, 1033. [Google Scholar] [CrossRef]
- Zannella, C.; Shinde, S.; Vitiello, M.; Falanga, A.; Galdiero, E.; Fahmi, A.; Santella, B.; Nucci, L.; Gasparro, R.; Galdiero, M.; et al. Antibacterial Activity of Indolicidin-Coated Silver Nanoparticles in Oral Disease. Appl. Sci. 2020, 10, 1837. [Google Scholar] [CrossRef]
- Willcox, M.D.P. Characterization of the normal microbiota of the ocular surface. Exp. Eye Res. 2013, 117, 99–105. [Google Scholar] [CrossRef]
- Hsu, H.Y.; Lind, J.T.; Tseng, L.; Miller, D. Ocular Flora and Their Antibiotic Resistance Patterns in the Midwest: Prospective Study of Patients Undergoing Cataract Surgery. Am. J. Ophthalmol. 2013, 155, 36–44.e2. [Google Scholar] [CrossRef]
- Graham, J.E.; Moore, J.E.; Jiru, X.; Moore, J.E.; Goodall, E.A.; Dooley, J.S.G.; Hayes, V.E.A.; Dartt, D.A.; Downes, C.S.; Moore, T.C.B. Ocular Pathogen or Commensal: A PCR-Based Study of Surface Bacterial Flora in Normal and Dry Eyes. Investig. Opthalmol. Vis. Sci. 2007, 48, 5616–5623. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Brulc, J.M.; Iovieno, A.; Bates, B.; Garoutte, A.; Miller, D.; Revanna, K.V.; Gao, X.; Antonopoulos, D.A.; Slepak, V.Z.; et al. Diversity of Bacteria at Healthy Human Conjunctiva. Investig. Opthalmol. Vis. Sci. 2011, 52, 5408–5413. [Google Scholar] [CrossRef]
- Shin, H.; Price, K.; Albert, L.; Dodick, J.; Park, L.; Dominguez-Bello, M.G. Changes in the Eye Microbiota Associated with Contact Lens Wearing. mBio 2016, 7, e00198-16. [Google Scholar] [CrossRef]
- Kautz, S.; Rubin, B.E.R.; Russell, J.A.; Moreau, C.S. Surveying the Microbiome of Ants: Comparing 454 Pyrosequencing with Traditional Methods To Uncover Bacterial Diversity. Appl. Environ. Microbiol. 2012, 79, 525–534. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, B.; Li, W. Defining the normal core microbiome of conjunctival microbial communities. Clin. Microbiol. Infect. 2016, 22, 643.e7–643.e12. [Google Scholar] [CrossRef] [PubMed]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A review of methods and databases for metagenomic classification and assembly. Briefings Bioinform. 2019, 20, 1125–1136. [Google Scholar] [CrossRef]
- Hsiao, Y.-T.; Fang, P.-C.; Chen, J.-L.; Hsu, S.-L.; Chao, T.-L.; Yu, H.-J.; Lai, Y.-H.; Huang, Y.-T.; Kuo, M.-T. Molecular Bioburden of the Lens Storage Case for Contact Lens-Related Keratitis. Cornea 2018, 37, 1542–1550. [Google Scholar] [CrossRef]
- Fang, P.-C.; Chien, C.-C.; Yu, H.-J.; Ho, R.-W.; Tseng, S.-L.; Lai, Y.-H.; Kuo, M.-T. A dot hybridization assay for the diagnosis of bacterial keratitis. Mol. Vis. 2017, 23, 306–317. [Google Scholar]
- Kuo, M.-T.; Chien, C.-C.; Lo, J.; Hsiao, C.-C.; Tseng, S.-L.; Lai, Y.-H.; Fang, P.-C.; Chang, T.C. A DNA Dot Hybridization Model for Assessment of Bacterial Bioburden in Orthokeratology Lens Storage Cases. Investig. Ophthalmol. Vis. Sci. 2014, 56, 445–450. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fang, P.-C.; Lo, J.; Chang, T.C.; Chien, C.-C.; Hsiao, C.-C.; Tseng, S.-L.; Lai, Y.-H.; Kuo, M.-T. Bacterial Bioburden Decrease in Orthokeratology Lens Storage Cases After Forewarning. Eye Contact Lens 2017, 43, 174–180. [Google Scholar] [CrossRef]
- Han, H.W.; Chang, H.-C.; Chang, T.C. Identification of Staphylococcus spp. and detection of mecA by an oligonucleotide array. Diagn. Microbiol. Infect. Dis. 2016, 86, 23–29. [Google Scholar] [CrossRef]
- Ko, W.-C.; Lee, N.-Y.; Su, S.C.; Dijkshoorn, L.; Vaneechoutte, M.; Wang, L.-R.; Yan, J.-J.; Chang, T.C. Oligonucleotide Array-Based Identification of Species in the Acinetobacter calcoaceticus-A. baumannii Complex in Isolates from Blood Cultures and Antimicrobial Susceptibility Testing of the Isolates. J. Clin. Microbiol. 2008, 46, 2052–2059. [Google Scholar] [CrossRef]
- Lin, Y.T.; Vaneechoutte, M.; Huang, A.H.; Teng, L.J.; Chen, H.-M.; Su, S.-L.; Chang, T.C. Identification of Clinically Important Anaerobic Bacteria by an Oligonucleotide Array. J. Clin. Microbiol. 2010, 48, 1283–1290. [Google Scholar] [CrossRef]
- Nejima, R.; Shimizu, K.; Ono, T.; Noguchi, Y.; Yagi, A.; Iwasaki, T.; Shoji, N.; Miyata, K. Effect of the administration period of perioperative topical levofloxacin on normal conjunctival bacterial flora. J. Cataract Refract. Surg. 2017, 43, 42–48. [Google Scholar] [CrossRef]
- Venugopal, R.; Satpathy, G.; Sangwan, S.; Kapil, A.; Aron, N.; Agarwal, T.; Pushker, N.; Sharma, N. Conjunctival Microbial Flora in Ocular Stevens-Johnson Syndrome Sequelae Patients at a Tertiary Eye Care Center. Cornea 2016, 35, 1117–1121. [Google Scholar] [CrossRef]
- Zhang, S.D.; Na He, J.; Niu, T.T.; Chan, C.Y.; Ren, C.Y.; Liu, S.S.; Qu, Y.; Chong, K.L.; Wang, H.L.; Tao, J.; et al. Bacteriological profile of ocular surface flora in meibomian gland dysfunction. Ocul. Surf. 2017, 15, 242–247. [Google Scholar] [CrossRef]
- Doan, T.; Akileswaran, L.; Andersen, D.; Johnson, B.; Ko, N.; Shrestha, A.; Shestopalov, V.; Lee, C.S.; Lee, A.Y.; Van Gelder, R.N. Paucibacterial Microbiome and Resident DNA Virome of the Healthy Conjunctiva. Investig. Opthalmol. Vis. Sci. 2016, 57, 5116–5126. [Google Scholar] [CrossRef]
- Suzuki, T.; Sutani, T.; Nakai, H.; Shirahige, K.; Kinoshita, S. The Microbiome of the Meibum and Ocular Surface in Healthy Subjects. Investig. Opthalmol. Vis. Sci. 2020, 61, 18. [Google Scholar] [CrossRef]
- Mshangila, B.; Paddy, M.; Kajumbula, H.; Ateenyi-Agaba, C.; Kahwa, B.; Seni, J. External ocular surface bacterial isolates and their antimicrobial susceptibility patterns among pre-operative cataract patients at Mulago National Hospital in Kampala, Uganda. BMC Ophthalmol. 2013, 13, 71. [Google Scholar] [CrossRef]
- Ratnumnoi, R.; Keorochana, N.; Sonthisombat, C. Normal flora of conjunctiva and lid margin, as well as its antibiotic sensitivity, in patients undergoing cataract surgery at Phramongkutklao Hospital. Clin. Ophthalmol. 2017, 11, 237–241. [Google Scholar] [CrossRef]
- Mamada, H.T.N.; Suto, C.; Morinaga, M.; Yagi, T.; Tsuji, C. Conjunctival sac bacterial flora isolated prior to cataract surgery. Infect. Drug Resist. 2012, 5, 37–41. [Google Scholar] [CrossRef][Green Version]
- Matsuura, K.; Miyazaki, D.; Sasaki, S.-I.; Inoue, Y.; Sasaki, Y.; Shimizu, Y. Conjunctival bacterial flora and antimicrobial susceptibility in bacterial pathogens isolated prior to cataract surgery. Jpn. J. Ophthalmol. 2020, 64, 423–428. [Google Scholar] [CrossRef]
- Goering, R.V.; Swartzendruber, E.A.; Obradovich, A.E.; Tickler, I.A.; Tenover, F.C. Emergence of Oxacillin Resistance in Stealth Methicillin-Resistant Staphylococcus aureus Due to mecA Sequence Instability. Antimicrob. Agents Chemother. 2019, 63, e00558-19. [Google Scholar] [CrossRef]
- Lakhundi, S.; Zhang, K. Methicillin-ResistantStaphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhao, F.; Hutchinson, D.S.; Sun, W.; Ajami, N.J.; Lai, S.; Wong, M.C.; Petrosino, J.F.; Fang, J.; Jiang, J.; et al. Conjunctival Microbiome Changes Associated With Soft Contact Lens and Orthokeratology Lens Wearing. Investig. Opthalmol. Vis. Sci. 2017, 58, 128. [Google Scholar] [CrossRef]
- Wen, X.; Miao, L.; Deng, Y.; Bible, P.W.; Hu, X.; Zou, Y.; Liu, Y.; Guo, S.; Liang, J.; Chen, T.; et al. The Influence of Age and Sex on Ocular Surface Microbiota in Healthy Adults. Investig. Opthalmol. Vis. Sci. 2017, 58, 6030–6037. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.S.; Grzybowski, A.; Michalska-Małecka, K.; Szaflik, J.P.; Kozioł, M.; Niemczyk, W.; Grabska-Liberek, I. Incidence and Characteristics of Endophthalmitis after Cataract Surgery in Poland, during 2010–2015. Int. J. Environ. Res. Public Health 2019, 16, 2188. [Google Scholar] [CrossRef]
- Kim, S.H.; Yu, M.H.; Lee, J.H.; Kim, S.W.; Rah, S.H. Endophthalmitis after Cataract Surgery in Korea: A Nationwide Study Evaluating Incidence and Risk Factors in a Korean Population. Yonsei Med. J. 2019, 60, 467–473. [Google Scholar] [CrossRef]
- Tan, C.S.; Wong, H.K.; Yang, F.P. Epidemiology of postoperative endophthalmitis in an Asian population: 11-year incidence and effect of intracameral antibiotic agents. J. Cataract. Refract. Surg. 2012, 38, 425–430. [Google Scholar] [CrossRef]
- Zhao, F.; Zhang, D.; Ge, C.; Zhang, L.; Reinach, P.S.; Tian, X.; Tao, C.; Zhao, Z.; Zhao, C.; Fu, W.; et al. Metagenomic Profiling of Ocular Surface Microbiome Changes in Meibomian Gland Dysfunction. Investig. Opthalmol. Vis. Sci. 2020, 61, 22. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, J.; Nielsen, S.; Diez-Vives, C.; Coroneo, M.; Thomas, T.; Willcox, M. Temporal Stability and Composition of the Ocular Surface Microbiome. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Watters, G.A.; Turnbull, P.R.; Swift, S.; Petty, A.; Craig, J.P. Ocular surface microbiome in meibomian gland dysfunction. Clin. Exp. Ophthalmol. 2016, 45, 105–111. [Google Scholar] [CrossRef]
- Murakami, K.; Minamide, W.; Wada, K.; Nakamura, E.; Teraoka, H.; Watanabe, S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 1991, 29, 2240–2244. [Google Scholar] [CrossRef]
- Hsiao, C.R.; Huang, L.; Bouchara, J.-P.; Barton, R.; Li, H.C.; Chang, T.C. Identification of Medically Important Molds by an Oligonucleotide Array. J. Clin. Microbiol. 2005, 43, 3760–3768. [Google Scholar] [CrossRef]

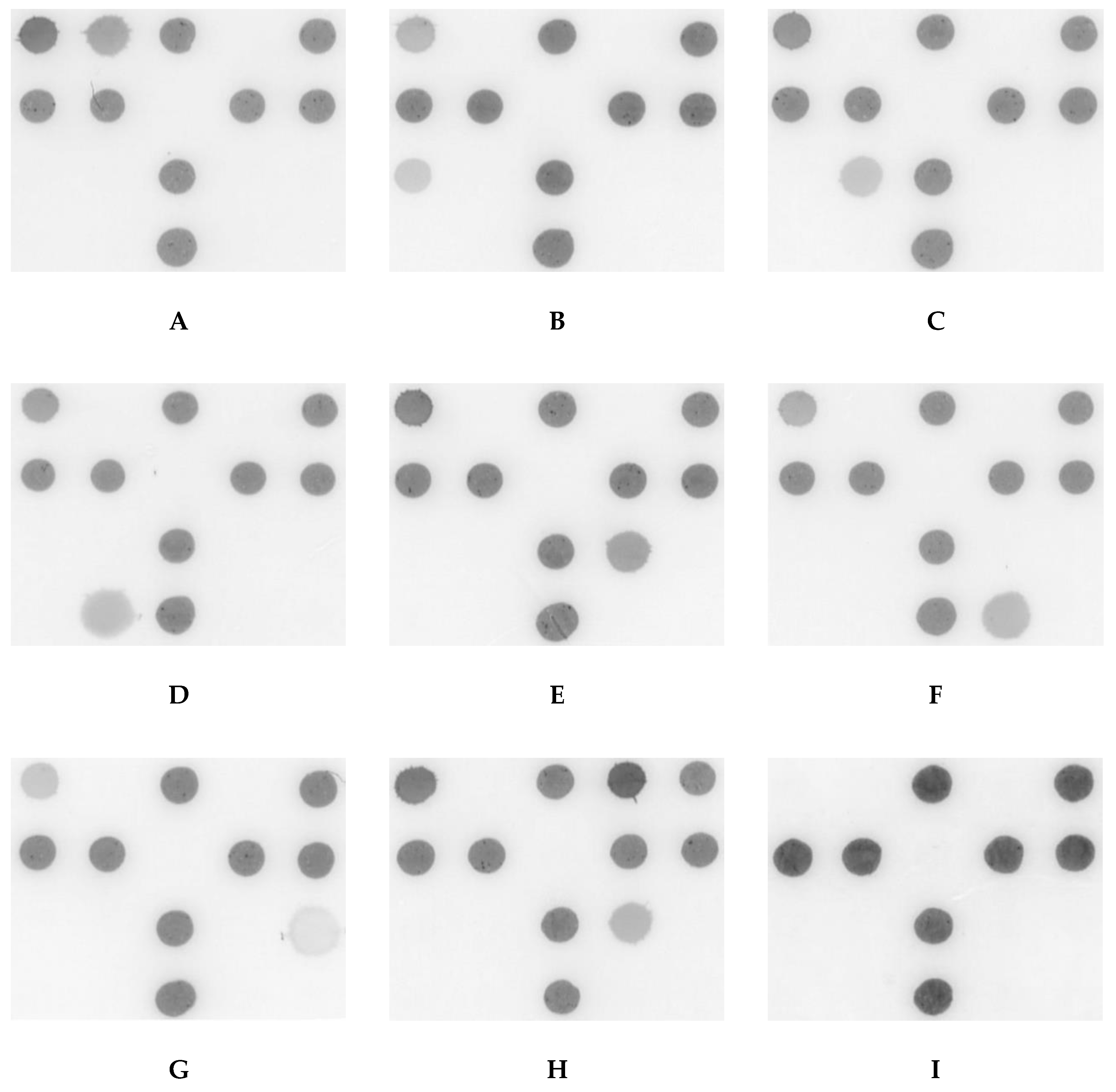



| Dot | Assessment Target | Probe Sequence(s) (5′–3′) | Conc. (μM) | Ref. |
|---|---|---|---|---|
| BPx | All bacteria | GGGGCTAAGTCGTAACAAGGTAGCCGTAtttttttttt b | 20 | [19,20] |
| Psu a | Pseudomonas aeruginosa | GTTCTTTAAAAATTTGGGTATGTGATAGAA CAGTGACCAGATTGCTTGGGGTTATATtttttttt b | 10 10 | [19,20] |
| Aci | Acinetobacterbaumannii | CGGTAATTAGTGTGATCTGACGAtttttttttt b | 10 | [23] |
| Klb | Klebsiella pneumoniae | CTTAAAGAACCTGCCTTTGTAGTGCTC | 20 | [19,20] |
| Sem a | Serratiamarcescens | AAGGTACTGCGCGTGACTGTATGGtttttttttt b,c; CATATAGTCCGGTATTTAATACTTCAGAGTtttttttttt b,c | 20 20 | - |
| Sta | Staphylococcus aureus | CGTTATTCCGCATCTTCTGAAGAAGAttttt | 20 | [22] |
| Ste | Staphylococcus epidermidis | TTGAATAACAATTCAAAATATGGTGGAttttttttttt b | 20 | [22] |
| Pro a | Propionibacterium acnes | TTGCTGTATGTGTTCGTGCGACtttttttttt b; GAGCATCTTATTTTTTGTGTGGCTTGTGttttttttttttttt b | 20 20 | [24] |
| mecA | Potentially resistant strain | TGATGGTATGCAACAAGTCG | 10 | [22] |
| M | Marker | 5′-digoxigenin-TCCTCCGCTTATTGATATGC | 10 | [19,20] |
| Isolated Microorganisms | Number of Isolates | Number of Oxacillin Resistant Strains |
|---|---|---|
| Gram Positive Microorganisms | ||
| Staphylococcus aureus | 5 | 1 |
| Staphylococcus epidermidis | 16 | 7 |
| Staphylococcus haemolyticus | 7 | 6 |
| Staphylococcus warneri | 2 | 0 |
| Staphylococcus sciuri | 1 | 1 |
| Unspecified CoNS | 3 | 0 |
| Streptococcus mitis | 1 | 0 |
| Streptococcus oralis | 2 | 0 |
| Streptococcus salivarius | 1 | 0 |
| Enterococcus faecalis | 3 | 0 |
| Bacillus cereus | 1 | 0 |
| Bacillus spp. | 8 | 0 |
| Paenibacillus spp. | 1 | 0 |
| Unspecified Gram-positive bacilli | 2 | 0 |
| Gram Negative Microorganisms | ||
| Microbacterium aurum | 1 | 0 |
| Gordonia spp. | 1 | 0 |
| Morganella spp. | 1 | 0 |
| Serratia marcescens | 1 | 0 |
| Citrobacter spp. | 1 | 0 |
| Brevundimonas spp. | 1 | 0 |
| Research | Sampling Sites | Culture System | No. (%) of CoNS | No. (%) of S. epidermidis | Ref. |
|---|---|---|---|---|---|
| Mshangila et al. (Uganda, n = 131) | Lower eyelid margin and inferior conjunctival sac | Brain–heart infusion broth | 91/138 (65.9%) | 70/138 (50.7%) | [31] |
| Ratnumnoi et al. (Thailand, n = 120) | Eyelid margin and conjunctiva | Blood agar and chocolate agar | 106/115 (92.2%) | N.A. | [32] |
| Suto et al. (Japan, n = 579) | Inferior conjunctiva sac | Blood agar and chocolate agar | 164/284 (57.7%) | 164/284 (57.7%) | [33] |
| In this study (Taiwan, n = 43) | Upper and lower lid margin | Pediatric blood bottle | 29/59 (49.2%) | 16/59 (27.1%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuo, M.-T.; Chao, T.-L.; Kuo, S.-F.; Chien, C.-C.; Chen, A.; Lai, Y.-H.; Huang, Y.-T. A Genomic Approach to Investigating Ocular Surface Microorganisms: Monitoring Core Microbiota on Eyelid Margin with a Dot hybridization Assay. Int. J. Mol. Sci. 2020, 21, 8299. https://doi.org/10.3390/ijms21218299
Kuo M-T, Chao T-L, Kuo S-F, Chien C-C, Chen A, Lai Y-H, Huang Y-T. A Genomic Approach to Investigating Ocular Surface Microorganisms: Monitoring Core Microbiota on Eyelid Margin with a Dot hybridization Assay. International Journal of Molecular Sciences. 2020; 21(21):8299. https://doi.org/10.3390/ijms21218299
Chicago/Turabian StyleKuo, Ming-Tse, Tsai-Ling Chao, Shu-Fang Kuo, Chun-Chih Chien, Alexander Chen, Yu-Hsuan Lai, and Yu-Ting Huang. 2020. "A Genomic Approach to Investigating Ocular Surface Microorganisms: Monitoring Core Microbiota on Eyelid Margin with a Dot hybridization Assay" International Journal of Molecular Sciences 21, no. 21: 8299. https://doi.org/10.3390/ijms21218299
APA StyleKuo, M.-T., Chao, T.-L., Kuo, S.-F., Chien, C.-C., Chen, A., Lai, Y.-H., & Huang, Y.-T. (2020). A Genomic Approach to Investigating Ocular Surface Microorganisms: Monitoring Core Microbiota on Eyelid Margin with a Dot hybridization Assay. International Journal of Molecular Sciences, 21(21), 8299. https://doi.org/10.3390/ijms21218299






