Cellular Reprogramming—A Model for Melanoma Cellular Plasticity
Abstract
:1. Introduction
2. Cellular Reprogramming and iPS Cells
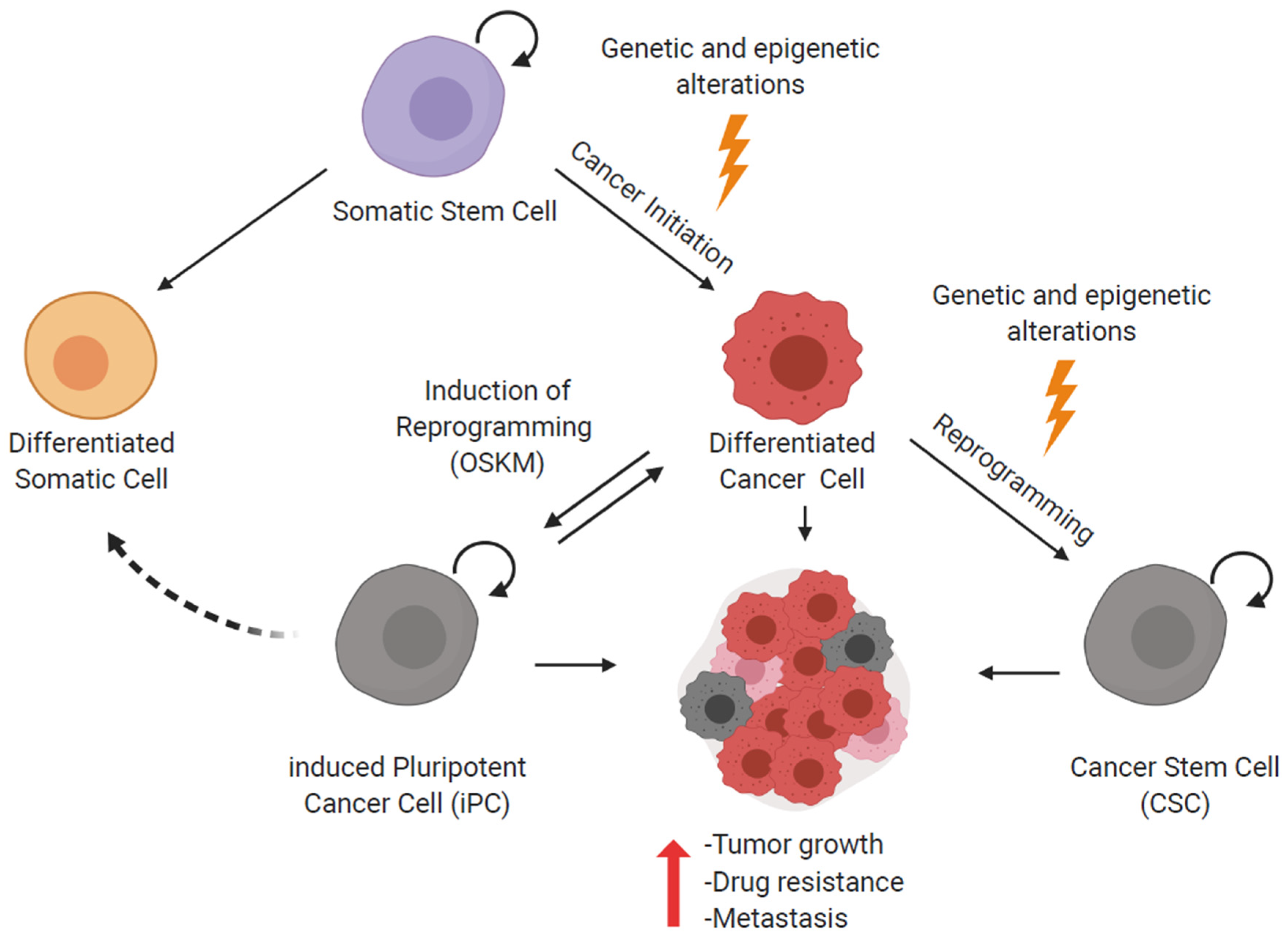
3. Reprogramming of Cancer Cells
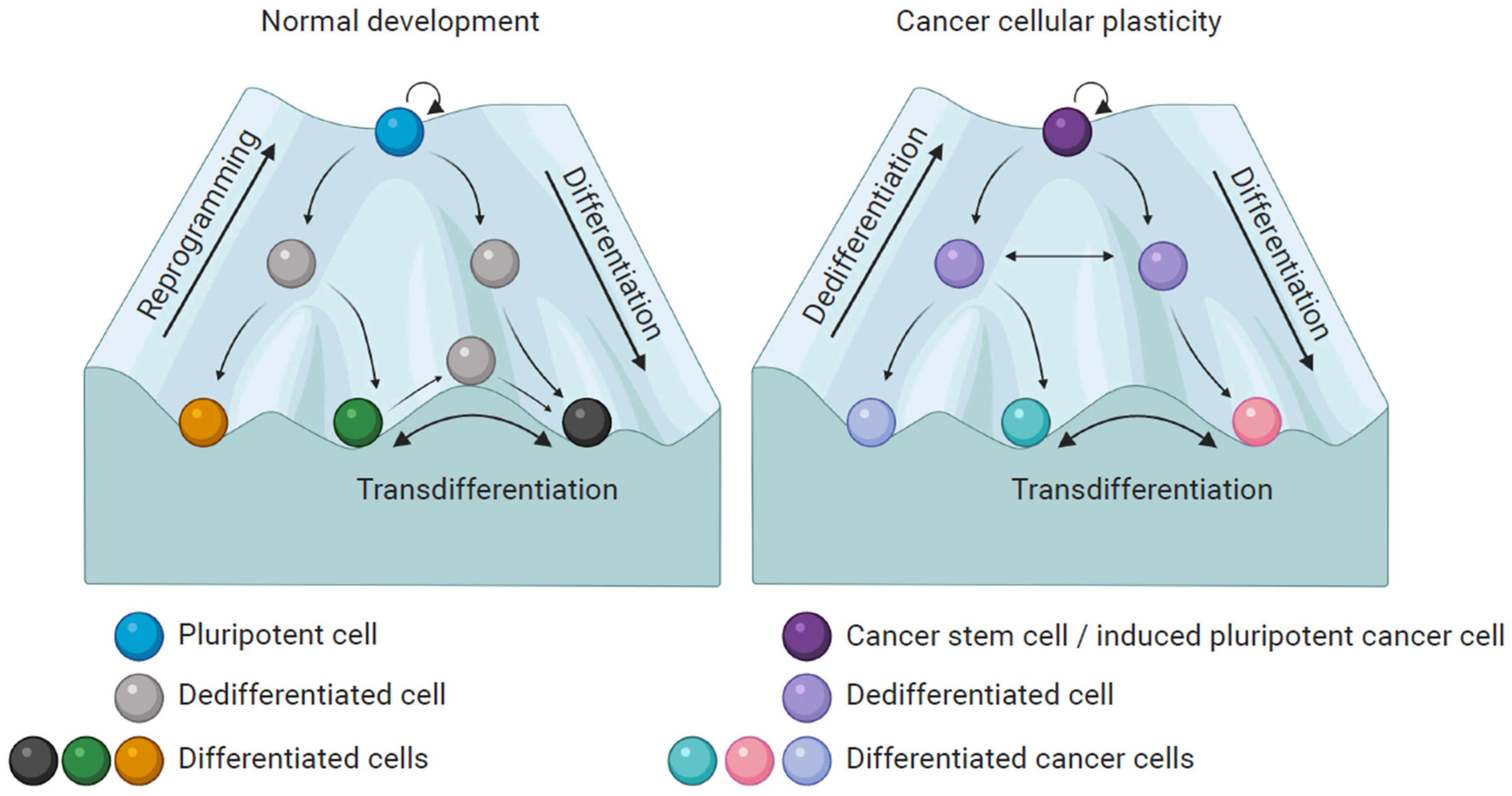
4. Melanoma Plasticity
5. Partial Reprogramming of Melanoma Cells
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Jopling, C.; Boue, S.; Belmonte, J.C.I. Dedifferentiation, transdifferentiation and reprogramming: Three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011, 12, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, J.; Robert, L.; Paraiso, K.; Galvan, C.; Sheu, K.M.; Lay, J.; Wong, D.J.L.; Atefi, M.; Shirazi, R.; Wang, X.; et al. Multi-stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress. Cancer Cell 2018, 33, 890–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, S.; Norgard, R.J.; Stanger, B.Z. Cellular plasticity in cancer. Cancer Discov. 2019, 9, 837–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.; Wang, C. Dedifferentiation: Inspiration for devising engineering strategies for regenerative medicine. NPJ Regen. Med. 2020, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Massi, D.; Mihic-Probst, D.; Schadendorf, D.; Dummer, R.; Mandalà, M. Dedifferentiated melanomas: Morpho-phenotypic profile, genetic reprogramming and clinical implications. Cancer Treat. Rev. 2020, 88, 102060. [Google Scholar] [CrossRef]
- Waddington, C. The Strategy of the Genes, 1st ed.; George Allen & Unwin LTD.: Crows Nest, Australia, 1957. [Google Scholar]
- Karagiannis, P.; Yamanaka, S. The fate of cell reprogramming. Nat. Methods 2014, 11, 1006–1008. [Google Scholar] [CrossRef] [PubMed]
- Friedmann-Morvinski, D.; Verma, I.M. Dedifferentiation and reprogramming: Origins of cancer stem cells. EMBO Rep. 2014, 15, 244–253. [Google Scholar] [CrossRef] [Green Version]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef]
- Nguyen, L.V.; Vanner, R.; Dirks, P.; Eaves, C.J. Cancer stem cells: An evolving concept. Nat. Rev. Cancer 2012, 12, 133–143. [Google Scholar] [CrossRef] [PubMed]
- van Neerven, S.M.; Tieken, M.; Vermeulen, L.; Bijlsma, M.F. Bidirectional interconversion of stem and non-stem cancer cell populations: A reassessment of theoretical models for tumor heterogeneity. Mol. Cell. Oncol. 2016, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Schwitalla, S.; Fingerle, A.A.; Cammareri, P.; Nebelsiek, T.; Göktuna, S.I.; Ziegler, P.K.; Canli, O.; Heijmans, J.; Huels, D.J.; Moreaux, G.; et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013, 152, 25–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez Alvarado, A.; Yamanaka, S. Rethinking differentiation: Stem cells, regeneration, and plasticity. Cell 2014, 157, 110–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jopling, C.; Sleep, E.; Raya, M.; Martí, M.; Raya, A.; Belmonte, J.C.I. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010, 464, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Chaffer, C.L.; Brueckmann, I.; Scheel, C.; Kaestli, A.J.; Wiggins, P.A.; Rodrigues, L.O.; Brooks, M.; Reinhardt, F.; Suc, Y.; Polyak, K.; et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl. Acad. Sci. USA 2011, 108, 7950–7955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Es, J.H.; Sato, T.; van de Wetering, M.; Luybimova, A.; Gregorieff, A.; Zeinstra, L.; van den Born, M.; Korving, J.; Martens, A.C.; van den Oudenaarden, A.; et al. Dll1 marks early secretory progenitors in gut crypts that can revert to stem cells upon tissue damage. Nat. Cell Biol. 2012, 14, 1099–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Gascard, P.; Dumont, N.; Zhao, J.; Pan, D.; Petrie, S.; Margeta, M.; Tlsty, T.D. Rare somatic cells from human breast tissue exhibit extensive lineage plasticity. Proc. Natl. Acad. Sci. USA 2013, 110, 4598–4603. [Google Scholar] [CrossRef] [Green Version]
- Tata, P.R.; Mou, H.; Pardo-Saganta, A.; Zhao, R.; Prabhu, M.; Law, B.M.; Vinarsky, V.; Cho, J.L.; Breton, S.; Sahay, A.; et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 2013, 503, 218–223. [Google Scholar] [CrossRef] [Green Version]
- Gong, L.; Yan, Q.; Zhang, Y.; Fang, X.; Liu, B.; Guan, X. Cancer cell reprogramming: A promising therapy converting malignancy to benignity. Cancer Commun. 2019, 39, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Dulak, J.; Szade, K.; Szade, A.; Nowak, W.; Józkowicz, A. Adult stem cells: Hopes and hypes of regenerative medicine. Acta Biochim. Pol. 2015, 62, 329–337. [Google Scholar] [CrossRef] [Green Version]
- Wyles, S.P.; Brandt, E.B.; Nelson, T.J. Stem cells: The pursuit of genomic stability. Int. J. Mol. Sci. 2014, 15, 20948–20967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daley, G.Q. Stem cells and the evolving notion of cellular identity. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; David, B.T.; Trawczynski, M.; Fessler, R.G. Advances in Pluripotent Stem Cells: History, Mechanisms, Technologies, and Applications. Stem Cell Rev. Rep. 2020, 16, 3–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurdon, J.B. Sexually mature individuals of xenopus laevis from the transplantation of single somatic nuclei. Nature 1958, 182, 800–801. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [Green Version]
- Utikal, J.; Maherali, N.; Kulalert, W.; Hochedlinger, K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J. Cell Sci. 2009, 122, 3502–3510. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science (80-.) 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.B.; Zaehres, H.; Wu, G.; Gentile, L.; Ko, K.; Sebastiano, V.; Araúzo-Bravo, M.J.; Ruau, D.; Han, D.W.; Zenke, M.; et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 2008, 454, 646–650. [Google Scholar] [CrossRef]
- Ambrosi, D.J.; Tanasijevic, B.; Kaur, A.; Obergfell, C.; O’Neill, R.J.; Krueger, W.; Rasmussen, T.P. Genome-Wide Reprogramming in Hybrids of Somatic Cells and Embryonic Stem Cells. Stem Cells 2007, 25, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Koga, C.; Kobayashi, S.; Nagano, H.; Tomimaru, Y.; Hama, N.; Wada, H.; Kawamoto, K.; Eguchi, H.; Konno, M.; Ishii, H.; et al. Reprogramming Using microRNA-302 Improves Drug Sensitivity in Hepatocellular Carcinoma Cells. Ann. Surg. Oncol. 2014, 21, 591–600. [Google Scholar] [CrossRef]
- Ma, X.; Kong, L.; Zhu, S. Reprogramming cell fates by small molecules. Protein Cell 2017, 8, 328–348. [Google Scholar] [CrossRef] [Green Version]
- Cimmino, L.; Neel, B.G.; Aifantis, I. Vitamin C in Stem Cell Reprogramming and Cancer. Trends Cell Biol. 2018, 28, 698–708. [Google Scholar] [CrossRef]
- Huangfu, D.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Snitow, M.; Chen, A.E.; Melton, D.A. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008, 26, 795–797. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, T.; Guan, J.; Zhang, X.; Fu, Y.; Ye, J.; Zhu, J.; Meng, G.; Ge, J.; Yang, S.; et al. A XEN-like State Bridges Somatic Cells to Pluripotency during Chemical Reprogramming. Cell 2015, 163, 1678–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plath, K.; Lowry, W.E. Progress in understanding reprogramming to the induced pluripotent state. Nat. Rev. Genet. 2011, 12, 253–265. [Google Scholar] [CrossRef] [Green Version]
- Buganim, Y.; Faddah, D.A.; Jaenisch, R. Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 2013, 14, 427–439. [Google Scholar] [CrossRef] [Green Version]
- Smith, O.K.; Kim, R.; Fu, H.; Martin, M.M.; Lin, C.M.; Utani, K.; Zhang, Y.; Marks, A.B.; Lalande, M.; Chamberlain, S.; et al. Distinct epigenetic features of differentiation-regulated replication origins. Epigenetics Chromatin 2016, 9, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Soufi, A. Mechanisms for enhancing cellular reprogramming. Curr. Opin. Genet. Dev. 2014, 25, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Prieto, J.; Seo, A.Y.; León, M.; Santacatterina, F.; Torresano, L.; Palomino-Schätzlein, M.; Giménez, K.; Vallet-Sánchez, A.; Ponsoda, X.; Pineda-Lucena, A.; et al. MYC Induces a Hybrid Energetics Program Early in Cell Reprogramming. Stem Cell Rep. 2018, 11, 1479–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimura, K.; Fukuda, A.; Hisatake, K. Mechanisms of the metabolic shift during somatic cell reprogramming. Int. J. Mol. Sci. 2019, 20, 2254. [Google Scholar] [CrossRef] [Green Version]
- Jaenisch, R.; Young, R. Stem Cells, the Molecular Circuitry of Pluripotency and Nuclear Reprogramming. Cell 2008, 132, 567–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takata, C.; Albright, J.F.; Yamada, T. Lens fiber differentiation and gamma crystallins: Immunofluorescent study of wolffian regeneration. Science (80-.) 1965, 147, 1299–1301. [Google Scholar] [CrossRef]
- Donati, G.; Rognoni, E.; Hiratsuka, T.; Liakath-Ali, K.; Hoste, E.; Kar, G.; Kayikci, M.; Russell, R.; Kretzschmar, K.; Mulder, K.W.; et al. Wounding induces dedifferentiation of epidermal Gata6 + cells and acquisition of stem cell properties. Nat. Cell Biol. 2017, 19, 603–613. [Google Scholar] [CrossRef] [Green Version]
- Xiong, S.; Feng, Y.; Cheng, L. Cellular Reprogramming as a Therapeutic Target in Cancer. Trends Cell Biol. 2019, 20, 1–12. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, C.A.; Kreso, A.; Jamieson, C.H.M. Cancer stem cells and self-renewal. Clin. Cancer Res. 2010, 16, 3113–3120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahn, M. Wnt Signaling in Stem Cells and Cancer Stem Cells: A Tale of Two Coactivators; Elsevier Inc.: Amsterdam, The Netherlands, 2018; Volume 153. [Google Scholar]
- Câmara, D.A.D.; Mambelli, L.I.; Porcacchia, A.S.; Kerkis, I. Advances and challenges on cancer cells reprogramming using induced pluripotent stem cells technologies. J. Cancer 2016, 7, 2296–2303. [Google Scholar] [CrossRef] [Green Version]
- Knappe, N.; Novak, D.; Weina, K.; Bernhardt, M.; Reith, M.; Larribere, L.; Hölzel, M.; Tüting, T.; Gebhardt, C.; Umansky, V.; et al. Directed Dedifferentiation Using Partial Reprogramming Induces Invasive Phenotype in Melanoma Cells. Stem Cells 2016, 34, 832–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernhardt, M.; Galach, M.; Novak, D.; Utikal, J. Mediators of induced pluripotency and their role in cancer cells-current scientific knowledge and future perspectives. Biotechnol. J. 2012, 7, 810–821. [Google Scholar] [CrossRef]
- Carette, J.E.; Pruszak, J.; Varadarajan, M.; Blomen, V.A.; Gokhale, S.; Camargo, F.D.; Wernig, M.; Jaenisch, R.; Brummelkamp, T.R. Generation of iPSCs from cultured human malignant cells. Blood 2010, 115, 4039–4042. [Google Scholar] [CrossRef] [Green Version]
- Miyoshi, N.; Ishii, H.; Nagai, K.I.; Hoshino, H.; Mimori, K.; Tanaka, F.; Nagano, H.; Sekimoto, M.; Doki, Y.; Mori, M. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc. Natl. Acad. Sci. USA 2010, 107, 40–45. [Google Scholar] [CrossRef] [Green Version]
- Choong, P.F.; Teh, H.X.; Teoh, H.K.; Ong, H.K.; Choo, K.B.; Sugii, S.; Cheong, S.K.; Kamarul, T. Heterogeneity of osteosarcoma cell lines led to variable responses in reprogramming. Int. J. Med. Sci. 2014, 11, 1154–1160. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Jeong, J.; Park, S.; Jin, Y.W.; Lee, S.S.; Lee, S.B.; Choi, D. Establishment of Hepatocellular Cancer Induced Pluripotent Stem Cells Using a Reprogramming Technique. Gut Liver 2017, 11, 261–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iskender, B.; Izgi, K.; Canatan, H. Reprogramming bladder cancer cells for studying cancer initiation and progression. Tumor Biol. 2016, 37, 13237–13245. [Google Scholar] [CrossRef]
- Islam, S.M.R.; Suenaga, Y.; Takatori, A.; Ueda, Y.; Kaneko, Y.; Kawana, H.; Itami, M.; Ohira, M.; Yokoi, S.; Nakagawara, A. Sendai virus-mediated expression of reprogramming factors promotes plasticity of human neuroblastoma cells. Cancer Sci. 2015, 106, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Hoffman, J.P.; Alpaugh, R.K.; Rhimm, A.D.; Reichert, M.; Stanger, B.Z.; Furth, E.E.; Sepulveda, A.R.; Yuan, C.X.; Won, K.J.; et al. An iPSC Line from Human Pancreatic Ductal Adenocarcinoma Undergoes Early to Invasive Stages of Pancreatic Cancer Progression. Cell Rep. 2013, 3, 2088–2099. [Google Scholar] [CrossRef] [Green Version]
- Kreso, A.; Dick, J.E. Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Tu, J.; Gingold, J.A.; Kong, C.S.L.; Lee, D.-F. Cancer in a dish: Progress using stem cells as a platform for cancer research. Am. J. Cancer Res. 2018, 8, 944–954. [Google Scholar]
- Rambow, F.; Rogiers, A.; Marin-Bejar, O.; Aibar, S.; Femel, J.; Dewaele, M.; Karras, P.; Brown, D.; Chang, Y.H.; Debiec-Rychter, M.; et al. Toward Minimal Residual Disease-Directed Therapy in Melanoma. Cell 2018, 174, 843–855.e19. [Google Scholar] [CrossRef] [Green Version]
- Czyz, M.; Sztiller-Sikorska, M.; Gajos-Michniewicz, A.; Osrodek, M.; Hartman, M.L. Plasticity of drug-naïve and vemurafenib- or trametinib-resistant melanoma cells in execution of differentiation/pigmentation program. J. Oncol. 2019, 2019. [Google Scholar] [CrossRef] [Green Version]
- Rowling, E.J.; Miskolczi, Z.; Nagaraju, R.; Wilcock, D.J.; Wang, P.; Telfer, B.; Li, Y.; Lasheras-Otero, I.; Redondo-Muñoz, M.; Sharrocks, A.D.; et al. Cooperative behaviour and phenotype plasticity evolve during melanoma progression. Pigment Cell Melanoma Res. 2020, 33, 695–708. [Google Scholar] [CrossRef]
- Castro-Pérez, E.; Rodríguez, C.I.; Mikheil, D.; Siddique, S.; McCarthy, A.; Newton, M.A.; Setaluri, V. Melanoma Progression Inhibits Pluripotency and Differentiation of Melanoma-Derived iPSCs Produces Cells with Neural-like Mixed Dysplastic Phenotype. Stem Cell Rep. 2019, 13, 177–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bettum, I.J.; Gorad, S.S.; Barkovskaya, A.; Pettersen, S.; Moestue, S.A.; Vasiliauskaite, K.; Tenstad, E.; Øyjord, T.; Risa, Ø.; Nygaard, V.; et al. Metabolic reprogramming supports the invasive phenotype in malignant melanoma. Cancer Lett. 2015, 366, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Mandalá, M.; Romano, E. Mechanisms of Drug Resistance in Cancer Therapy; Springer International Publishing: Berlin, Germany, 2018; Volume 1249, ISBN 9783030105068. [Google Scholar]
- Vandamme, N.; Berx, G. Melanoma Cells Revive an Embryonic Transcriptional Network to Dictate Phenotypic Heterogeneity. Front. Oncol. 2014, 4, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Woods, K.; Pasam, A.; Jayachandran, A.; Andrews, M.C.; Cebon, J. Effects of Epithelial to Mesenchymal Transition on T Cell Targeting of Melanoma Cells. Front. Oncol. 2014, 4, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.E.; Leung, E.; Baguley, B.C.; Finlay, G.J. Heterogeneity of expression of epithelial-mesenchymal transition markers in melanocytes and melanoma cell lines. Front. Genet. 2013, 4, 1–8. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, M.P.; Weeraratna, A.T. Change is in the air: The hypoxic induction of phenotype switching in melanoma. J. Investig. Dermatol. 2013, 133, 2316–2317. [Google Scholar] [CrossRef] [Green Version]
- Widmer, D.S.; Hoek, K.S.; Cheng, P.F.; Eichhoff, O.M.; Biedermann, T.; Raaijmakers, M.I.G.; Hemmi, S.; Dummer, R.; Levesque, M.P. Hypoxia contributes to melanoma heterogeneity by triggering HIF1α-dependent phenotype switching. J. Investig. Dermatol. 2013, 133, 2436–2443. [Google Scholar] [CrossRef] [Green Version]
- Monaghan-Benson, E.; Burridge, K. Mutant B-RAF regulates a Rac-dependent cadherin switch in melanoma. Oncogene 2013, 32, 4836–4844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, K.S.; Kim, M. Decision to grow or to invade is at the flick of metabolic switch, PGC1α. Pigment Cell Melanoma Res. 2017, 30, 179–180. [Google Scholar] [CrossRef]
- Ratnikov, B.I.; Scott, D.A.; Osterman, A.L.; Smith, J.W.; Ronai, Z.A. Metabolic rewiring in melanoma. Oncogene 2017, 36, 147–157. [Google Scholar] [CrossRef] [Green Version]
- Bristot, I.J.; Kehl Dias, C.; Chapola, H.; Parsons, R.B.; Klamt, F. Metabolic rewiring in melanoma drug-resistant cells. Crit. Rev. Oncol. Hematol. 2020, 153, 102995. [Google Scholar] [CrossRef] [PubMed]
- Boumahdi, S.; de Sauvage, F.J. The great escape: Tumour cell plasticity in resistance to targeted therapy. Nat. Rev. Drug Discov. 2020, 19, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Chapman, A.; del Ama, L.F.; Ferguson, J.; Kamarashev, J.; Wellbrock, C.; Hurlstone, A. Heterogeneous tumor subpopulations cooperate to drive invasion. Cell Rep. 2014, 8, 688–695. [Google Scholar] [CrossRef]
- Hoek, K.S.; Eichhoff, O.M.; Schlegel, N.C.; Döbbeling, U.; Kobert, N.; Schaerer, L.; Hemmi, S.; Dummer, R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008, 68, 650–656. [Google Scholar] [CrossRef] [Green Version]
- Roesch, A.; Paschen, A.; Landsberg, J.; Helfrich, I.; Becker, J.C.; Schadendorf, D. Phenotypic tumour cell plasticity as a resistance mechanism and therapeutic target in melanoma. Eur. J. Cancer 2016, 59, 109–112. [Google Scholar] [CrossRef]
- Wellbrock, C.; Arozarena, I. The Complexity of the ERK/MAP-Kinase Pathway and the Treatment of Melanoma Skin Cancer. Front. Cell Dev. Biol. 2016, 4, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Kemeny, L.V.; Fisher, D.E. Targeting the (Un)differentiated State of Cancer. Cancer Cell 2018, 33, 793–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goding, C.R. A picture of Mitf in melanoma immortality. Oncogene 2011, 30, 2304–2306. [Google Scholar] [CrossRef] [PubMed]
- Granados, K.; Hüser, L.; Federico, A.; Sachindra, S.; Wolff, G.; Hielscher, T.; Novak, D. T-type calcium channel inhibition restores sensitivity to MAPK inhibitors in dedifferentiated and adaptive melanoma cells. Br. J. Cancer 2020. [Google Scholar] [CrossRef] [Green Version]
- Konieczkowski, D.J.; Johannessen, C.M.; Abudayyeh, O.; Kim, J.W.; Cooper, Z.A.; Piris, A.; Frederick, D.T.; Barzily-Rokni, M.; Straussman, R.; Haq, R.; et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014, 4, 816–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, J.; Krijgsman, O.; Tsoi, J.; Robert, L.; Hugo, W.; Song, C.; Kong, X.; Possik, P.A.; Cornelissen-Steijger, P.D.M.; Foppen, M.H.G.; et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun. 2014, 5, 1–15. [Google Scholar] [CrossRef]
- Kemper, K.; De Goeje, P.L.; Peeper, D.S.; Van Amerongen, R. Phenotype switching: Tumor cell plasticity as a resistance mechanism and target for therapy. Cancer Res. 2014, 74, 5937–5941. [Google Scholar] [CrossRef] [Green Version]
- Landsberg, J.; Kohlmeyer, J.; Renn, M.; Bald, T.; Rogava, M.; Cron, M.; Fatho, M.; Lennerz, V.; Wölfel, T.; Hölzel, M.; et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature 2012, 490, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Johannessen, C.M.; Johnson, L.A.; Piccioni, F.; Townes, A.; Frederick, D.T.; Donahue, M.K.; Narayan, R.; Flaherty, K.T.; Wargo, J.A.; Root, D.E.; et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature 2013, 504, 138–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Hugo, W.; Kong, X.; Hong, A.; Koya, R.C.; Moriceau, G.; Chodon, T.; Guo, R.; Johnson, D.B.; Dahlman, K.B.; et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014, 4, 80–93. [Google Scholar] [CrossRef] [Green Version]
- Wellbrock, C.; Arozarena, I. Microphthalmia-associated transcription factor in melanoma development and MAP-kinase pathway targeted therapy. Pigment Cell Melanoma Res. 2015, 28, 390–406. [Google Scholar] [CrossRef] [Green Version]
- Van Allen, E.M.; Wagle, N.; Sucker, A.; Treacy, D.J.; Johannessen, C.M.; Goetz, E.M.; Place, C.S.; Taylor-Weiner, A.; Whittaker, S.; Kryukov, G.V.; et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014, 4, 94–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Câmara, D.A.D.; Porcacchia, A.S.; Costa, A.S.; Azevedo, R.A.; Kerkis, I. Murine melanoma cells incomplete reprogramming using non-viral vector. Cell Prolif. 2017, 50, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Hiew, M.S.Y.; Cheng, H.P.; Huang, C.J.; Chong, K.Y.; Cheong, S.K.; Choo, K.B.; Kamarul, T. Incomplete cellular reprogramming of colorectal cancer cells elicits an epithelial/mesenchymal hybrid phenotype. J. Biomed. Sci. 2018, 25, 1–13. [Google Scholar] [CrossRef]
- Long, G.V.; Flaherty, K.T.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; de Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; et al. Erratum: Dabrafenib plus trametinib versus dabrafenib monotherapy in patients with metastatic BRAF V600E/K-mutant melanoma: Long-term survival and safety analysis of a phase 3 study (Annals of Oncology (2017) 28(7) (1631–1639), (S0923753419322707), 10.10. Ann. Oncol. 2019, 30, 1848. [Google Scholar] [CrossRef] [PubMed]
- Kraft, A.; Rubin, B.P. Changing cells: An analysis of the concept of plasticity in the context of cellular differentiation. Biosocieties 2016, 11, 497–525. [Google Scholar] [CrossRef] [Green Version]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 1–15. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Granados, K.; Poelchen, J.; Novak, D.; Utikal, J. Cellular Reprogramming—A Model for Melanoma Cellular Plasticity. Int. J. Mol. Sci. 2020, 21, 8274. https://doi.org/10.3390/ijms21218274
Granados K, Poelchen J, Novak D, Utikal J. Cellular Reprogramming—A Model for Melanoma Cellular Plasticity. International Journal of Molecular Sciences. 2020; 21(21):8274. https://doi.org/10.3390/ijms21218274
Chicago/Turabian StyleGranados, Karol, Juliane Poelchen, Daniel Novak, and Jochen Utikal. 2020. "Cellular Reprogramming—A Model for Melanoma Cellular Plasticity" International Journal of Molecular Sciences 21, no. 21: 8274. https://doi.org/10.3390/ijms21218274




