Analysis of Melanoma Secretome for Factors That Directly Disrupt the Barrier Integrity of Brain Endothelial Cells
Abstract
1. Introduction
2. Results
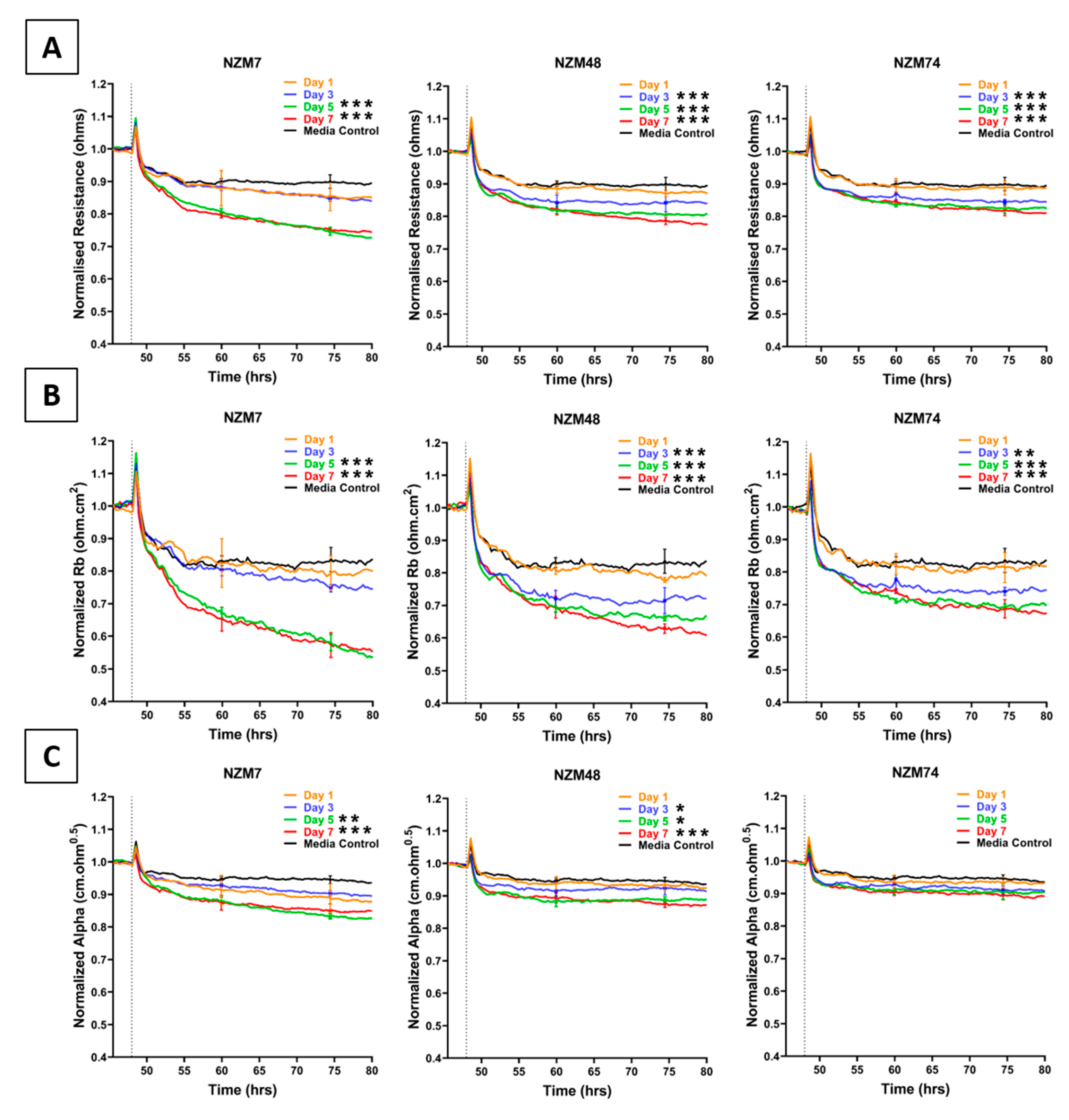
2.1. Effect of Melanoma Conditioned Media on Brain Endothelium
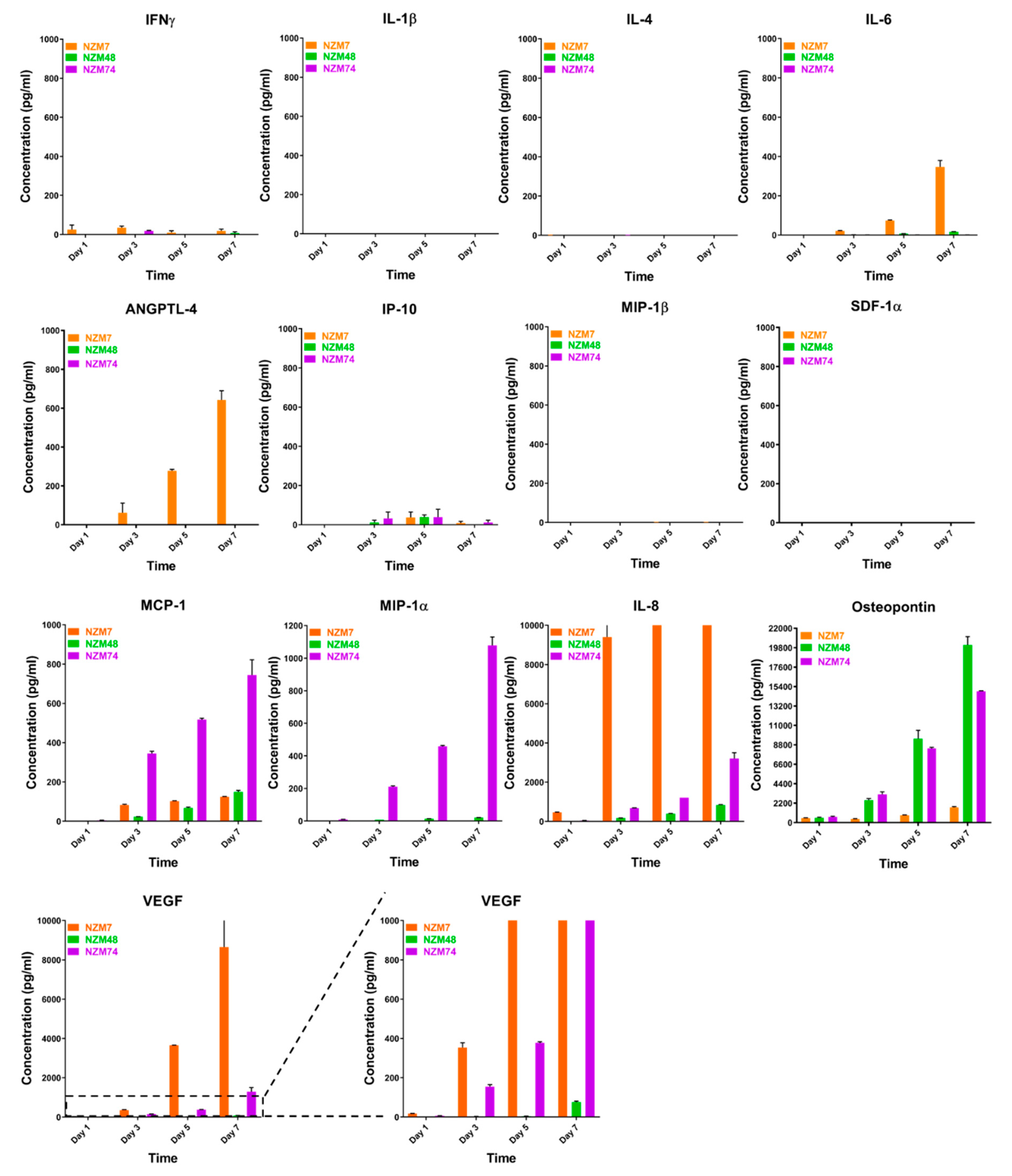
2.2. Characterisation of Melanoma Conditioned Media
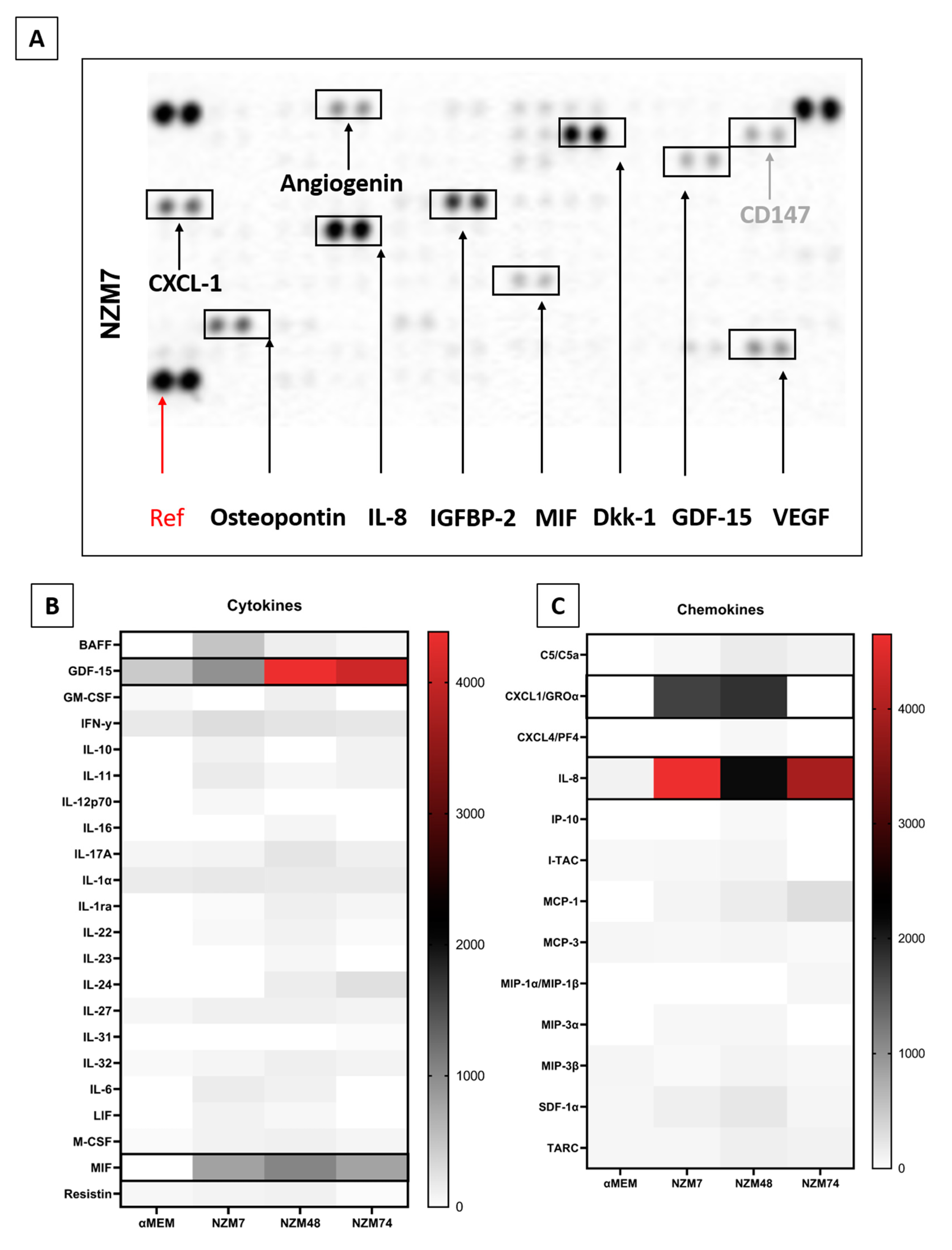
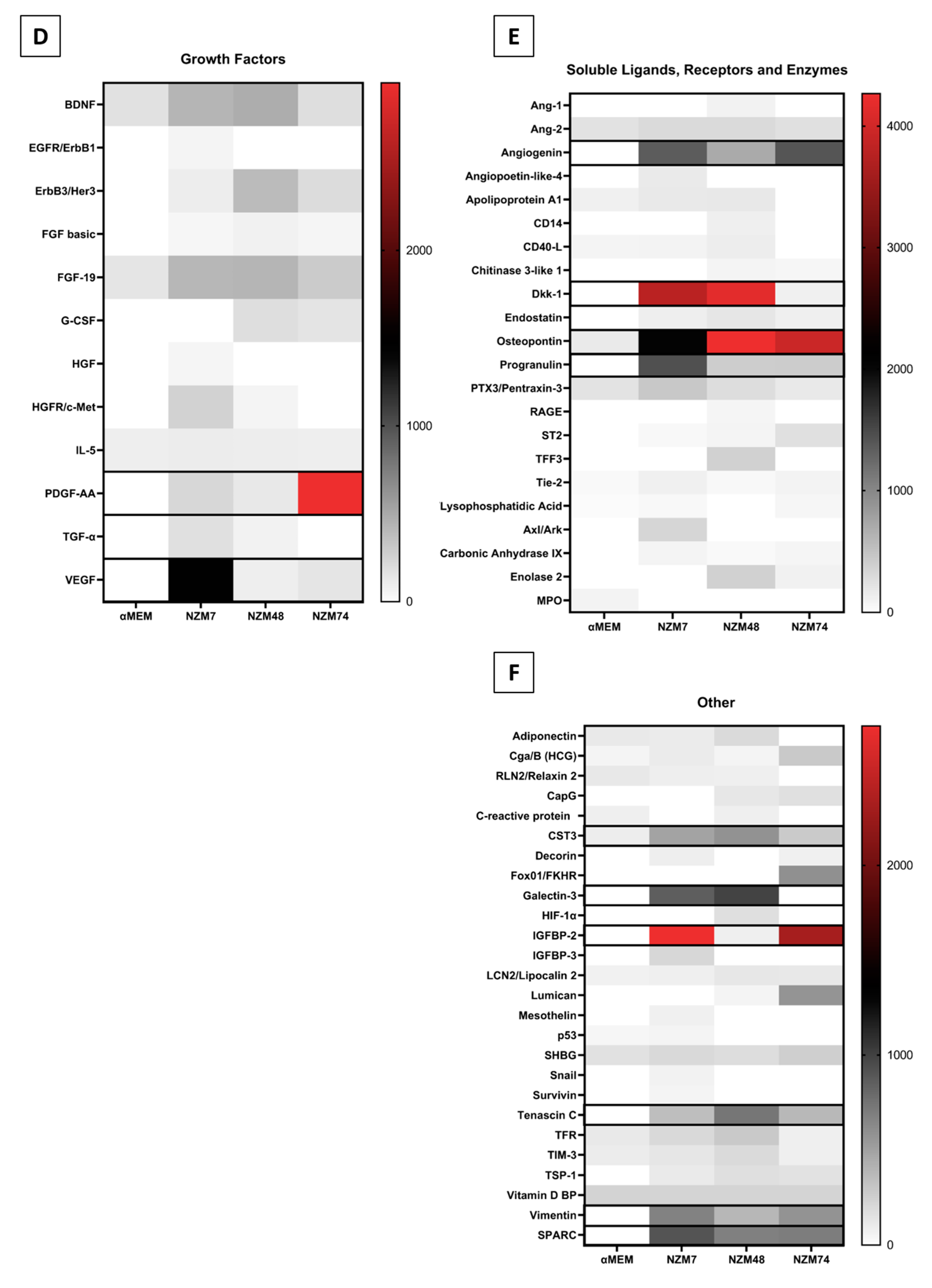
2.3. Common Mediators in Oncology and Inflammation
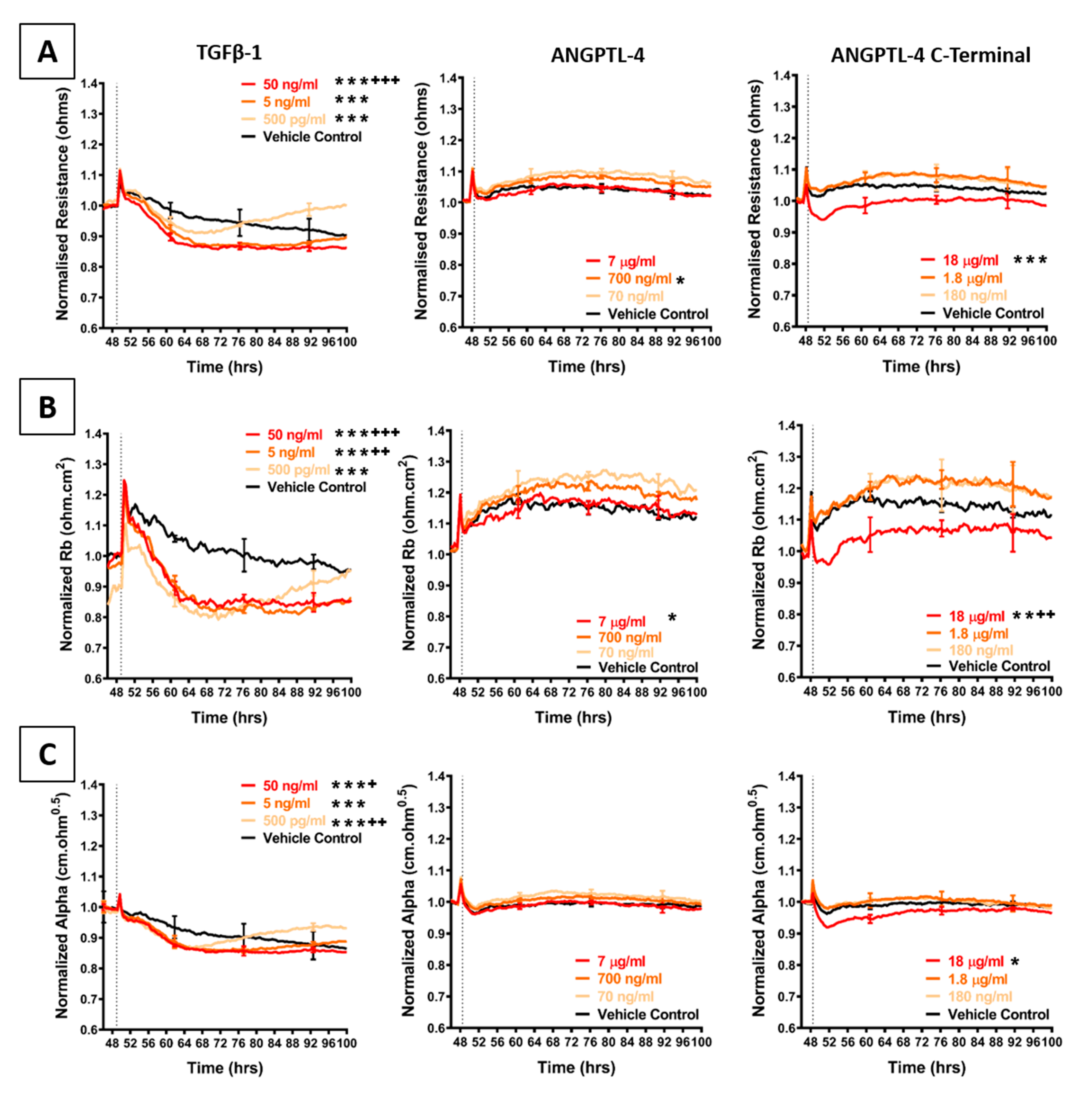
2.4. Effect of Selected Secretory Molecules on the Brain Endothelium
3. Materials and Methods
3.1. Culture of the Melanoma Cells
3.2. Collection of the Conditioned Media
3.3. Culture of the Brain Endothelial Cells
3.4. ECIS Technology Assessment of Conditioned Media
3.5. Cytokine Measurements Using CBA and Luminex
3.6. NanoString nCounter Analysis
3.7. Cytokine Screening Using Proteome Profiler Arrays
3.8. Assessment of Recombinant Cytokines on Barrier Integrity
3.9. Statistical Analysis
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Array/Set | Aliases | Assay | Company | Catalogue No. | Standard Curve (pg/mL) |
|---|---|---|---|---|---|
| Secreted proteins tested | |||||
| Human IL-8 | CXCL8 | CBA | BD Bioscience | 558277 | 0–5000 |
| Human MCP-1 | CCL2 | CBA | BD Bioscience | 558287 | 0–5000 |
| Human MIP-1α | CCL3 | CBA | BD Bioscience | 558325 | 0–5000 |
| Human VEGF | - | CBA | BD Bioscience | 558336 | 0–5000 |
| Human IL-4 | BSF-1 | CBA | BD Bioscience | 558272 | 0–5000 |
| Human MIP-1β | CCL4 | CBA | BD Bioscience | 558288 | 0–5000 |
| Human IL-6 | BSF-2/HGF | CBA | BD Bioscience | 558276 | 0–5000 |
| Human IP-10 | CXCL10 | CBA | BD Bioscience | 558280 | 0–5000 |
| Human ANGPTL-4 | HFARP | Luminex | R&D Systems | LXSAH-4 | 0–360810 |
| Human Osteopontin | OPN/SPP-1 | Luminex | R&D Systems | LXSAH-4 | 0–636310 |
| Human SDF-1α | CXCl12 | Luminex | R&D Systems | LXSAH-4 | 0–36870 |
| Human IFNγ | IFG | CBA | BD Bioscience | 558269 | 0–5000 |
| Human IL-1β | - | CBA | BD Bioscience | 558279 | 0–5000 |
| TGFβ-1 | CBA | BD Bioscience | 560429 | 0–10000 | |
| Secreted proteins tested | |||||
| Human XL Cytokine Array Kit | Proteome Profiler | R&D Systems | ARY022B | ||
| Human XL Oncology Array Kit | Proteome Profiler | R&D Systems | ARY026 | ||
| Protein | Gene | Probe NSID |
|---|---|---|
| TGFβ-1 | TGFB1 | NM_000660.3:1260 |
| VEGF-A | VEGFA | NM_001025366.1:1325 |
| Osteopontin | SPP1 | NM_000582.2:760 |
| MCP-1 | CCL2 | NM_002982.3:123 |
| IL-8 | CXCL8 | NM_000584.2:25 |
| Treatment | Company | Catalogue No. |
|---|---|---|
| Angiogenin | R&D | 265-AN-050/CF |
| GDF-15 | R&D | 8146-GD/CF |
| Progranulin | R&D | 2420-PG-050 |
| SPARC | R&D | 941-SP-050 |
| SPARC Like-1 | R&D | 2728-SL-050 |
| Tenascin C/Cytotactin | R&D | 3358-TC-050 |
| IGFBP-2 | R&D | 674-B2-025 |
| Cystatin C | Biolegend | 756202 |
| DKK-1 | Biolegend | 778602 |
| Galectin-3 | Biolegend | 599704 |
| MIF | Biolegend | 599404 |
| Osteopontin | Biolegend | 557102 |
| TGFβ-1 | Biolegend | 580702 |
| CXCL-1 | Biolegend | 574402 |
| ANGPTL-4 | R&D | RDS4487AN050 |
| cANGPTL-4 | R&D | RDS3485AN050 |
| IL-8 | PrimeGene | 101-08A |
| VEGF | PrimeGene | 105-16Y |
References
- Anchan, A.; Kalogirou-Baldwin, P.; Johnson, R.; Kho, D.T.; Joseph, W.; Hucklesby, J.; Finlay, G.J.; O’Carroll, S.J.; Angel, C.E.; Graham, E.S. Real-Time measurement of melanoma cell-mediated human brain endothelial barrier disruption using electric cell-substrate impedance sensing technology. Biosensors 2019, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.I.; Cayrol, R.; Prat, A. Disruption of central nervous system barriers in multiple sclerosis. Biochim. Biophys. Acta 2011, 1812, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Nagyoszi, P.; Wilhelm, I.; Farkas, A.E.; Fazakas, C.; Dung, N.T.; Hasko, J.; Krizbai, I.A. Expression and regulation of toll-like receptors in cerebral endothelial cells. Neurochem. Int. 2010, 57, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Larochelle, C.; Alvarez, J.I.; Prat, A. How do immune cells overcome the blood-brain barrier in multiple sclerosis? FEBS Lett. 2011, 585, 3770–3780. [Google Scholar] [CrossRef] [PubMed]
- O’Carroll, S.J.; Kho, D.T.; Wiltshire, R.; Nelson, V.; Rotimi, O.; Johnson, R.; Angel, C.E.; Graham, E.S. Pro-inflammatory TNFalpha and IL-1beta differentially regulate the inflammatory phenotype of brain microvascular endothelial cells. J. Neuroinflamm. 2015, 12, 131. [Google Scholar] [CrossRef] [PubMed]
- Bryan, B.A.; Dennstedt, E.; Mitchell, D.C.; Walshe, T.E.; Noma, K.; Loureiro, R.; Saint-Geniez, M.; Campaigniac, J.P.; Liao, J.K.; D’Amore, P.A. RhoA/ROCK signaling is essential for multiple aspects of VEGF-mediated angiogenesis. FASEB J. 2010, 24, 3186–3195. [Google Scholar] [CrossRef]
- Hobbs, S.K.; Monsky, W.L.; Yuan, F.; Roberts, W.G.; Griffith, L.; Torchilin, V.P.; Jain, R.K. Regulation of transport pathways in tumor vessels: Role of tumor type and microenvironment. Proc. Natl. Acad. Sci. USA 1998, 95, 4607–4612. [Google Scholar] [CrossRef]
- Monsky, W.L.; Fukumura, D.; Gohongi, T.; Ancukiewcz, M.; Weich, H.A.; Torchilin, V.P.; Yuan, F.; Jain, R.K. Augmentation of transvascular transport of macromolecules and nanoparticles in tumors using vascular endothelial growth factor. Cancer Res. 1999, 59, 4129–4135. [Google Scholar]
- Claffey, K.P.; Brown, L.F.; del Aguila, L.F.; Tognazzi, K.; Yeo, K.T.; Manseau, E.J.; Dvorak, H.F. Expression of vascular permeability factor/vascular endothelial growth factor by melanoma cells increases tumor growth, angiogenesis, and experimental metastasis. Cancer Res. 1996, 56, 172–181. [Google Scholar]
- Yu, J.L.; Rak, J.W.; Klement, G.; Kerbel, R.S. Vascular endothelial growth factor isoform expression as a determinant of blood vessel patterning in human melanoma xenografts. Cancer Res. 2002, 62, 1838–1846. [Google Scholar]
- Ugurel, S.; Rappl, G.; Tilgen, W.; Reinhold, U. Increased serum concentration of angiogenic factors in malignant melanoma patients correlates with tumor progression and survival. J. Clin. Oncol. 2001, 19, 577–583. [Google Scholar] [CrossRef]
- Papageorgis, P. TGFbeta signaling in tumor initiation, epithelial-to-mesenchymal transition, and metastasis. J. Oncol. 2015, 2015, 587193. [Google Scholar] [CrossRef] [PubMed]
- Zavadil, J.; Bottinger, E.P. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene 2005, 24, 5764–5774. [Google Scholar] [CrossRef] [PubMed]
- Berking, C.; Takemoto, R.; Schaider, H.; Showe, L.; Satyamoorthy, K.; Robbins, P.; Herlyn, M. Transforming growth factor-beta1 increases survival of human melanoma through stroma remodeling. Cancer Res. 2001, 61, 8306–8316. [Google Scholar]
- Xouri, G.; Christian, S. Origin and function of tumor stroma fibroblasts. Semin. Cell Dev. Biol. 2010, 21, 40–46. [Google Scholar] [CrossRef]
- Fuxe, J.; Karlsson, M.C.I. TGF-β-induced epithelial-mesenchymal transition: A link between cancer and inflammation. Semin. Cancer Biol. 2012, 22, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Settleman, J. EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene 2010, 29, 4741–4751. [Google Scholar] [CrossRef]
- van Meeteren, L.A.; ten Dijke, P. Regulation of endothelial cell plasticity by TGF-beta. Cell Tissue Res. 2012, 347, 177–186. [Google Scholar] [CrossRef]
- Zeisberg, E.M.; Potenta, S.; Xie, L.; Zeisberg, M.; Kalluri, R. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007, 67, 10123–10128. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, F.; Lee, J.; Dong, Z. Selective induction of interleukin-8 expression in metastatic melanoma cells by transforming growth factor-beta 1. Cytokine 2005, 31, 241–249. [Google Scholar] [CrossRef]
- Wang, J.M.; Taraboletti, G.; Matsushima, K.; Van Damme, J.; Mantovani, A. Induction of haptotactic migration of melanoma cells by neutrophil activating protein/interleukin-8. Biochem. Biophys. Res. Commun. 1990, 169, 165–170. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Patel, K.D. Regulation of matrix metalloproteinase-9 release from IL-8-stimulated human neutrophils. J. Leukoc Biol. 2005, 78, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Robilliard, L.D.; Kho, D.T.; Johnson, R.H.; Anchan, A.; O’Carroll, S.J.; Graham, E.S. The importance of multifrequency impedance sensing of endothelial barrier formation using ECIS technology for the generation of a strong and durable paracellular barrier. Biosensors 2018, 8, 64. [Google Scholar] [CrossRef]
- Huang, R.-L.; Teo, Z.; Chong, H.C.; Zhu, P.; Tan, M.J.; Tan, C.K.; Lam, C.R.I.; Sng, M.K.; Leong, D.T.W.; Tan, S.M.; et al. ANGPTL4 modulates vascular junction integrity by integrin signaling and disruption of intercellular VE-cadherin and claudin-5 clusters. Blood 2011, 118, 3990–4002. [Google Scholar] [CrossRef] [PubMed]
- Kebir, H.; Ifergan, I.; Alvarez, J.I.; Bernard, M.; Poirier, J.; Arbour, N.; Duquette, P.; Prat, A. Preferential recruitment of interferon-γ–expressing TH17 cells in multiple sclerosis. Ann. Neurol. 2009, 66, 390–402. [Google Scholar] [CrossRef]
- Argaw, A.T.; Zhang, Y.; Snyder, B.J.; Zhao, M.L.; Kopp, N.; Lee, S.C.; Raine, C.S.; Brosnan, C.F.; John, G.R. IL-1beta regulates blood-brain barrier permeability via reactivation of the hypoxia-angiogenesis program. J. Immunol. 2006, 177, 5574–5584. [Google Scholar] [CrossRef]
- Zhu, W.; London, N.R.; Gibson, C.C.; Davis, C.T.; Tong, Z.; Sorensen, L.K.; Shi, D.S.; Guo, J.; Smith, M.C.; Grossmann, A.H.; et al. Interleukin receptor activates a MYD88-ARNO-ARF6 cascade to disrupt vascular stability. Nature 2012, 492, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Skaria, T.; Burgener, J.; Bachli, E.; Schoedon, G. IL-4 causes hyperpermeability of vascular endothelial cells through Wnt5A signaling. PLoS ONE 2016, 11, e0156002. [Google Scholar] [CrossRef]
- Colombo, M.P.; Maccalli, C.; Mattei, S.; Melani, C.; Radrizzani, M.; Parmiani, G. Expression of cytokine genes, including IL-6, in human malignant melanoma cell lines. Melanoma Res. 1992, 2, 181–189. [Google Scholar] [CrossRef]
- Hoejberg, L.; Bastholt, L.; Schmidt, H. Interleukin-6 and melanoma. Melanoma Res. 2012, 22, 327–333. [Google Scholar] [CrossRef]
- Bar-Eli, M. Role of interleukin-8 in tumor growth and metastasis of human melanoma. Pathobiology 1999, 67, 12–18. [Google Scholar] [CrossRef]
- Holman, D.W.; Klein, R.S.; Ransohoff, R.M. The blood-brain barrier, chemokines and multiple sclerosis. Biochim. Biophys. Acta 2011, 1812, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Stamatovic, S.M.; Keep, R.F.; Kunkel, S.L.; Andjelkovic, A.V. Potential role of MCP-1 in endothelial cell tight junction ‘opening’: Signaling via Rho and Rho kinase. J. Cell Sci. 2003, 116, 4615–4628. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.J.; Hoos, A.; Bauer, J.; Boettcher, S.; Knust, M.; Weber, A.; Simonavicius, N.; Schneider, C.; Lang, M.; Sturzl, M.; et al. Endothelial CCR2 signaling induced by colon carcinoma cells enables extravasation via the JAK2-Stat5 and p38MAPK pathway. Cancer Cell 2012, 22, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Zozulya, A.L.; Reinke, E.; Baiu, D.C.; Karman, J.; Sandor, M.; Fabry, Z. Dendritic cell transmigration through brain microvessel endothelium is regulated by MIP-1α chemokine and matrix metalloproteinases. J. Immunol. 2007, 178, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Quandt, J.; Dorovini-Zis, K. The beta chemokines CCL4 and CCL5 enhance adhesion of specific CD4+ T cell subsets to human brain endothelial cells. J. Neuropathol. Exp. Neurol. 2004, 63, 350–362. [Google Scholar] [CrossRef] [PubMed]
- Alonso, S.R.; Tracey, L.; Ortiz, P.; Perez-Gomez, B.; Palacios, J.; Pollan, M.; Linares, J.; Serrano, S.; Saez-Castillo, A.I.; Sanchez, L.; et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007, 67, 3450–3460. [Google Scholar] [CrossRef]
- Zhou, Y.; Dai, D.L.; Martinka, M.; Su, M.; Zhang, Y.; Campos, E.I.; Dorocicz, I.; Tang, L.; Huntsman, D.; Nelson, C.; et al. Osteopontin expression correlates with melanoma invasion. J. Investig. Dermatol. 2005, 124, 1044–1052. [Google Scholar] [CrossRef]
- Bartolome, R.A.; Galvez, B.G.; Longo, N.; Baleux, F.; Van Muijen, G.N.; Sanchez-Mateos, P.; Arroyo, A.G.; Teixido, J. Stromal cell-derived factor-1alpha promotes melanoma cell invasion across basement membranes involving stimulation of membrane-type 1 matrix metalloproteinase and Rho GTPase activities. Cancer Res. 2004, 64, 2534–2543. [Google Scholar] [CrossRef]
- Mohammad, K.S.; Javelaud, D.; Fournier, P.G.; Niewolna, M.; McKenna, C.R.; Peng, X.H.; Duong, V.; Dunn, L.K.; Mauviel, A.; Guise, T.A. TGF-beta-RI kinase inhibitor SD-208 reduces the development and progression of melanoma bone metastases. Cancer Res. 2011, 71, 175–184. [Google Scholar] [CrossRef]
- Dvorak, H.F.; Brown, L.F.; Detmar, M.; Dvorak, A.M. Vascular permeability factor/vascular endothelial growth factor, microvascular hyperpermeability, and angiogenesis. Am. J. Pathol. 1995, 146, 1029–1039. [Google Scholar]
- Gavard, J.; Gutkind, J.S. VEGF controls endothelial-cell permeability by promoting the beta-arrestin-dependent endocytosis of VE-cadherin. Nat. Cell Biol. 2006, 8, 1223–1234. [Google Scholar] [CrossRef]
- Hoeben, A.; Landuyt, B.; Highley, M.S.; Wildiers, H.; Van Oosterom, A.T.; de Bruijn, E.A. Vascular endothelial growth factor and angiogenesis. Pharmacol. Rev. 2004, 56, 549–580. [Google Scholar] [CrossRef] [PubMed]
- Rajabi, P.; Neshat, A.; Mokhtari, M.; Rajabi, M.A.; Eftekhari, M.; Tavakoli, P. The role of VEGF in melanoma progression. J. Res. Med. Sci. 2012, 17, 534–539. [Google Scholar] [PubMed]
- Guo, L.; Li, S.Y.; Ji, F.Y.; Zhao, Y.F.; Zhong, Y.; Lv, X.J.; Wu, X.L.; Qian, G.S. Role of Angptl4 in vascular permeability and inflammation. Inflamm. Res. 2014, 63, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Kaddatz, K.; Adhikary, T.; Finkernagel, F.; Meissner, W.; Muller-Brusselbach, S.; Muller, R. Transcriptional profiling identifies functional interactions of TGF beta and PPAR beta/delta signaling: Synergistic induction of ANGPTL4 transcription. J. Biol. Chem. 2010, 285, 29469–29479. [Google Scholar] [CrossRef] [PubMed]
- Padua, D.; Zhang, X.H.; Wang, Q.; Nadal, C.; Gerald, W.L.; Gomis, R.R.; Massague, J. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell 2008, 133, 66–77. [Google Scholar] [CrossRef]
- Wilson, C.A.; Cajulis, E.E.; Green, J.L.; Olsen, T.M.; Chung, Y.A.; Damore, M.A.; Dering, J.; Calzone, F.J.; Slamon, D.J. HER-2 overexpression differentially alters transforming growth factor-beta responses in luminal versus mesenchymal human breast cancer cells. Breast Cancer Res. 2005, 7, R1058–R1079. [Google Scholar] [CrossRef]
- Izraely, S.; Ben-Menachem, S.; Sagi-Assif, O.; Meshel, T.; Marzese, D.M.; Ohe, S.; Zubrilov, I.; Pasmanik-Chor, M.; Hoon, D.S.B.; Witz, I.P. ANGPTL4 promotes the progression of cutaneous melanoma to brain metastasis. Oncotarget 2017, 8, 75778–75796. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Brekken, R.A.; Sage, E.H. SPARC, a matricellular protein: At the crossroads of cell–matrix. Matrix Biol. 2000, 19, 569–580. [Google Scholar] [CrossRef]
- Hartmann, A.; Kunz, M.; Kostlin, S.; Gillitzer, R.; Toksoy, A.; Brocker, E.B.; Klein, C.E. Hypoxia-induced up-regulation of angiogenin in human malignant melanoma. Cancer Res. 1999, 59, 1578–1583. [Google Scholar] [PubMed]
- Moshe, A.; Izraely, S.; Sagi-Assif, O.; Prakash, R.; Telerman, A.; Meshel, T.; Carmichael, T.; Witz, I.P. Cystatin C takes part in melanoma-microglia cross-talk: Possible implications for brain metastasis. Clin. Exp. Metastasis 2018, 35, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, P.; Richmond, A. Role of CXCL1 in tumorigenesis of melanoma. J. Leukoc. Biol. 2002, 72, 9–18. [Google Scholar]
- Wu, F.; Zhao, Y.; Jiao, T.; Shi, D.; Zhu, X.; Zhang, M.; Shi, M.; Zhou, H. CXCR2 is essential for cerebral endothelial activation and leukocyte recruitment during neuroinflammation. J. Neuroinflamm. 2015, 12, 98. [Google Scholar] [CrossRef]
- Chen, J.; Li, H.; Chen, H.; Hu, D.; Xing, Q.; Ren, G.; Luo, X. Dickkopf-1 inhibits the invasive activity of melanoma cells. Clin. Exp. Dermatol. 2012, 37, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Ueland, T.; Otterdal, K.; Lekva, T.; Halvorsen, B.; Gabrielsen, A.; Sandberg, W.J.; Paulsson-Berne, G.; Pedersen, T.M.; Folkersen, L.; Gullestad, L.; et al. Dickkopf-1 enhances inflammatory interaction between platelets and endothelial cells and shows increased expression in atherosclerosis. Arter. Thromb Vasc. Biol. 2009, 29, 1228–1234. [Google Scholar] [CrossRef] [PubMed]
- Braeuer, R.R.; Zigler, M.; Kamiya, T.; Dobroff, A.S.; Huang, L.; Choi, W.; McConkey, D.J.; Shoshan, E.; Mobley, A.K.; Song, R.; et al. Galectin-3 contributes to melanoma growth and metastasis via regulation of NFAT1 and autotaxin. Cancer Res. 2012, 72, 5757–5766. [Google Scholar] [CrossRef]
- Nangia-Makker, P.; Honjo, Y.; Sarvis, R.; Akahani, S.; Hogan, V.; Pienta, K.J.; Raz, A. Galectin-3 induces endothelial cell morphogenesis and angiogenesis. Am. J. Pathol. 2000, 156, 899–909. [Google Scholar] [CrossRef]
- Weide, B.; Schafer, T.; Martens, A.; Kuzkina, A.; Uder, L.; Noor, S.; Garbe, C.; Harter, P.N.; Mittelbronn, M.; Wischhusen, J. High GDF-15 serum levels independently correlate with poorer overall survival of patients with tumor-free stage III and unresectable stage IV melanoma. J. Investig. Dermatol. 2016, 136, 2444–2452. [Google Scholar] [CrossRef]
- Wang, H.; Shen, S.S.; Wang, H.; Diwan, A.H.; Zhang, W.; Fuller, G.N.; Prieto, V.G. Expression of insulin-like growth factor-binding protein 2 in melanocytic lesions. J. Cutan. Pathol. 2003, 30, 599–605. [Google Scholar] [CrossRef]
- Chen, H.R.; Chao, C.H.; Liu, C.C.; Ho, T.S.; Tsai, H.P.; Perng, G.C.; Lin, Y.S.; Wang, J.R.; Yeh, T.M. Macrophage migration inhibitory factor is critical for dengue NS1-induced endothelial glycocalyx degradation and hyperpermeability. PLoS Pathog. 2018, 14, e1007033. [Google Scholar] [CrossRef] [PubMed]
- Tanese, K.; Hashimoto, Y.; Berkova, Z.; Wang, Y.; Samaniego, F.; Lee, J.E.; Ekmekcioglu, S.; Grimm, E.A. Cell Surface CD74-MIF Interactions Drive Melanoma Survival in Response to Interferon-gamma. J. Investig. Dermatol. 2015, 135, 2901. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, R.; Nakano, T.; Wakabayashi, I. Progranulin and granulin-like protein as novel VEGF-independent angiogenic factors derived from human mesothelioma cells. Oncogene 2017, 36, 714–722. [Google Scholar] [CrossRef]
- Voshtani, R.; Song, M.; Wang, H.; Li, X.; Zhang, W.; Tavallaie, M.S.; Yan, W.; Sun, J.; Wei, F.; Ma, X. Progranulin promotes melanoma progression by inhibiting natural killer cell recruitment to the tumor microenvironment. Cancer Lett. 2019, 465, 24–35. [Google Scholar] [CrossRef]
- Girotti, M.R.; Fernandez, M.; Lopez, J.A.; Camafeita, E.; Fernandez, E.A.; Albar, J.P.; Benedetti, L.G.; Valacco, M.P.; Brekken, R.A.; Podhajcer, O.L.; et al. SPARC promotes cathepsin B-mediated melanoma invasiveness through a collagen I/alpha2beta1 integrin axis. J. Investig. Dermatol. 2011, 131, 2438–2447. [Google Scholar] [CrossRef]
- Robert, G.; Gaggioli, C.; Bailet, O.; Chavey, C.; Abbe, P.; Aberdam, E.; Sabatie, E.; Cano, A.; de Herreros, A.G.; Ballotti, R.; et al. SPARC represses E-cadherin and induces mesenchymal transition during melanoma development. Cancer Res. 2006, 66, 7516–7523. [Google Scholar] [CrossRef]
- Grahovac, J.; Becker, D.; Wells, A. Melanoma cell invasiveness is promoted at least in part by the epidermal growth factor–like repeats of tenascin-C. J. Investig. Dermatol. 2013, 133, 210–220. [Google Scholar] [CrossRef]
- Lo, C.M.; Keese, C.R.; Giaever, I. pH changes in pulsed CO2 incubators cause periodic changes in cell morphology. Exp. Cell Res. 1994, 213, 391–397. [Google Scholar] [CrossRef]
- Tiruppathi, C.; Malik, A.B.; Del Vecchio, P.J.; Keese, C.R.; Giaever, I. Electrical method for detection of endothelial cell shape change in real time: Assessment of endothelial barrier function. Proc. Natl. Acad. Sci. USA 1992, 89, 7919–7923. [Google Scholar] [CrossRef] [PubMed]
- Giaever, I.; Keese, C.R. Micromotion of mammalian cells measured electrically. Proc. Natl. Acad. Sci. USA 1991, 88, 7896. [Google Scholar] [CrossRef] [PubMed]
- Burkert, K.; Moodley, K.; Angel, C.E.; Brooks, A.; Graham, E.S. Detailed analysis of inflammatory and neuromodulatory cytokine secretion from human NT2 astrocytes using multiplex bead array. Neurochem. Int. 2012, 60, 573–580. [Google Scholar] [CrossRef]
- Chao, C.H.; Chen, H.R.; Chuang, Y.C.; Yeh, T.M. Macrophage migration inhibitory factor-induced autophagy contributes to thrombin-triggered endothelial hyperpermeability in sepsis. Shock 2018, 50, 103–111. [Google Scholar] [CrossRef]
- Lei, X.; Shi, F.; Basu, D.; Huq, A.; Routhier, S.; Day, R.; Jin, W. Proteolytic processing of angiopoietin-like protein 4 by proprotein convertases modulates its inhibitory effects on lipoprotein lipase activity. J. Biol. Chem. 2011, 286, 15747–15756. [Google Scholar] [CrossRef]
- Stawowy, P.; Kallisch, H.; Stawowy, N.B.P.; Stibenz, D.; Veinot, J.P.; Gräfe, M.; Seidah, N.G.; Chrétien, M.; Fleck, E.; Graf, K. Immunohistochemical localization of subtilisin/kexin-like proprotein convertases in human atherosclerosis. Virchows Arch. 2005, 446, 351–359. [Google Scholar] [CrossRef] [PubMed]



| Secretory Molecule | Common Alias | Function (Relevant to Metastasis or Vascular Modulation) | References |
|---|---|---|---|
| ANGPTL-4 | HFARP | TJ disruption | [24] |
| IFN-γ | - | Lymphocyte extravasation (Thelper17) | [2,3,25] |
| IL-1β | - | Influences VE-Cadherin arrangement and increases vascular permeability | [26,27] |
| IL-4 | BSF-1 | Vascular hyperpermeability | [28] |
| IL-6 | BSF-2 | Elevated in metastatic melanoma | [29,30] |
| IL-8 | CXCL8 | Elevated expression closely associated with melanoma invasion and metastasis | [20,31] |
| IP-10 | CXCL10 | Lymphocyte extravasation (T-cell) | [4,32] |
| MCP-1 | CCL2 | Induces endothelial TJ opening. Vascular permeability | [33,34] |
| MIP-1α | CCL3 | Leukocyte extravasation (Dendritic cell) | [4,35] |
| MIP-1β | CCL4 | Lymphocyte extravasation (T-Cell CD4+) | [4,36] |
| Osteopontin (OPN) | BSP-1 | Expression associated with tumour invasion. | [37,38] |
| SDF-1α | CXCL12 | Melanoma matrix invasion | [39] |
| TGFβ-1 | - | Cancer progression, angiogenesis, invasion and metastasis. Osteolytic metastasis in melanoma | [12,40] |
| VEGF | VPF | Elevated expression closely associated with metastasis. Increases endothelial permeability. Reversible VE-Cadherin endocytosis | [26,41,42,43,44] |
| Secretory Molecule | Function (Relevant to Metastasis or Vascular Modulation) | References |
|---|---|---|
| Angiogenin | Associated with metastatic potential | [52] |
| Cystatin C | Implicated in melanoma brain metastasis | [53] |
| CXCL-1 | Tumorigenesis Endothelial activation and leukocyte recruitment | [54,55] |
| DKK-1 | Inhibits melanoma invasiveness Increases platelet mediated endothelial activation | [56,57] |
| Galectin-3 | Contributes to metastasis. Induces endothelial angiogenesis | [58,59] |
| GDF-15 | High expression correlated with reduced overall survival in patients with melanoma | [60] |
| IGFBP-2 | Increased in malignancy | [61] |
| MIF | Tumour survival Endothelial permeability | [62,63] |
| Progranulin | VEGF independent angiogenesis. Correlated with melanoma survival | [64,65] |
| SPARC | Highly implicated in EMT processes and overexpression in melanoma Mediates invasiveness | [66,67] |
| Tenascin C | Upregulated in melanoma but likely supportive only to cellular adhesion and ECM movement. | [68] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anchan, A.; Martin, O.; Hucklesby, J.J.W.; Finlay, G.; Johnson, R.H.; Robilliard, L.D.; O’Carroll, S.J.; Angel, C.E.; Graham, E.S. Analysis of Melanoma Secretome for Factors That Directly Disrupt the Barrier Integrity of Brain Endothelial Cells. Int. J. Mol. Sci. 2020, 21, 8193. https://doi.org/10.3390/ijms21218193
Anchan A, Martin O, Hucklesby JJW, Finlay G, Johnson RH, Robilliard LD, O’Carroll SJ, Angel CE, Graham ES. Analysis of Melanoma Secretome for Factors That Directly Disrupt the Barrier Integrity of Brain Endothelial Cells. International Journal of Molecular Sciences. 2020; 21(21):8193. https://doi.org/10.3390/ijms21218193
Chicago/Turabian StyleAnchan, Akshata, Olivia Martin, James J. W. Hucklesby, Graeme Finlay, Rebecca H. Johnson, Laverne D. Robilliard, Simon J. O’Carroll, Catherine E. Angel, and E Scott Graham. 2020. "Analysis of Melanoma Secretome for Factors That Directly Disrupt the Barrier Integrity of Brain Endothelial Cells" International Journal of Molecular Sciences 21, no. 21: 8193. https://doi.org/10.3390/ijms21218193
APA StyleAnchan, A., Martin, O., Hucklesby, J. J. W., Finlay, G., Johnson, R. H., Robilliard, L. D., O’Carroll, S. J., Angel, C. E., & Graham, E. S. (2020). Analysis of Melanoma Secretome for Factors That Directly Disrupt the Barrier Integrity of Brain Endothelial Cells. International Journal of Molecular Sciences, 21(21), 8193. https://doi.org/10.3390/ijms21218193






