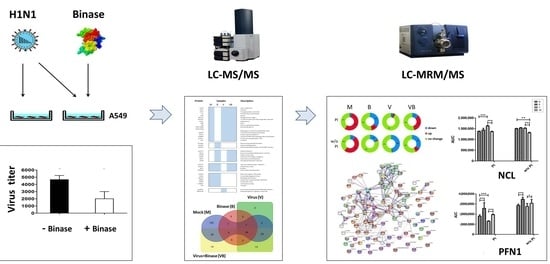
Anti-Influenza Activity of the Ribonuclease Binase: Cellular Targets Detected by Quantitative Proteomics
Abstract
:1. Introduction
2. Results
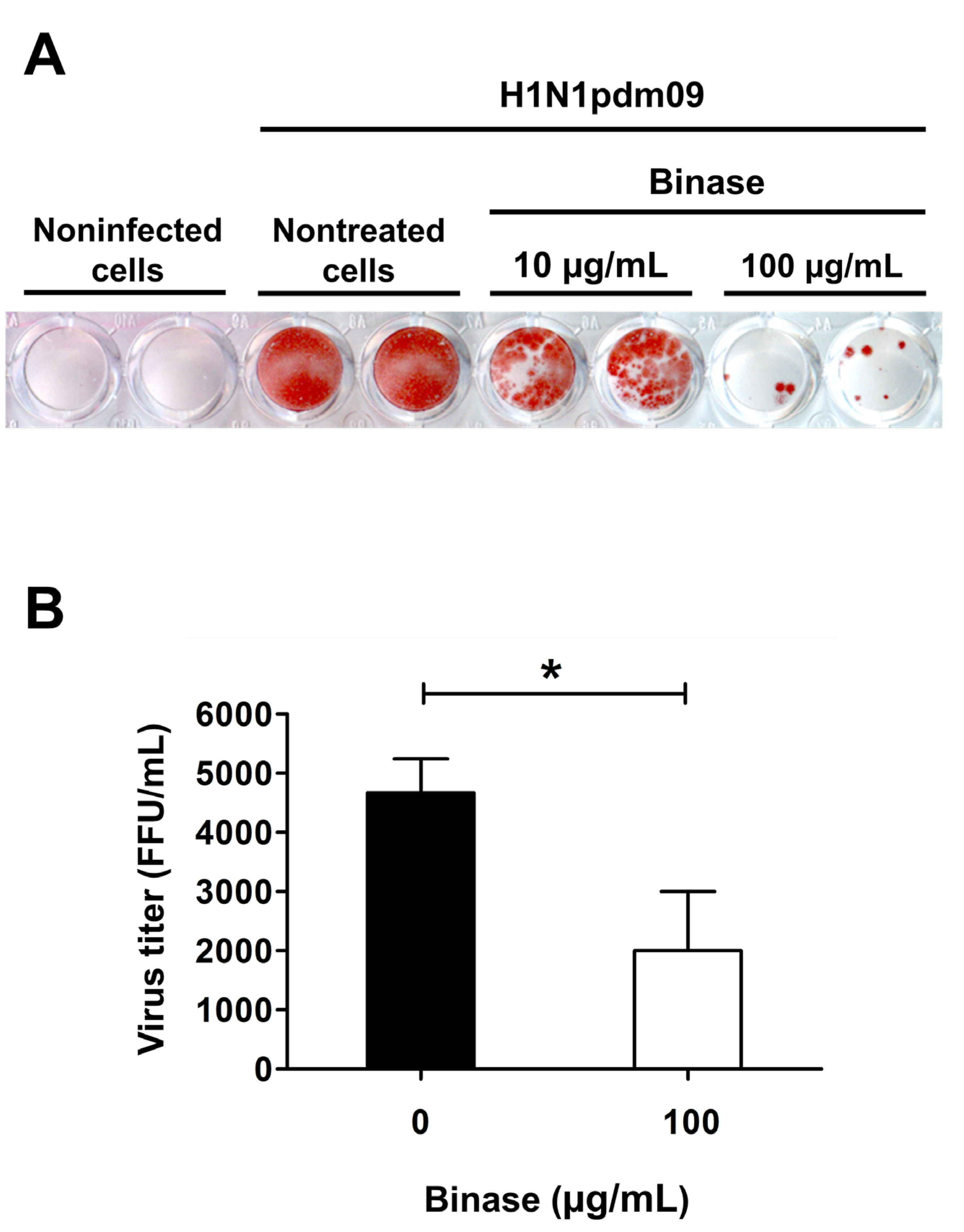
2.1. Confirmation of Viral Infection of A549 Cells and Antiviral Action of Binase
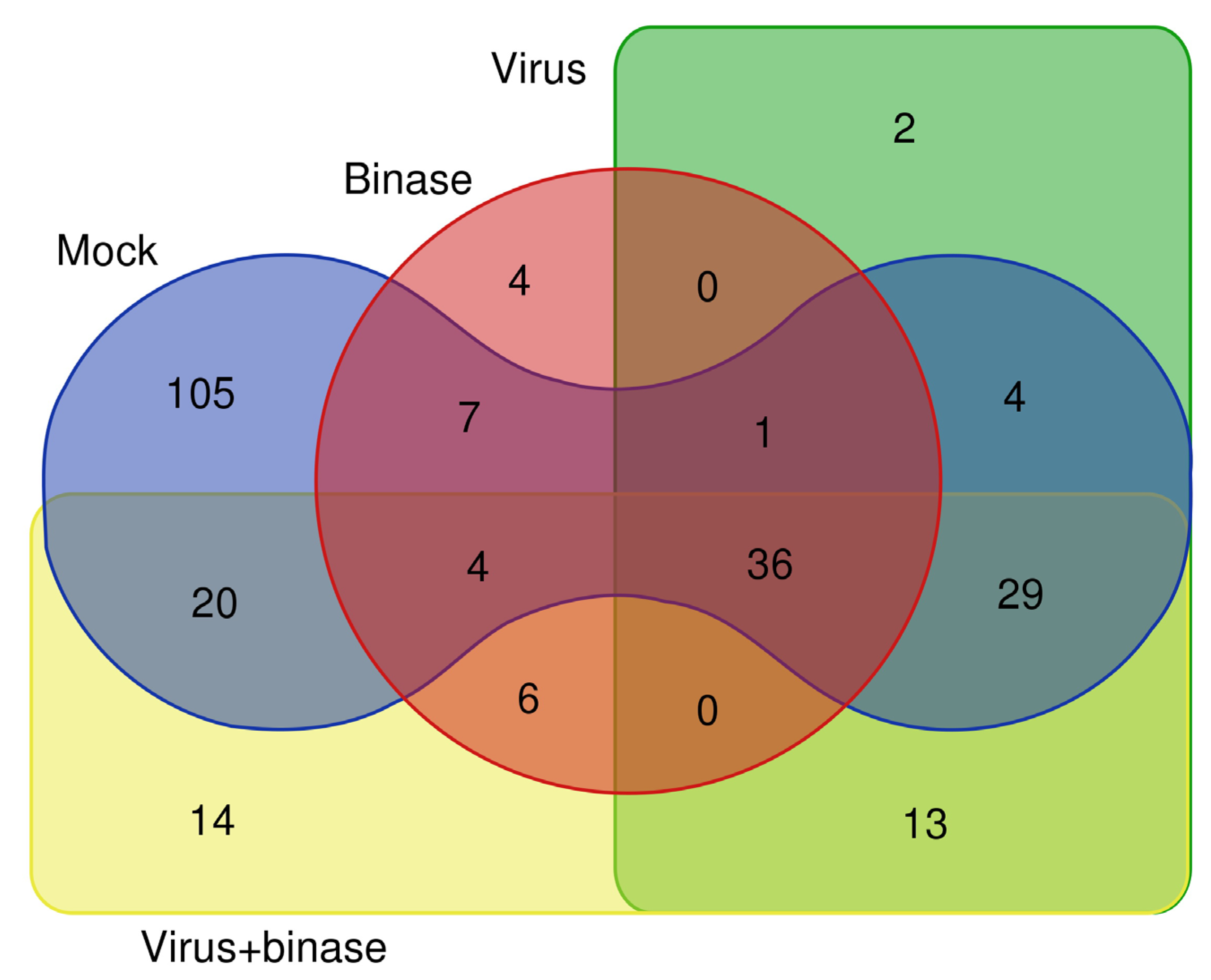
2.2. Qualitative Shotgun Proteomic Profiling of Virus-Infected and Binase-Treated Cells
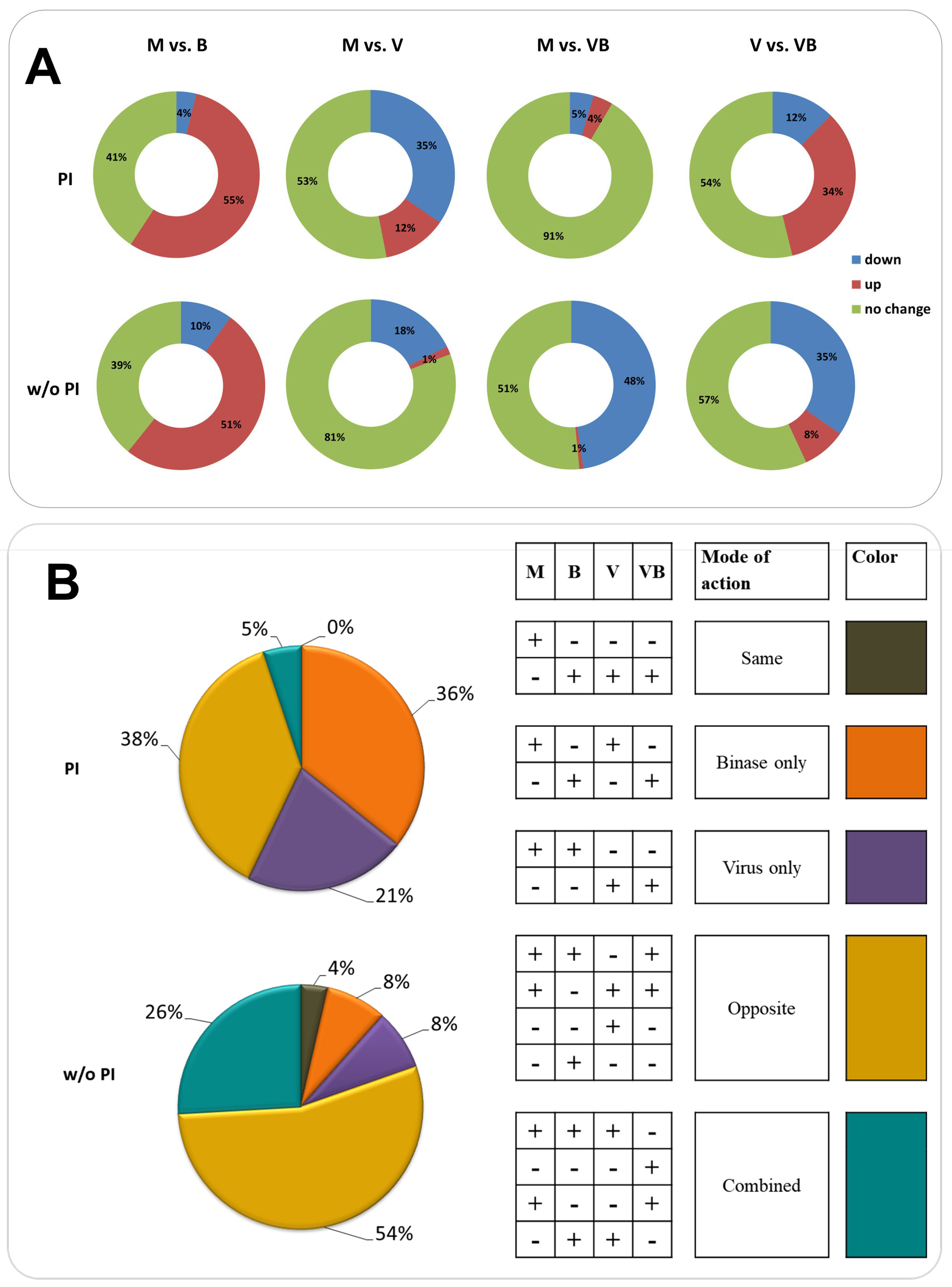
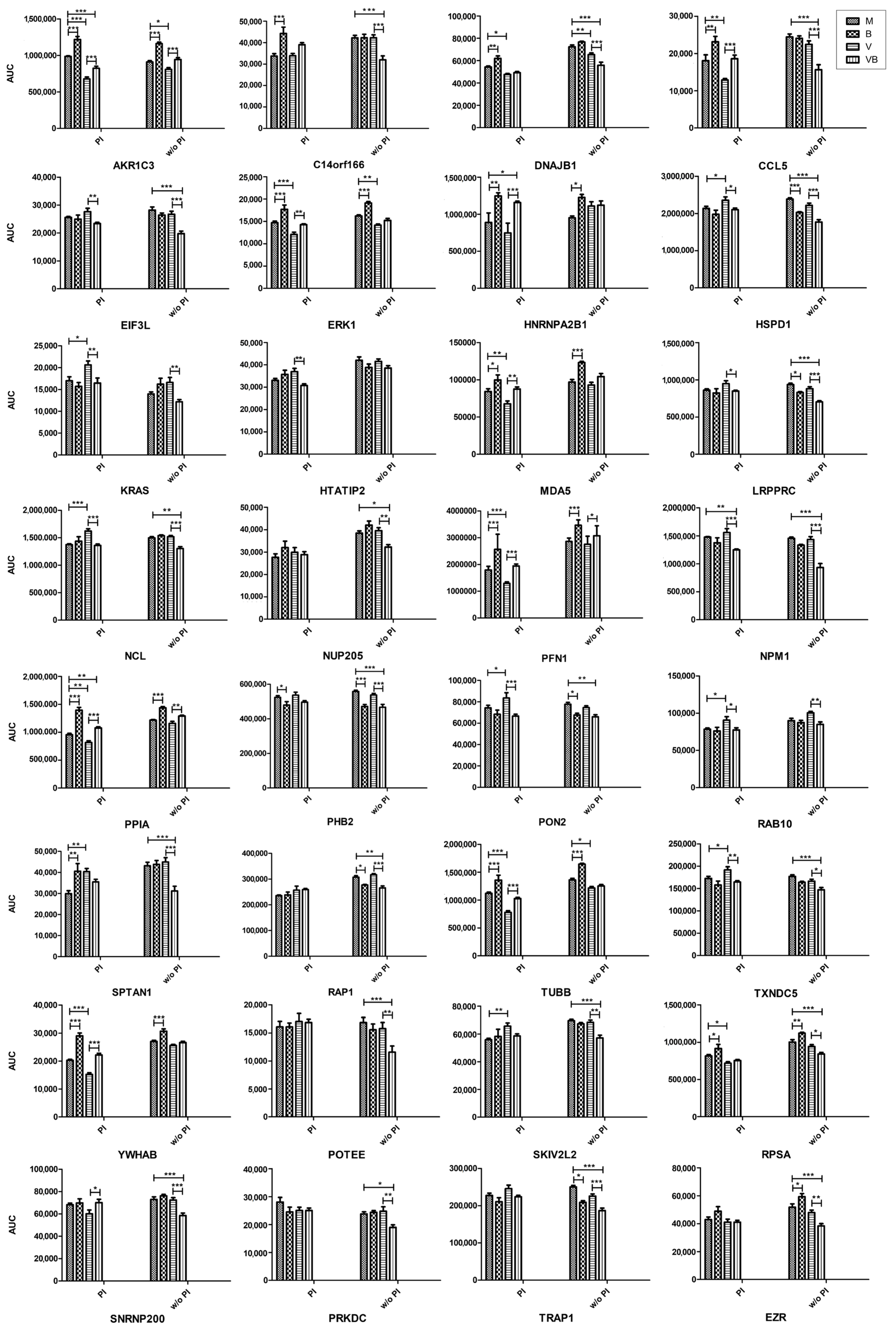
2.3. Quantitative Proteomic Profiling of Virus-Infected and Binase-Treated Cells
3. Discussion
4. Materials and Methods
4.1. Materials and Methods Cell Culture, Virus, and Binase
4.2. Determination of Virus Titers by Focus Forming Assay (FFA)
4.3. Cell Treatment and Lysis
4.4. Qualitative Shotgun Proteomics
4.4.1. “In-Solution” Protein Digestion
4.4.2. LC-MS/MS Analysis
4.4.3. Database Search
4.5. Quantitative Targeted Proteomics
4.5.1. “In-gel” Protein Digestion
4.5.2. “In-Silico” Protein Digestion
4.5.3. Targeted LC-MS/MS Analysis (LC-MRM/MS)
4.6. Protein Networks and Pathway Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mostafa, A.; Abdelwhab, E.M.; Mettenleiter, T.C.; Pleschka, S. Zoonotic Potential of Influenza A Viruses: A Comprehensive Overview. Viruses 2018, 10, 497. [Google Scholar] [CrossRef] [Green Version]
- Reperant, L.A.; Grenfell, B.T.; Osterhaus, A. Quantifying the Risk of Pandemic Influenza Virus Evolution by Mutation and Re-Assortment. Vaccine 2015, 33, 6955–6966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, H.; Mostafa, A.; Tantawy, M.A.; Iqbal, A.A.; Hoffmann, D.; Tallam, A.; Selvakumar, B.; Pessler, F.; Beer, M.; Rautenschlein, S.; et al. NS Segment of A 1918 Influenza A Virus-Descendent Enhances Replication of H1N1pdm09 and Virus-Induced Cellular Immune Response in Mammalian and Avian Systems. Front. Microbiol. 2018, 9, 526. [Google Scholar] [CrossRef]
- Stincarelli, M.; Arvia, R.; De Marco, M.A.; Clausi, V.; Corcioli, F.; Cotti, C.; Delogu, M.; Donatelli, I.; Azzi, A.; Giannecchini, S. Reassortment Ability of The 2009 Pandemic H1n1 Influenza Virus with Circulating Human and Avian Influenza Viruses: Public Health Risk Implications. Virus Res. 2013, 175, 151–154. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, A.; Abdelwhab, E.M.; Slanina, H.; Hussein, M.A.; Kuznetsova, I.; Schüttler, C.G.; Ziebuhr, J.; Pleschka, S. Phylogenetic Analysis of Human Influenza A/H3n2 Viruses Isolated in 2015 in Germany Indicates Significant Genetic Divergence from Vaccine Strains. Arch. Virol. 2016, 161, 1505–1515. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, A.; Pleschka, S. Influenza H3N2 Vaccines: Recent Challenges. Trends Microbiol. 2018, 26, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Nachbagauer, R.; Krammer, F. Universal Influenza Virus Vaccines and Therapeutic Antibodies. Clin. Microbiol. Infect. 2017, 23, 222–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Shesheny, R.; Bagato, O.; Kandeil, A.; Mostafa, A.; Mahmoud, S.H.; Hassanneen, H.M.; Webby, R.J.; Ali, M.A.; Kayali, G. Re-emergence of Amantadine-Resistant Variants Among Highly Pathogenic Avian Influenza H5N1 Viruses in Egypt. Infect. Genet. Evol. 2016, 46, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-S.; Jiao, X.-Y.; Wang, S.; Su, W.-Z.; Jiang, L.-Z.; Zhang, X.; Ke, C.; Xiong, P. Susceptibility of Influenza A(H1N1)/pdm2009, Seasonal A(H3N2) and B Viruses to Oseltamivir in Guangdong, China between 2009 and 2014. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Han, H.; Qiu, H.; Lin, H.; Yu, L.; Zhu, W.; Qi, J.; Yang, R.; Pang, Y.; Wang, X.-M.; et al. Antiviral Activity of A Synthesized Shikonin Ester against Influenza A (H1n1) Virus and Insights into Its Mechanism. Biomed. Pharmacother. 2017, 93, 636–645. [Google Scholar] [CrossRef]
- Vaidya, B.; Cho, S.-Y.; Oh, K.-S.; Kim, S.H.; O Kim, Y.; Jeong, E.-H.; Nguyen, T.T.; Kim, S.H.; Kim, I.S.; Kwon, J.; et al. Effectiveness of Periodic Treatment of Quercetin against Influenza A Virus H1N1 through Modulation of Protein Expression. J. Agric. Food Chem. 2016, 64, 4416–4425. [Google Scholar] [CrossRef] [PubMed]
- Timin, A.S.; Muslimov, A.R.; Petrova, A.V.; Lepik, K.V.; Okilova, M.V.; Vasin, A.V.; Afanasyev, B.V.; Sukhorukov, G.B. Hybrid Inorganic-Organic Capsules for Efficient Intracellular Delivery of Novel siRNAs against Influenza A (H1N1) Virus Infection. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Koszalka, P.; Tilmanis, D.; Hurt, A.C. Influenza Antivirals Currently in Late-Phase Clinical Trial. Influ. Other Respir. Viruses 2017, 11, 240–246. [Google Scholar] [CrossRef]
- Amarelle, L.; Lecuona, E.; Sznajder, J. Anti-Influenza Treatment: Drugs Currently Used and Under Development. Archivos de Bronconeumología 2017, 53, 19–26. [Google Scholar] [CrossRef]
- Scheuch, G.; Canisius, S.; Nocker, K.; Hofmann, T.; Naumann, R.; Pleschka, S.; Ludwig, S.; Welte, T.; Planz, O. Targeting Intracellular Signaling as An Antiviral Strategy: Aerosolized LASAG for the Treatment of Influenza in Hospitalized Patients. Emerg. Microbes. Infect. 2018, 7, 21. [Google Scholar] [CrossRef]
- Haasbach, E.; Müller, C.; Ehrhardt, C.; Schreiber, A.; Pleschka, S.; Ludwig, S.; Planz, O. The MEK-Inhibitor CI-1040 Displays A Broad Anti-Influenza Virus Activity in Vitro and Provides A Prolonged Treatment Window Compared to Standard of Care In Vivo. Antivir. Res. 2017, 142, 178–184. [Google Scholar] [CrossRef]
- Ardelt, W.; Ardelt, B.; Darzynkiewicz, Z. Ribonucleases as Potential Modalities in Anticancer Therapy. Eur. J. Pharmacol. 2009, 625, 181–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilinskaya, O.N.; Shah Mahmud, R. Ribonucleases as Antiviral Agents. Mol Biol. 2014, 48, 615–623. [Google Scholar] [CrossRef]
- Koczera, P.; Martin, L.; Marx, G.; Schuerholz, T. The Ribonuclease A Superfamily in Humans: Canonical RNases as the Buttress of Innate Immunity. Int. J. Mol. Sci. 2016, 17, 1278. [Google Scholar] [CrossRef]
- Aguado, L.C.; Schmid, S.; May, J.; Sabin, L.R.; Panis, M.; Blanco-Melo, D.; Shim, J.V.; Sachs, D.; Cherry, S.; Simon, A.E.; et al. RNase III Nucleases from Diverse Kingdoms Serve as Antiviral Effectors. Nat. Cell Biol. 2017, 547, 114–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drappier, M.; Michiels, T. Inhibition of the OAS/RNase L Pathway by Viruses. Curr. Opin. Virol. 2015, 15, 19–26. [Google Scholar] [CrossRef]
- Thomas, S.P.; Kim, E.; Kim, J.-S.; Raines, R.T. Knockout of the Ribonuclease Inhibitor Gene Leaves Human Cells Vulnerable to Secretory Ribonucleases. Biochemistry 2016, 55, 6359–6362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulyanova, V.; Vershinina, V.; Ilinskaya, O.N.; Harwood, C.R. Binase-Like Guanyl-Preferring Ribonucleases Are New Members of Bacillus PhoP Regulon. Microbiol. Res. 2015, 170, 131–138. [Google Scholar] [CrossRef]
- Ulyanova, V.; Vershinina, V.; Ilinskaya, O. Barnase and Binase: Twins with Distinct Fates. FEBS J. 2011, 278, 3633–3643. [Google Scholar] [CrossRef]
- Shah Mahmud, R.; Garifulina, K.I.; Ulyanova, V.V.; Evtugyn, V.G.; Mindubaeva, L.N.; Khazieva, L.R.; Dudkina, E.V.; Vershinina, V.I.; Kolpakov, A.I.; Ilinskaya, O.N. Bacteriophages of Soil Bacilli: A New Multivalent Phage of Bacillus altitudinis. Mol. Genet. Microbiol. Virol. 2017, 32, 87–93. [Google Scholar] [CrossRef]
- Alekseeva, I.I.; Kurinenko, B.M.; Kleiner, G.I.; Skuia, A.Z.; Penzikova, G.A. [Comparative Study of the Antiviral Activity of Pancreatic and Microbial RNAse]. Antibiot. 1981, 26, 527–532. [Google Scholar]
- Gribencha, S.V.; A Potselueva, L.; Barinskiĭ, I.F.; Balandin, T.G.; Deev, S.M.; Leshchinskaia, I.B. [The Antiviral Activity of RNAse Bacillus Intermedius in Experiments with Mice Preinfected with Street Rabies Virus]. Vopr. Virusol. 2004, 49, 38–41. [Google Scholar]
- Shah Mahmud, R.; Efimova, M.A.; Ulyanova, V.; Ravilov, R.K.; Shuralev, E.A.; Kolpakov, A.; Ilinskaya, O. Bacillus Pumilus Ribonuclease Rescues Mice Infected by Double-Stranded RNA-Containing Reovirus Serotype 1. Virus Res. 2020, 286, 198086. [Google Scholar] [CrossRef]
- Efimova, M.; Shah Mahmud, R.; Zelenikhin, P.V.; Sabirova, M.I.; Kolpakov, A.I.; Ilinskaya, O.N. Exogenous Bacillus Pumilus RNase (binase) Suppresses the Reproduction of Reovirus Serotype 1. Mol. Biol. 2017, 51, 96–101. [Google Scholar] [CrossRef]
- Shah Mahmud, R.; Efimova, M.; Mostafa, A.; Ulyanova, V.; Ilinskaya, O. Antiviral Activity of Bacterial Extracellular Ribonuclease against Single-, Double-Stranded RNA and DNA Containing Viruses in Cell Cultures. BioNanoScience 2016, 6, 561–563. [Google Scholar] [CrossRef]
- Müller, C.; Ulyanova, V.; Ilinskaya, O.; Pleschka, S.; Shah Mahmud, R. A Novel Antiviral Strategy against MERS-CoV and HCoV-229E Using Binase to Target Viral Genome Replication. BioNanoScience 2016, 7, 294–299. [Google Scholar] [CrossRef]
- Shah Mahmud, R.; Müller, C.; Romanova, Y.; Mostafa, A.; Ulyanova, V.; Pleschka, S.; Ilinskaya, O. Ribonuclease from Bacillus Acts as an Antiviral Agent against Negative- and Positive-Sense Single Stranded Human Respiratory RNA Viruses. BioMed Res. Int. 2017, 2017, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Shah Mahmud, R.; Ilinskaya, O.N. Antiviral Activity of Binase against the Pandemic Influenza A (H1N1) Virus. Acta Naturae 2013, 5, 44–51. [Google Scholar] [CrossRef]
- Shah Mahmud, R.; Mostafa, A.; Müller, C.; Kanrai, P.; Ulyanova, V.; Sokurenko, Y.; Dzieciolowski, J.; Kuznetsova, I.; Ilinskaya, O.; Pleschka, S. Bacterial Ribonuclease Binase Exerts An Intra-Cellular Anti-Viral Mode of Action Targeting Viral Rnas in Influenza A Virus-Infected MDCK-II cells. Virol. J. 2018, 15, 5. [Google Scholar] [CrossRef] [Green Version]
- Cabrera-Fuentes, H.A.; Aslam, M.; Saffarzadeh, M.; Kolpakov, A.; Zelenikhin, P.; Preissner, K.T.; Ilinskaya, O.N. Internalization of Bacillus Intermedius Ribonuclease (BINASE) Induces Human Alveolar Adenocarcinoma Cell Death. Toxicon 2013, 69, 219–226. [Google Scholar] [CrossRef]
- Yang, P.; Deng, J.; Li, C.; Zhang, P.; Xing, L.; Li, Z.; Wang, W.; Zhao, Y.; Yan, Y.; Gu, H.; et al. Characterization of the 2009 Pandemic A/Beijing/501/2009 H1N1 Influenza Strain in Human Airway Epithelial Cells and Ferrets. PLoS ONE 2012, 7, e46184. [Google Scholar] [CrossRef]
- Simon, P.F.; McCorrister, S.; Hu, P.; Chong, P.; Silaghi, A.; Westmacott, G.; Coombs, K.M.; Kobasa, D. Highly Pathogenic H5N1 and Novel H7N9 Influenza A Viruses Induce More Profound Proteomic Host Responses than Seasonal and Pandemic H1N1 Strains. J. Proteome Res. 2015, 14, 4511–4523. [Google Scholar] [CrossRef]
- Mindaye, S.T.; Ilyushina, N.A.; Fantoni, G.; Alterman, M.A.; Donnelly, R.P.; Eichelberger, M.C. Impact of Influenza A Virus Infection on the Proteomes of Human Bronchoepithelial Cells from Different Donors. J. Proteome Res. 2017, 16, 3287–3297. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Lu, J.; Yu, R.; Wang, X.; Wang, T.; Dong, F.; Peng, B.; Wu, W.; Liu, H.; Geng, Y.; et al. Preliminary Proteomic Analysis of A549 Cells Infected with Avian Influenza Virus H7N9 and Influenza A Virus H1N1. PLoS ONE 2016, 11, e0156017. [Google Scholar] [CrossRef]
- Li, Y.; Ming, F.; Huang, H.; Guo, K.; Chen, H.; Jin, M.; Zhou, H. Proteome Response of Chicken Embryo Fibroblast Cells to Recombinant H5N1 Avian Influenza Viruses with Different Neuraminidase Stalk Lengths. Sci. Rep. 2017, 7, 40698. [Google Scholar] [CrossRef]
- Sokurenko, Y.V.; Zelenikhin, P.V.; Ulyanova, V.; Kolpakov, A.I.; Muller, D.; Ilinskaya, O.N. Identification of 2′,3′-cGMP as An Intermediate of RNA Catalytic Cleavage by Binase and Evaluation of Its Biological Action. Russ. J. Bioorganic Chem. 2015, 41, 31–36. [Google Scholar] [CrossRef]
- Jackson, E.K.; Mi, Z.; Janesko-Feldman, K.; Jackson, T.C.; Kochanek, P.M. 2′,3′-cGMP Exists In Vivo and Comprises a 2′,3′-cGMP-Guanosine pathway. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2019, 316, R783–R790. [Google Scholar] [CrossRef]
- Mitkevich, V.A.; Tchurikov, N.A.; Zelenikhin, P.V.; Petrushanko, I.Y.; Makarov, A.A.; Ilinskaya, O. Binase Cleaves Cellular Noncoding RNAs and Affects Coding mRNAs. FEBS J. 2009, 277, 186–196. [Google Scholar] [CrossRef]
- Ilinskaya, O.N.; Singh, I.; Dudkina, E.; Ulyanova, V.; Kayumov, A.; Barreto, G. Direct inhibition of Oncogenic KRAS by Bacillus Pumilus Ribonuclease (binase). Biochim. Biophys. Acta (BBA)—Bioenerg. 2016, 1863, 1559–1567. [Google Scholar] [CrossRef]
- Wyżewski, Z.; Gregorczyk, K.P.; Szczepanowska, J.; Szulc-Dąbrowska, L. Functional Role of Hsp60 as a Positive Regulator of Human Viral Infection Progression. Acta Virol. 2018, 62, 33–40. [Google Scholar] [CrossRef]
- Li, J.; Chen, C.; Wong, G.; Dong, W.; Zheng, W.; Li, Y.; Sun, L.; Zhang, L.; Gao, G.F.; Bi, Y.; et al. Cyclophilin A Protects Mice against Infection by Influenza A Virus. Sci. Rep. 2016, 6, 28978. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Sun, L.; Yu, M.; Wang, Z.; Xu, C.; Xue, Q.; Zhang, K.; Ye, X.; Kitamura, Y.; Liu, W. Cyclophilin A Interacts with Influenza A Virus M1 Protein and Impairs the Early Stage of the Viral Replication. Cell. Microbiol. 2009, 11, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, A.; Guo, J.; Yang, J.; Zhou, H.; Chen, H.; Jin, M. Identification of Human Host Proteins Contributing to H5N1 Influenza Virus Propagation by Membrane Proteomics. J. Proteome Res. 2012, 11, 5396–5405. [Google Scholar] [CrossRef]
- Benitez, A.A.; Panis, M.; Xue, J.; Varble, A.; Shim, J.V.; Frick, A.L.; López, C.B.; Sachs, D.H.; Tenoever, B.R. In Vivo RNAi Screening Identifies MDA5 as a Significant Contributor to the Cellular Defense against Influenza A Virus. Cell Rep. 2015, 11, 1714–1726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herold, S.; Ludwig, S.; Pleschka, S.; Wolff, T. Apoptosis Signaling in Influenza Virus Propagation, Innate Host Defense, and Lung Injury. J. Leukoc. Biol. 2012, 92, 75–82. [Google Scholar] [CrossRef]
- Mühlbauer, D.; Dzieciolowski, J.; Hardt, M.; Hocke, A.; Schierhorn, K.L.; Mostafa, A.; Müller, C.; Wisskirchen, C.; Herold, S.; Wolff, T.; et al. Influenza Virus-Induced Caspase-Dependent Enlargement of Nuclear Pores Promotes Nuclear Export of Viral Ribonucleoprotein Complexes. J. Virol. 2015, 89, 6009. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, I.; Thompson, W.E.; Thomas, K. Prohibitins Role in Cellular Survival through Ras-Raf-MEK-ERK Pathway. J. Cell. Physiol. 2014, 229, 998–1004. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Li, B.; He, Q.-Y. Significance of Prohibitin Domain Family in Tumorigenesis and Its Implication in Cancer Diagnosis and Treatment. Cell Death Dis. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Pleschka, S.; Wolff, T.; Ehrhardt, C.; Hobom, G.; Planz, O.; Rapp, U.R.; Ludwig, S. Influenza Virus Propagation Is Impaired by Inhibition of the Raf/MEK/ERK signalling Cascade. Nat. Cell Biol. 2001, 3, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Terrier, O.; Carron, C.; De Chassey, B.; Dubois, J.; Traversier, A.; Julien, T.; Cartet, G.; Proust, A.; Hacot, S.; Ressnikoff, D.; et al. Nucleolin Interacts with Influenza A Nucleoprotein and Contributes to Viral Ribonucleoprotein Complexes Nuclear Trafficking and Efficient Influenza Viral Replication. Sci. Rep. 2016, 6, 29006. [Google Scholar] [CrossRef] [PubMed]
- Hovanessian, A.G.; Puvion-Dutilleul, F.; Nisole, S.; Svaba, J.; Perretb, E.; Dengd, J.-S.; Krusta, B. The Cell-Surface-Expressed Nucleolin Is Associated with the Actin Cytoskeleton. Exp. Cell Res. 2000, 261, 312–328. [Google Scholar] [CrossRef]
- Chan, C.-M.; Chu, H.; Zhang, A.J.; Leung, L.-H.; Sze, K.-H.; Kao, R.Y.-T.; Chik, K.K.-H.; To, K.K.-W.; Chan, J.F.-W.; Chen, H.; et al. Hemagglutinin of Influenza A Virus Binds Specifically to Cell Surface Nucleolin and Plays A Role in Virus Internalization. Virology 2016, 494, 78–88. [Google Scholar] [CrossRef]
- Bortz, E.; Westera, L.; Maamary, J.; Steel, J.; Albrecht, R.A.; Manicassamy, B.; Chase, G.; Martínez-Sobrido, L.; Schwemmle, M.; García-Sastre, A. Host- and Strain-Specific Regulation of Influenza Virus Polymerase Activity by Interacting Cellular Proteins. mBio 2011, 2, 00151–00211. [Google Scholar] [CrossRef] [Green Version]
- Mayer, D.; Molawi, K.; Martínez-Sobrido, L.; Ghanem, A.; Thomas, S.; Baginsky, S.; Grossmann, J.; García-Sastre, A.; Schwemmle, M. Identification of Cellular Interaction Partners of the Influenza Virus Ribonucleoprotein Complex and Polymerase Complex Using Proteomic-Based Approaches. J. Proteome Res. 2007, 6, 672–682. [Google Scholar] [CrossRef] [Green Version]
- Westera, L.; Jennings, A.M.; Maamary, J.; Schwemmle, M.; García-Sastre, A.; Bortz, E. Poly-ADP Ribosyl Polymerase 1 (PARP1) Regulates Influenza A Virus Polymerase. Adv. Virol. 2019, 2019, 8512363-11. [Google Scholar] [CrossRef]
- Ranadheera, C.; Coombs, K.M.; Kobasa, D. Comprehending a Killer: The Akt/mTOR Signaling Pathways Are Temporally High-Jacked by the Highly Pathogenic 1918 Influenza Virus. EBioMedicine 2018, 32, 142–163. [Google Scholar] [CrossRef] [PubMed]
- McLean, J.E.; Datan, E.; Matassov, D.; Zakeri, Z. Lack of Bax Prevents Influenza A Virus-Induced Apoptosis and Causes Diminished Viral Replication. J. Virol. 2009, 83, 8233–8246. [Google Scholar] [CrossRef] [Green Version]
- Yeganeh, B.; Ghavami, S.; Rahim, N.; Klonisch, T.; Halayko, A.; Coombs, K. Autophagy Activation Is Required for Influenza A Virus-Induced Apoptosis and Replication. Biochim. Biophys. Acta (BBA)—Bioenerg. 2018, 1865, 364–378. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, J.; Du, Y. hnRNP A2/B1 Interacts with Influenza A Viral Protein Ns1 and Inhibits Virus Replication Potentially Through Suppressing NS1 RNA/Protein Levels and NS1 mRNA Nuclear Export. Virology 2014, 449, 53–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedi, S.; Ono, A. Friend or Foe: The Role of the Cytoskeleton in Influenza A Virus Assembly. Viruses 2019, 11, 46. [Google Scholar] [CrossRef] [Green Version]
- Kumakura, M.; Kawaguchi, A.; Nagata, K. Actin-Myosin Network Is Required for Proper Assembly of Influenza Virus Particles. Virology 2015, 476, 141–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Digard, P.; Elton, D.; Bishop, K.; Medcalf, E.; Weeds, A.; Pope, B. Modulation of Nuclear Localization of the Influenza Virus Nucleoprotein through Interaction with Actin Filaments. J. Virol. 1999, 73, 2222–2231. [Google Scholar] [CrossRef] [Green Version]
- Mehrbod, P.; Hair-Bejo, M.; Ibrahim, T.A.T.; Omar, A.R.; El Zowalaty, M.E.; Ajdari, Z.; Ideris, A. Simvastatin Modulates Cellular Components in Influenza A Virus-Infected Cells. Int. J. Mol. Med. 2014, 34, 61–73. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Kim, J.H.; Ranjan, P.; Metcalfe, M.G.; Cao, W.; Mishina, M.; Gangappa, S.; Guo, Z.; Boyden, E.S.; Zaki, S.; et al. Influenza Virus Exploits Tunneling Nanotubes for Cell-to-Cell Spread. Sci. Rep. 2017, 7, 40360. [Google Scholar] [CrossRef]
- Dudkina, E.; Ulyanova, V.; Shah Mahmud, R.; Khodzhaeva, V.; Dao, L.; Vershinina, V.; Kolpakov, A.; Ilinskaya, O. Three-Step Procedure for Preparation of Pure Bacillus Altitudinis Ribonuclease. FEBS Open Bio 2016, 6, 24–32. [Google Scholar] [CrossRef]
- Erde, J.; Loo, R.R.O.; Loo, J.A. Enhanced FASP (eFASP) to Increase Proteome Coverage and Sample Recovery for Quantitative Proteomic Experiments. J. Proteome Res. 2014, 13, 1885–1895. [Google Scholar] [CrossRef]
- Okuda, S.; Watanabe, Y.; Moriya, Y.; Kawano, S.; Yamamoto, T.; Matsumoto, M.; Takami, T.; Kobayashi, D.; Araki, N.; Yoshizawa, A.C.; et al. jPOSTrepo: An International Standard Data Repository for Proteomes. Nucleic Acids Res. 2017, 45, D1107–D1111. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2018, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Binase or IAV Effect | GO Term | Biological Process or Cellular Component | Proteins |
|---|---|---|---|
| Downregulated by IAV | |||
| GO:0051186 | Cofactor metabolic process | PKM, ENO1, PGK1, LDHA, GPI, PGAM, G6PD, TKT, AKR1B1, AKR1B10, AKR1C3, AKR1C1, HSP90AA1 | |
| GO:0044281 | Small-molecule metabolic process | SORL1, ALDH1A1, ACSS3, RAN, HSPA8, TARS, MAPK1, MAPK3 | |
| GO:0006955 | Immune response | CCL5, MDA5, PPIA, HSPA8, HSP90AA1, EEF1A1, MAPK1, MAPK3, TUBB4B, TUBB, PKM, ACTB, GPI, PGAM1, G6PD | |
| GO:0007010 | cytoskeleton organization | TUBB3, TUBA4A, TUBB4B, ACTB, PFN1 | |
| GO:0006810, GO:0051179 | transport and localization | SORL1, RDX, RAN, YWHAB, TOMM5 | |
| Upregulated by IAV | |||
| GO:0045321 | leucocyte activation | SPTAN1, DDX3X, HSPD1, RAB10, ANXA2, TXNDC5 | |
| GO:0065003 | protein-containing complex assembly | ATL3, EIF3A, NDUFS8, ANXA2, HSPD1, DDX3X | |
| Downregulated by binase | |||
| GO:0016020 | Membrane | LRPPRC, TRAP1, HSPD1, PHB2, SURF4, NDUFS8, KRT10, PON2, MYOF, RAP1A | |
| Upregulated by binase | |||
| GO:0010468 | regulation of gene expression | PFN1, DNAJB1, EEF1A1, PPP2CA, PRKDC, XRCC6, NCL, RDX, MSN, ACTB, MAPK1, MAPK2, YWHAB, HSPA8, RAN, RPS2, RPSA, RPL15, EIF3K, EIF2A, EIF3E, EIF3H, HNRNPA2B1, CAND1, CCL5, TRAP1, C14ORF166, ENO1 | |
| GO:0006887 | exocytosis | CAND1, CCL5, PGAM1, GPI, PKM, ANXA2, PYGB, CAP1, TUBB, TUBB4B, TUBB4A, TXNDC5, PPI, MAPK1, RAP1A, HSP90AA1, HSPA8, ANXA5, EEF1A1, SPTAN, PSMD13, XRCC6 | |
| GO:0006897 | endocytosis | MAPK3, MAPK1, HSP90AA1, CLTC, ACTB, CAP1, SORL1, TXNDC5 | |
| GO:0051641 | cellular localization | RAN, IPO4, HNRNPA2B1, HNRNPA1L2, KIF5B, C14orf166, RDX, MSN, SRP68, RPSA, RPS2, RPL25, BAX, YWHAB, TUBB4A, TUBB, TUBB4B, ACTB, CLTC, ANXA2, ANXA5, SORL1, AKR1C1, AKR1C3, TXNDC5, PPIA | |
| GO:0042786 | immune process | MDA5, CCL5, PGAM1, PKM, HSPA8, PSMD3, CAND1, KIF5B, XRCC6, RAP1A, HSP90AA1, EEE1A1, MAPK1, PPIA, GPI, PYGB, CAP1, ANXA2, SPTAN1, BAX, TUBB4B, TXNDC5, TUBB, MSN | |
| GO:0055114, GO:0008152 | oxidation-reduction and metabolic processes | AKR1C3, AKR1C1, AKR1B10, ALDH16A1, ALDH18A1, ALDH1A1, ALDH3A1, FASN, PYGB, GPI, G6PD, LDHA, ENO1, PGAM1, PKM, PGK1 | |
| Upregulated by binase in IAV-infected cells | |||
| GO:0051186, GO:1901135, GO:0001523, GO:0006629, GO:0050896 | cofactor metabolic process, carbohydrate derivative metabolic process, retinoid metabolic process, lipid metabolic process, response to stimulus | AKR1B1, AKR1B10, AKR1C1, AKR1C3, ANXA5, ENO1, PFN1, PGAM1, PGK1, PPIA | |
| GO:0022406 | membrane docking | TUBB, TUBB4A, TUBB4B, HSP90AA1 | |
| GO:0006955 | immune response | MDA5, CCL5, CAP1, KIF5B, MAPK3, TUBB, TUBB4A, TUBB4B, HSP90AA1 | |
| Downregulated by binase in IAV-infected cells | |||
| GO:0016032 | viral process | NPM1, HSPD1, EIF3L, EIF3A | |
| GO:0007029 | endoplasmic reticulum organization | RAB10, ATL3 | |
| GO:0043066 | negative regulation of apoptosis | TXNDC5, KRAS, NPM1, HSPD1, and HTATIP2 | |
| GO:0061028 | establishment of endothelial barrier | RDX, EZR, RAP1A, FASN | |
| GO:0006413 | translational initiation | RPL15, RPSA, EIF3H, EIF3E, RPS2 | |
| GO:0032272 | negative regulation of protein polymerization | MAPRE1, SRTAN1, RDX | |
| GO:0034470 | ncRNA processing | SSB, C14orf166, RPSA, RPS2, SKIV2L2 | |
| GO:0045321, GO:0042119 | leukocyte and neutrophil activation | BAX, CCL5, SPTAN1, RAP1A, CAP1, SURF4, PRKDC | |
| GO:0044056 | symbiont process | CCL5, NUP205, BAX, C14orf166, RPSA, SSB | |
| GO:0006913 | nucleocytoplasmic transport | PHB2, RPSA, NUP205, SSB | |
| GO:0070372 | regulation of ERK1 and ERK2 cascade | PHB2, RAP1A, CCL5, EZR | |
| GO:0010467, GO:0006349 | gene expression and its regulation | PRKDC, TRAP1, DNAJB1, YLPM1, C14orf166, SSB, RDX, EZR, CCL5, RPSA, RPL15, RPS2, EIF3H, EIF3E, PHB2, SNRNP200, SKIV2L2, BAX, NUP205 | |
| GO:0051234 | establishment of localization | CLINT1, CCL5, NUP205, BAX, MAPRE1, C14orf166, RPL15, RPSA, RPS2, SSB, RAP1A, SPTAN1, EZR, RDX, CAP1, SURF4, PHB2 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulyanova, V.; Shah Mahmud, R.; Laikov, A.; Dudkina, E.; Markelova, M.; Mostafa, A.; Pleschka, S.; Ilinskaya, O. Anti-Influenza Activity of the Ribonuclease Binase: Cellular Targets Detected by Quantitative Proteomics. Int. J. Mol. Sci. 2020, 21, 8294. https://doi.org/10.3390/ijms21218294
Ulyanova V, Shah Mahmud R, Laikov A, Dudkina E, Markelova M, Mostafa A, Pleschka S, Ilinskaya O. Anti-Influenza Activity of the Ribonuclease Binase: Cellular Targets Detected by Quantitative Proteomics. International Journal of Molecular Sciences. 2020; 21(21):8294. https://doi.org/10.3390/ijms21218294
Chicago/Turabian StyleUlyanova, Vera, Raihan Shah Mahmud, Alexander Laikov, Elena Dudkina, Maria Markelova, Ahmed Mostafa, Stephan Pleschka, and Olga Ilinskaya. 2020. "Anti-Influenza Activity of the Ribonuclease Binase: Cellular Targets Detected by Quantitative Proteomics" International Journal of Molecular Sciences 21, no. 21: 8294. https://doi.org/10.3390/ijms21218294
APA StyleUlyanova, V., Shah Mahmud, R., Laikov, A., Dudkina, E., Markelova, M., Mostafa, A., Pleschka, S., & Ilinskaya, O. (2020). Anti-Influenza Activity of the Ribonuclease Binase: Cellular Targets Detected by Quantitative Proteomics. International Journal of Molecular Sciences, 21(21), 8294. https://doi.org/10.3390/ijms21218294