Transcriptomic Analysis of Age-Associated Periventricular Lesions Reveals Dysregulation of the Immune Response
Abstract
:1. Introduction
2. Results
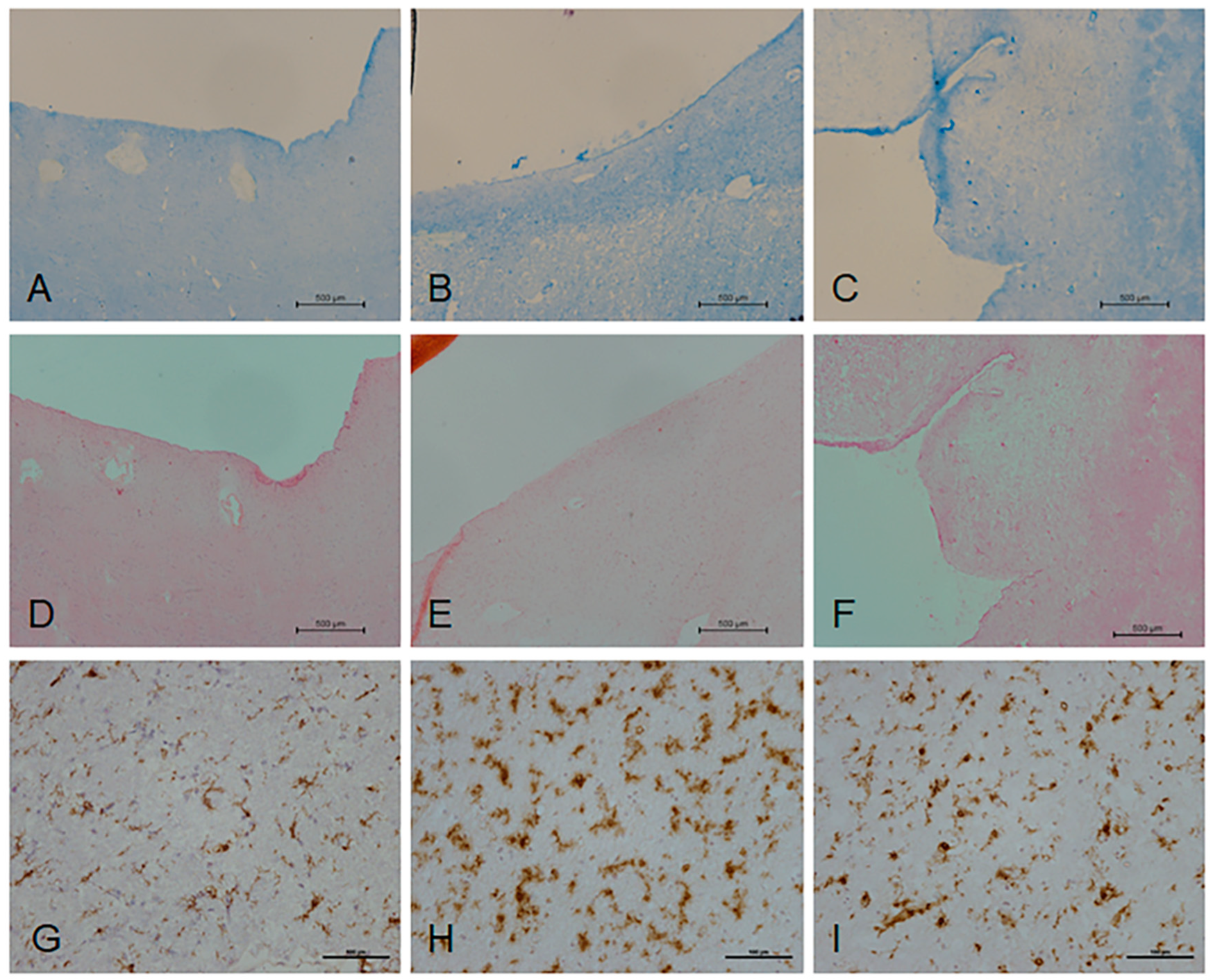
2.1. Histological Characterisation of Periventricular White Matter
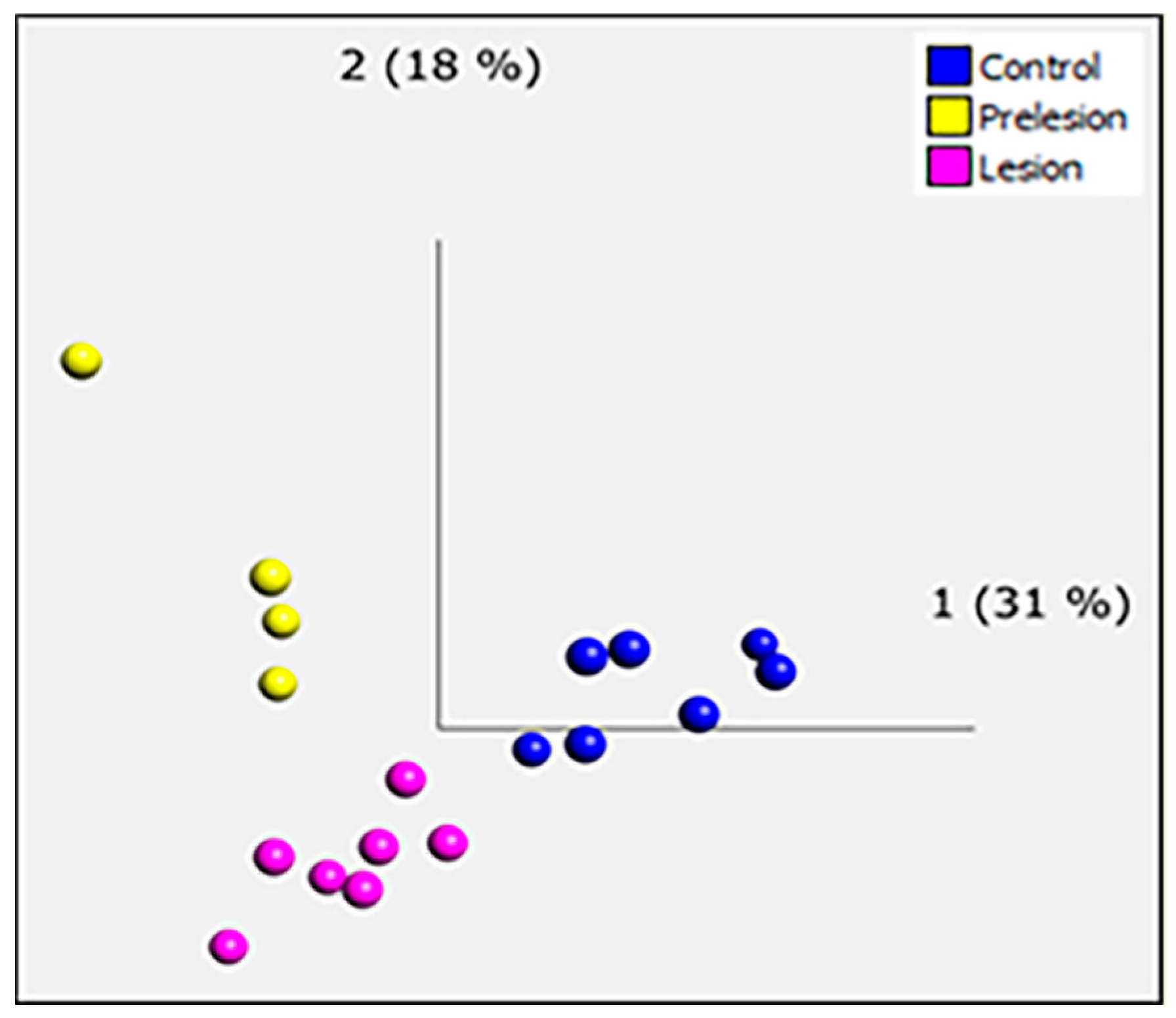
2.2. Microarray Analysis
2.3. EnrichR
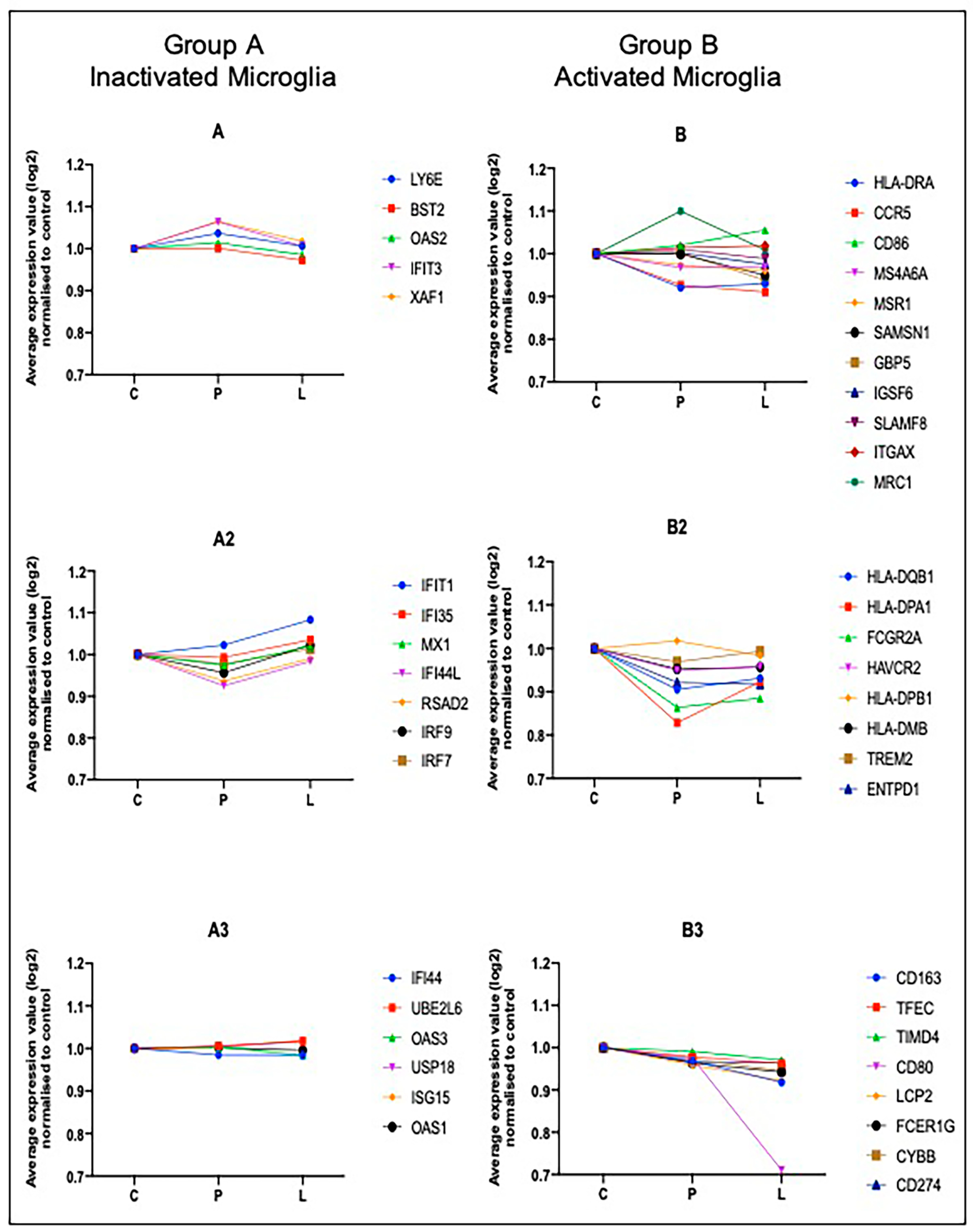
2.4. Computational Deconvolution of Microglial Gene Expression
2.5. Validation of Candidate Gene Expression by NanoString nCounter
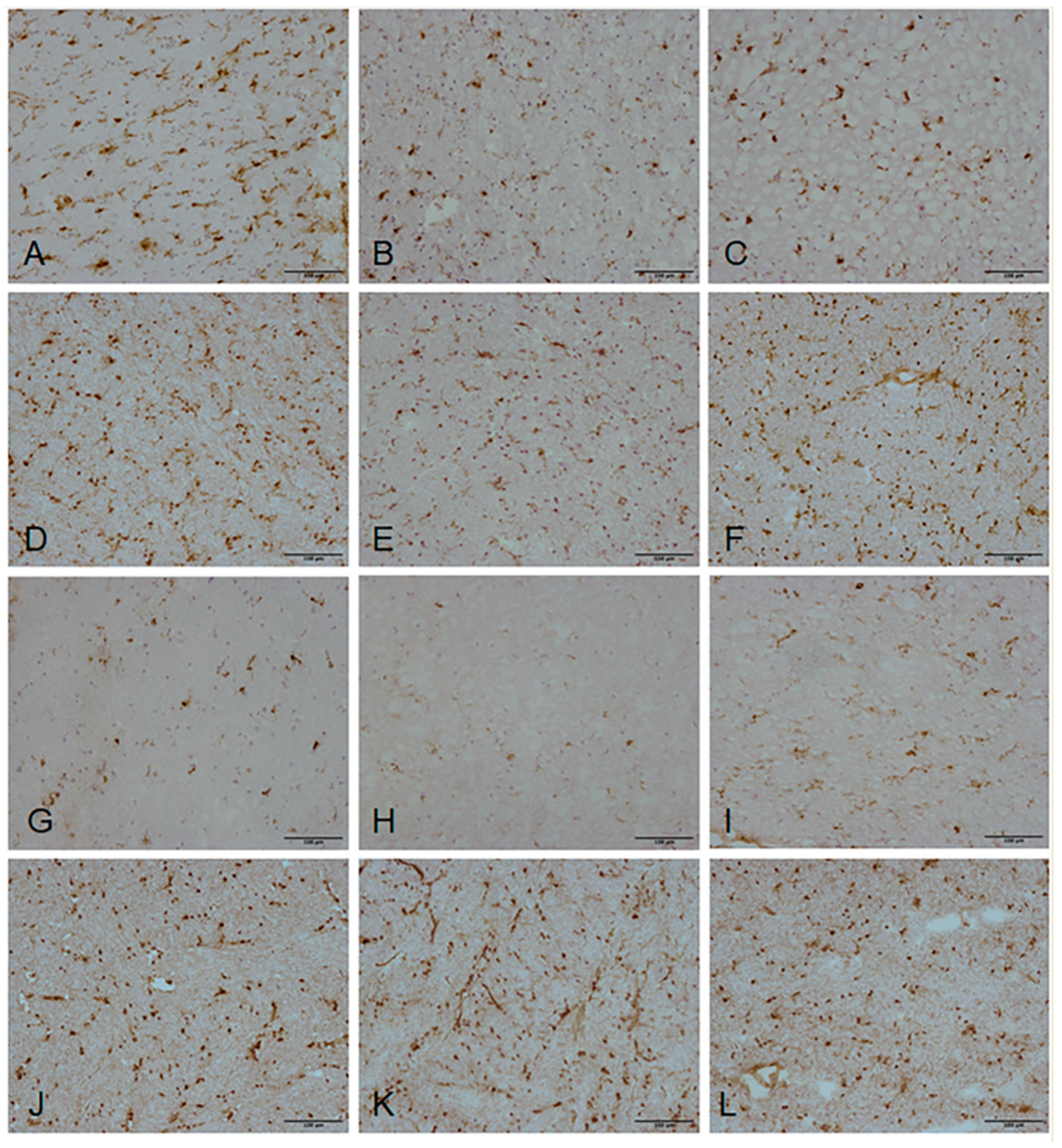
2.6. Validation of Candidate Gene Expression by Immunohistochemistry
3. Discussion
4. Materials and Methods
4.1. Human CNS Tissue
4.2. Histological Characterisation
4.3. Laser Capture Microdissection (LCM)
4.4. Microarray
4.5. Transcriptomic Analysis of Microarrays
4.6. Deconvolution of the Microglial Transcriptome
4.7. Validation of Gene Expression Changes—NanoString
4.8. Validation of Gene Expression Changes—Immunohistochemistry
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABC-HRP | avidin-biotin complex-horse radish peroxidase |
| ACTB | actin cytoplasmic 1 |
| AKT3 | V-akt murine thymoma viral oncogene homolog 3 |
| BBB | blood brain-barrier |
| BST2 | CD317, tethrin |
| CACNA1E | calcium voltage-gated channel subunit alpha 1E |
| CALM | calmodulin |
| CAMK2A | calcium/calmodulin-dependent protein kinase II alpha |
| CAMK4 | calcium/calmodulin-dependent protein kinase IV |
| cAMP | cyclic adenosine monophosphateP |
| CCL2 | C-C motif chemokine ligand 2 |
| CCL3 | C-C motif chemokine ligand 3 |
| CCL4 | C-C motif chemokine ligand 4 |
| CCR5 | C-C motif chemokine receptor 5 (gene/pseudogene) |
| CD163 | CD163 molecule, Scavenger Receptor Cysteine-Rich Type 1 Protein M130 |
| CD274 | cluster of differentiation 274, programmed death-ligand 1 |
| CD74 | CD74 molecule, HLA class II histocompatibility antigen gamma chain |
| CD80 | co-stimulatory molecule B7-2 |
| CD86 | cluster of differentiation 86, B7-2 |
| CD8A | CD8a molecule, T-cell surface glycoprotein CD8 alpha chain |
| cDNA | complementary DNA |
| CFAS | Cognitive Function and Ageing Study |
| CNS | central nervous system |
| COL6A3 | collagen type VI alpha 3 chain |
| CX3CR1 | C-X3-C motif chemokine receptor 1 |
| CYBB | cytochrome b-245, beta polypeptide |
| DAVID | Database for Annotation, Visualization and Integrated Discovery |
| DEPC | diethylpyrocarbonate |
| DSCL | deep subcortical lesions |
| ENTPD1 | ectonucleoside triphosphate diphosphohydrolase 1 |
| EPSTI1 | epithelial stromal interaction protein 1 |
| FCER1G | Fc fragment of IgE receptor Ig |
| FCGR1A | Fc fragment of immunoglobulin gamma Fc receptor 1A |
| FCGR2A | Fc fragment of immunoglobulin gamma Fc receptor 2A |
| FGL2 | fibrinogen like 2 |
| FPR1 | formyl peptide receptor 1 |
| GABA | gamma-aminobutyric acid |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| GBP5 | guanylate-binding protein 5 |
| GEO | gene expression omnibus |
| GRIA | glutamate receptor, ionotropic, AMPA |
| GRIN1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| H&E | haematoxylin and eosin |
| HAVCR2 | hepatitis A virus cellular receptor 2 |
| HLA-DMB | major histocompatibility complex, class II, DM beta |
| HLA-DPA1 | major histocompatibility complex, class II, DP alpha 1 |
| HLA-DPB1 | major histocompatibility complex, class II, DP beta 1 |
| HLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 |
| HLA-DRA | major histocompatibility complex, class II, DR alpha |
| IFI35 | interferon induced protein 35 |
| IFI44 | interferon induced protein 44 |
| IFI44L | interferon induced protein 44-like |
| IFIT1 | interferon induced protein with tetratricopeptide repeats 1 |
| IFIT3 | interferon induced protein with tetratricopeptide repeats 3 |
| IFNAR1 | interferon alpha receptor subunit 1 |
| IGSF6 | immunoglobulin superfamily member 6 |
| IHC | immunohistochemistry |
| IKBKB | inhibitor of nuclear factor kappa B kinase subunit beta |
| IL1A | interleukin 1 alpha |
| IL1B | interleukin 1 beta |
| IRF7 | interferon regulatory factor 7 |
| IRF9 | interferon regulatory factor 9 |
| ISG15 | interferon-stimulated gene 15 |
| ITGAX | integrin, alpha X (complement component 3 receptor 4 subunit), CD11c |
| IVT | in vitro transcription |
| JUN | jun proto-oncogene, AP-1 transcription factor subunit |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LCM | laser capture microdissection |
| LCP2 | lymphocyte cytosolic protein 2 |
| LFB | luxol fast blue |
| LY6E | lymphocyte antigen 6E |
| MAPK | mitogen-activated protein kinase |
| MHC-II | major histocompatibility complex class II |
| MRC1 | mannose receptor C-type 1 |
| MRI | magnetic resnance imaging |
| mRNA | messenger ribonucleic acid |
| MS4A6A | membrane spanning 4-domains A6A |
| MSR1 | macrophage scavenger receptor 1 |
| MX1 | MX dynamin like GTPase 1 |
| OAS1 | 2’-5’-oligoadenylate synthetase 1 |
| OAS2 | 2’-5’-oligoadenylate synthetase 2 |
| OAS3 | 2’-5’-oligoadenylate synthetase 3 |
| PCA | principal component analysis |
| PHF21A | PHD finger protein 21A |
| PI3K-Akt | phosphatidylinositol 3-kinase-protein kinase |
| PTGS2 | prostaglandin-endoperoxide synthase 2 |
| PVL | periventricular lesions |
| QC | quality control |
| REC | research ethics committee |
| RPLP0 | 60S acidic ribosomal protein P0 |
| RSAD2 | radical S-adenosyl methionine domain containing 2 |
| RT | room temperature |
| SAMSN1 | SAM domain, SH3 domain and nuclear localisation signals 1 |
| SLAMF8 | SLAM family member 8 |
| TAC | transcriptome analysis console |
| TFEC | transcription factor EC |
| TREM2 | triggering receptor expressed on myeloid cells 2 |
| UBE2L6 | ubiquitin conjugating enzyme E2 L6 |
| USP18 | ubiquitin specific peptidase 18 |
| WM | white matter |
| WML | white matter lesions |
| XAF1 | XIAP-associated factor 1 |
References
- Prins, N.D.; Scheltens, P. White matter hyperintensities, cognitive impairment and dementia: An update. Nat. Rev. Neurol. 2015, 11, 157–165. [Google Scholar] [CrossRef]
- Fernando, M.S.; Ince, P.G.; Function, M.R.C.C. Ageing Neuropathology Study G: Vascular pathologies and cognition in a population-based cohort of elderly people. J. Neurol. Sci. 2004, 226, 13–17. [Google Scholar] [CrossRef]
- Matthews, F.E.; Brayne, C.; Lowe, J.; McKeith, I.; Wharton, S.B.; Ince, P. Epidemiological pathology of dementia: Attributable-risks at death in the Medical Research Council Cognitive Function and Ageing Study. PLoS Med. 2009, 6, e1000180. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.L.; Chen, W.; Cai, W.J.; Hu, H.; Xu, W.; Wang, Z.T.; Cao, X.P.; Tan, L.; Yu, J.T. Alzheimer’s Disease Neuroimaging I: Associations of White Matter Hyperintensities with Cognitive Decline: A Longitudinal Study. J. Alzheimers Dis. 2020, 73, 759–768. [Google Scholar] [CrossRef]
- Biesbroek, J.M.; Weaver, N.A.; Biessels, G.J. Lesion location and cognitive impact of cerebral small vessel disease. Clin. Sci. 2017, 131, 715–728. [Google Scholar] [CrossRef]
- Gouw, A.A.; Seewann, A.; van der Flier, W.M.; Barkhof, F.; Rozemuller, A.M.; Scheltens, P.; Geurts, J.J. Heterogeneity of small vessel disease: A systematic review of MRI and histopathology correlations. J. Neurol. Neurosurg. Psychiatry 2011, 82, 126–135. [Google Scholar] [CrossRef]
- Wharton, S.B.; Simpson, J.E.; Brayne, C.; Ince, P.G. Age-associated white matter lesions: The MRC Cognitive Function and Ageing Study. Brain Pathol. 2015, 25, 35–43. [Google Scholar] [CrossRef]
- Alber, J.; Alladi, S.; Bae, H.J.; Barton, D.A.; Beckett, L.A.; Bell, J.M.; Berman, S.E.; Biessels, G.J.; Black, S.E.; Bos, I.; et al. White matter hyperintensities in vascular contributions to cognitive impairment and dementia (VCID): Knowledge gaps and opportunities. Alzheimers Dement. 2019, 5, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Hashemiaghdam, A.; Mroczek, M. Microglia heterogeneity and neurodegeneration: The emerging paradigm of the role of immunity in Alzheimer’s disease. J. Neuroimmunol. 2020, 341, 577185. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.E.; Ince, P.G.; Higham, C.E.; Gelsthorpe, C.H.; Fernando, M.S.; Matthews, F.; Forster, G.; O’Brien, J.T.; Barber, R.; Kalaria, R.N.; et al. Microglial activation in white matter lesions and nonlesional white matter of ageing brains. Neuropathol. Appl. Neurobiol. 2007, 33, 670–683. [Google Scholar] [CrossRef]
- Simpson, J.E.; Fernando, M.S.; Clark, L.; Ince, P.G.; Matthews, F.; Forster, G.; O’Brien, J.T.; Barber, R.; Kalaria, R.N.; Brayne, C.; et al. White matter lesions in an unselected cohort of the elderly: Astrocytic, microglial and oligodendrocyte precursor cell responses. Neuropathol. Appl. Neurobiol. 2007, 33, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Waller, R.; Baxter, L.; Fillingham, D.J.; Coelho, S.; Pozo, J.M.; Mozumder, M.; Frangi, A.F.; Ince, P.G.; Simpson, J.E.; Highley, J.R. Iba-1-/CD68+ microglia are a prominent feature of age-associated deep subcortical white matter lesions. PLoS ONE 2019, 14, e0210888. [Google Scholar] [CrossRef]
- Fernando, M.S.; Simpson, J.E.; Matthews, F.; Brayne, C.; Lewis, C.E.; Barber, R.; Kalaria, R.N.; Forster, G.; Esteves, F.; Wharton, S.B.; et al. White matter lesions in an unselected cohort of the elderly: Molecular pathology suggests origin from chronic hypoperfusion injury. Stroke 2006, 37, 1391–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, S.M.; Jansen, J.F.A.; Zhang, C.E.; Hoff, E.I.; Staals, J.; van Oostenbrugge, R.J.; Backes, W.H. Blood-brain barrier impairment and hypoperfusion are linked in cerebral small vessel disease. Neurology 2019, 92, e1669–e1677. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.E.; Wharton, S.B.; Cooper, J.; Gelsthorpe, C.; Baxter, L.; Forster, G.; Shaw, P.J.; Savva, G.; Matthews, F.E.; Brayne, C.; et al. Alterations of the blood-brain barrier in cerebral white matter lesions in the ageing brain. Neurosci. Lett. 2010, 486, 246–251. [Google Scholar] [CrossRef]
- Zhang, C.E.; Wong, S.M.; Uiterwijk, R.; Backes, W.H.; Jansen, J.F.A.; Jeukens, C.; van Oostenbrugge, R.J.; Staals, J. Blood-brain barrier leakage in relation to white matter hyperintensity volume and cognition in small vessel disease and normal aging. Brain Imaging Behav. 2019, 13, 389–395. [Google Scholar] [CrossRef] [Green Version]
- Freeze, W.M.; Jacobs, H.I.L.; de Jong, J.J.; Verheggen, I.C.M.; Gronenschild, E.; Palm, W.M.; Hoff, E.I.; Wardlaw, J.M.; Jansen, J.F.A.; Verhey, F.R.; et al. White matter hyperintensities mediate the association between blood-brain barrier leakage and information processing speed. Neurobiol. Aging 2020, 85, 113–122. [Google Scholar] [CrossRef]
- McAleese, K.E.; Walker, L.; Graham, S.; Moya, E.L.J.; Johnson, M.; Erskine, D.; Colloby, S.J.; Dey, M.; Martin-Ruiz, C.; Taylor, J.P.; et al. Parietal white matter lesions in Alzheimer’s disease are associated with cortical neurodegenerative pathology, but not with small vessel disease. Acta Neuropathol. 2017, 134, 459–473. [Google Scholar] [CrossRef] [Green Version]
- Simpson, J.E.; Hosny, O.; Wharton, S.B.; Heath, P.R.; Holden, H.; Fernando, M.S.; Matthews, F.; Forster, G.; O’Brien, J.T.; Barber, R.; et al. Microarray RNA expression analysis of cerebral white matter lesions reveals changes in multiple functional pathways. Stroke 2009, 40, 369–375. [Google Scholar] [CrossRef] [Green Version]
- Gupta, N.; Simpkins, A.N.; Hitomi, E.; Dias, C.; Leigh, R. Investigators NIHNHoS: White Matter Hyperintensity-Associated Blood-Brain Barrier Disruption and Vascular Risk Factors. J. Stroke Cerebrovasc. Dis. 2018, 27, 466–471. [Google Scholar] [CrossRef]
- Kloppenborg, R.P.; Nederkoorn, P.J.; Geerlings, M.I.; van den Berg, E. Presence and progression of white matter hyperintensities and cognition: A meta-analysis. Neurology 2014, 82, 2127–2138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fournier, M.L.; Paulson, A.; Pavelka, N.; Mosley, A.L.; Gaudenz, K.; Bradford, W.D.; Glynn, E.; Li, H.; Sardiu, M.E.; Fleharty, B.; et al. Delayed correlation of mRNA and protein expression in rapamycin-treated cells and a role for Ggc1 in cellular sensitivity to rapamycin. Mol. Cell Proteom. 2010, 9, 271–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ajami, B.; Samusik, N.; Wieghofer, P.; Ho, P.P.; Crotti, A.; Bjornson, Z.; Prinz, M.; Fantl, W.J.; Nolan, G.P.; Steinman, L. Single-cell mass cytometry reveals distinct populations of brain myeloid cells in mouse neuroinflammation and neurodegeneration models. Nat. Neurosci. 2018, 21, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Polyak, M.J.; Vivithanaporn, P.; Maingat, F.G.; Walsh, J.G.; Branton, W.; Cohen, E.A.; Meeker, R.; Power, C. Differential type 1 interferon-regulated gene expression in the brain during AIDS: Interactions with viral diversity and neurovirulence. FASEB J. 2013, 27, 2829–2844. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Gomez, J.A.; Kavanagh, E.; Engskog-Vlachos, P.; Engskog, M.K.R.; Herrera, A.J.; Espinosa-Oliva, A.M.; Joseph, B.; Hajji, N.; Venero, J.L.; Burguillos, M.A. Microglia: Agents of the CNS Pro-Inflammatory Response. Cells 2020, 9, 1717. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Raj, D.; Saiepour, N.; Van Dam, D.; Brouwer, N.; Holtman, I.R.; Eggen, B.J.L.; Moller, T.; Tamm, J.A.; Abdourahman, A.; et al. Immune hyperreactivity of Abeta plaque-associated microglia in Alzheimer’s disease. Neurobiol. Aging 2017, 55, 115–122. [Google Scholar] [CrossRef]
- Baron, R.; Babcock, A.A.; Nemirovsky, A.; Finsen, B.; Monsonego, A. Accelerated microglial pathology is associated with Abeta plaques in mouse models of Alzheimer’s disease. Aging Cell 2014, 13, 584–595. [Google Scholar] [CrossRef]
- Busse, S.; Steiner, J.; Alter, J.; Dobrowolny, H.; Mawrin, C.; Bogerts, B.; Hartig, R.; Busse, M. Expression of HLA-DR, CD80, and CD86 in Healthy Aging and Alzheimer’s Disease. J. Alzheimers Dis. 2015, 47, 177–184. [Google Scholar] [CrossRef]
- Li, T.; Zhu, J. Entanglement of CCR5 and Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 209. [Google Scholar] [CrossRef]
- Roy, E.R.; Wang, B.; Wan, Y.W.; Chiu, G.; Cole, A.; Yin, Z.; Propson, N.E.; Xu, Y.; Jankowsky, J.L.; Liu, Z.; et al. Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J. Clin. Investig. 2020, 130, 1912–1930. [Google Scholar] [CrossRef] [PubMed]
- Sebastian Monasor, L.; Muller, S.A.; Colombo, A.V.; Tanrioever, G.; Konig, J.; Roth, S.; Liesz, A.; Berghofer, A.; Piechotta, A.; Prestel, M.; et al. Fibrillar Abeta triggers microglial proteome alterations and dysfunction in Alzheimer mouse models. Elife 2020, 9, e54083. [Google Scholar] [CrossRef]
- Askew, K.; Gomez-Nicola, D. A story of birth and death: Insights into the formation and dynamics of the microglial population. Brain Behav. Immun. 2018, 69, 9–17. [Google Scholar] [CrossRef]
- Friedman, B.A.; Srinivasan, K.; Ayalon, G.; Meilandt, W.J.; Lin, H.; Huntley, M.A.; Cao, Y.; Lee, S.H.; Haddick, P.C.G.; Ngu, H.; et al. Diverse Brain Myeloid Expression Profiles Reveal Distinct Microglial Activation States and Aspects of Alzheimer’s Disease Not Evident in Mouse Models. Cell Rep. 2018, 22, 832–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eastel, J.M.; Lam, K.W.; Lee, N.L.; Lok, W.Y.; Tsang, A.H.F.; Pei, X.M.; Chan, A.K.C.; Cho, W.C.S.; Wong, S.C.C. Application of NanoString technologies in companion diagnostic development. Expert Rev. Mol. Diagn. 2019, 19, 591–598. [Google Scholar] [CrossRef]
- Simpson, J.E.; Newcombe, J.; Cuzner, M.L.; Woodroofe, M.N. Expression of monocyte chemoattractant protein-1 and other beta-chemokines by resident glia and inflammatory cells in multiple sclerosis lesions. J. Neuroimmunol. 1998, 84, 238–249. [Google Scholar] [CrossRef]
- Buschmann, J.P.; Berger, K.; Awad, H.; Clarner, T.; Beyer, C.; Kipp, M. Inflammatory response and chemokine expression in the white matter corpus callosum and gray matter cortex region during cuprizone-induced demyelination. J. Mol. Neurosci. 2012, 48, 66–76. [Google Scholar] [CrossRef] [Green Version]
- Cui, L.Y.; Chu, S.F.; Chen, N.H. The role of chemokines and chemokine receptors in multiple sclerosis. Int. Immunopharmacol. 2020, 83, 106314. [Google Scholar] [CrossRef] [PubMed]
- Baltan, S. Ischemic injury to white matter: An age-dependent process. Neuroscientist 2009, 15, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Doyle, S.; Hansen, D.B.; Vella, J.; Bond, P.; Harper, G.; Zammit, C.; Valentino, M.; Fern, R. Vesicular glutamate release from central axons contributes to myelin damage. Nat. Commun. 2018, 9, 1032. [Google Scholar] [CrossRef] [Green Version]
- Karadottir, R.; Cavelier, P.; Bergersen, L.H.; Attwell, D. NMDA receptors are expressed in oligodendrocytes and activated in ischaemia. Nature 2005, 438, 1162–1166. [Google Scholar] [CrossRef] [Green Version]
- Fields, R.D. A new mechanism of nervous system plasticity: Activity-dependent myelination. Nat. Rev. Neurosci. 2015, 16, 756–767. [Google Scholar] [CrossRef]
- Schurch, N.J.; Schofield, P.; Gierlinski, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.G.; Owen-Hughes, T.; et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef] [Green Version]
- Scheltens, P.; Barkhof, F.; Leys, D.; Pruvo, J.P.; Nauta, J.J.; Vermersch, P.; Steinling, M.; Valk, J. A semiquantative rating scale for the assessment of signal hyperintensities on magnetic resonance imaging. J. Neurol. Sci. 1993, 114, 7–12. [Google Scholar] [CrossRef]
- Avila Cobos, F.; Vandesompele, J.; Mestdagh, P.; De Preter, K. Computational deconvolution of transcriptomics data from mixed cell populations. Bioinformatics 2018, 34, 1969–1979. [Google Scholar] [CrossRef] [Green Version]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Zhang, Y.; Sloan, S.A.; Clarke, L.E.; Caneda, C.; Plaza, C.A.; Blumenthal, P.D.; Vogel, H.; Steinberg, G.K.; Edwards, M.S.; Li, G.; et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 2016, 89, 37–53. [Google Scholar] [CrossRef] [Green Version]

| Pathway | Count | p-Value |
|---|---|---|
| hsa04612: Antigen processing and presentation | 20 | 9.17 × 10−6 |
| hsa04145: Phagosome | 24 | 0.003 |
| hsa04672: Intestinal immune network for IgA production | 11 | 0.004 |
| hsa04662: B cell receptor signalling pathway | 12 | 0.026 |
| Pathway | Count | p-Value |
|---|---|---|
| hsa04020:Calcium signalling pathway | 42 | 3.38 × 10−9 |
| hsa04728:Dopaminergic synapse | 34 | 5.29 ×10−9 |
| hsa04724:Glutamatergic synapse | 30 | 6.28 × 10−8 |
| hsa04725:Cholinergic synapse | 29 | 1.29 × 10−7 |
| hsa04727:GABAergic synapse | 23 | 1.82 × 10−6 |
| hsa04612:Antigen processing and presentation | 20 | 1.58 × 10−5 |
| hsa04024:cAMP signalling pathway | 34 | 1.54 × 10−4 |
| hsa04010:MAPK signalling pathway | 40 | 2.23 × 10−4 |
| hsa04145:Phagosome | 25 | 0.002 |
| hsa04062:Chemokine signalling pathway | 28 | 0.005 |
| hsa04151:PI3K-Akt signalling pathway | 45 | 0.005 |
| hsa04660:T cell receptor signalling pathway | 17 | 0.011 |
| hsa04662:B cell receptor signalling pathway | 13 | 0.014 |
| hsa04068:FoxO signalling pathway | 19 | 0.038 |
| Pathway | Count | p-Value |
|---|---|---|
| hsa04020:Calcium signalling pathway | 46 | 4.71 × 10−11 |
| hsa04724:Glutamatergic synapse | 29 | 3.97 ×10−7 |
| hsa04727:GABAergic synapse | 21 | 3.58 × 10−5 |
| hsa04725:Cholinergic synapse | 23 | 2.30 × 10−4 |
| hsa04310:Wnt signalling pathway | 26 | 3.83 × 10−4 |
| hsa04024:cAMP signalling pathway | 33 | 5.36 × 10−4 |
| hsa04022:cGMP-PKG signalling pathway | 28 | 5.96 × 10−4 |
| hsa04728:Dopaminergic synapse | 22 | 0.0039 |
| hsa04010:MAPK signalling pathway | 34 | 0.0143 |
| Pathway/Functional Group | p-Value |
|---|---|
| KEGG pathway | |
| hsa04612: Antigen processing and presentation | 6.77 × 10−6 |
| hsa05169: Epstein-Barr virus infection | 1.06 × 10−4 |
| hsa05215: Prostate cancer | 1.49 × 10−4 |
| hsa05140: Leishmaniasis | 4.58 × 10−4 |
| hsa05168: Herpes simplex infection | 8.04 × 10−4 |
| hsa05332: Graft-versus-host disease | 8.61 × 10−4 |
| hsa04940: Type I diabetes mellitus | 0.0012 |
| hsa04915: Estrogen signalling pathway | 0.0015 |
| hsa05330: Allograft rejection | 0.003 |
| hsa05164: Influenza A | 0.0031 |
| Panther pathway | |
| P00031: Inflammation mediated by chemokine and cytokine signalling | 1.20 × 10−4 |
| P00053: T cell activation | 0.0068 |
| P02756: N-acetylglucosamine metabolism | 0.0199 |
| P02776: Serine glycine biosynthesis | 0.0199 |
| P00010: B cell activation | 0.0217 |
| P00007: Axon guidance mediated by semaphorins | 0.0257 |
| P00049: Parkinson disease | 0.0359 |
| P00054: Toll receptor signalling pathway | 0.0391 |
| P00006: Apoptosis signalling pathway | 0.0426 |
| P04386: Histamine H2 receptor mediated signalling pathway | 0.0443 |
| Gene Ontology (cellular component) | |
| GO:0042613: MHC class II protein complex | 1.24 × 10−4 |
| GO:0016607: nuclear speck | 3.02 × 10−4 |
| GO:0045121: membrane raft | 4.73 × 10−4 |
| GO:0012507: ER to Golgi transport vesicle membrane | 5.67 × 10−4 |
| GO:0030671: clathrin-coated phagocytic vesicle membrane | 9.54 × 10−4 |
| GO:0016606: LYSP100-associated nuclear domain | 0.0022 |
| GO:0010445: nuclear dicing body | 0.0022 |
| GO:0071601: sphere organelle | 0.0022 |
| GO:0016604: nuclear body | 0.0022 |
| GO:0035363: histone locus body | 0.0023 |
| Group A (Inactivated Microglia) | Group B (Activated Microglia) |
|---|---|
| ISG15 | HLA-DPB1 |
| MX1 | HLA-DQB1 |
| IFI44L | HLA-DMB |
| OAS2 | HLA-DPA1 |
| IFI35 | LCP2 |
| OAS1 | FCER1G |
| LY6E | GBP5 |
| IFIT1 | CYBB |
| IFI44 | SAMSN1 |
| UBE2L6 | SLAMF8 |
| IFIT3 | MS4A6A |
| IRF7 | IGSF6 |
| XAF1 | HAVCR2 |
| RSAD2 | TFEC |
| OAS3 | CD163 |
| IRF9 | FCGR2A |
| USP18 | MSR1 |
| Microarray | NanoString | |||
|---|---|---|---|---|
| Gene | Fold Change | p-Value | Fold Change | p-Value |
| AKT3 | 1.33 | 0.002 ** | 1.52 | 0.127 |
| CCL2 | −1.7 | 0.05 * | −1.88 | 0.290 |
| CCL3 | −1.31 | 0.05 * | −16.11 | 0.008 ** |
| CCL4 | −1.16 | 0.060 | −5.14 | 0.098 |
| CCR5 | −1.49 | 0.01 * | −2.17 | 0.0003 *** |
| CD163 | −3.27 | 0.315 | −1.07 | 0.928 |
| CD74 | −2.67 | 0.006 ** | −1.54 | 0.013 * |
| CD8A | −1.28 | 0.016 * | −1.23 | 0.567 |
| COL6A3 | −1.62 | 0.01 * | −1.72 | 0.074 |
| CX3CR1 | −1.5 | 0.005 ** | −1.64 | 0.01 * |
| CYBB | −1.62 | 0.013 * | −1.17 | 0.577 |
| EPSTI1 | −1.31 | 0.839 | −1.39 | 0.101 |
| FCER1G | −1.52 | 0.076 | −1.28 | 0.265 |
| FGL2 | −1.77 | 0.012 * | −1.29 | 0.190 |
| FPR1 | −2.39 | 0.017 * | −2.82 | 0.066 |
| HLA-DRA | −1.71 | 0.004 ** | −1.51 | 0.071 |
| IFNAR1 | 1.79 | 0.004 ** | 1.25 | 0.190 |
| IGSF6 | −1.27 | 0.257 | −1.12 | 0.585 |
| IKBKB | 1.38 | 0.839 | 1.88 | 0.097 |
| IL1A | −1.06 | 0.206 | −1.71 | 0.061 |
| IL1B | −1.34 | 0.01 * | −4.72 | 0.007 ** |
| IRF7 | 1.04 | 0.673 | −1.04 | 0.846 |
| JUN | 1.33 | 0.008 ** | 1.98 | 0.008 ** |
| MSR1 | −1.95 | 0.023 * | −1.06 | 0.877 |
| OAS1 | −1.12 | 0.83 | 1.14 | 0.644 |
| PHF21A | 1.36 | 0.008 ** | 1.5 | 0.121 |
| PTGS2 | −1.3 | 0.011 * | −2.59 | 0.007 ** |
| RSAD2 | −1.2 | 0.065 | 1.09 | 0.757 |
| SLAMF8 | −1.03 | 0.679 | −1.48 | 0.327 |
| WM Group | Control | Pre-Lesional | PVL |
|---|---|---|---|
| CD74 | 3.47 (3.08–4.15) | 1.66 (0.9–2.14) | 1.3 (1.13–1.66) |
| IL-1β | 3.87 (1.91–4.86) | 2.76 (1.91–4.07) | 3.43 (2.48–4.22) |
| CD163 | 0.93 (0.75–1.21) | 0.39 (0.29–1.08) | 0.95 (0.59–1.32) |
| CD86 | 4.55 (4.52–5.66) | 5.34 (3.98–6.23) | 5.04 (3.35–6.03) |
| Experimental Approach | Classification | Median Age (y) (Range) | Gender (M/F) | PMD (h) (Range) |
|---|---|---|---|---|
| Microarray | Control (n = 11) | 84 (70–89) | 4/7 | 11 (6–42) |
| PVL (n = 7) | 89 (85–95) | 3/4 | 28 (7–72) | |
| NanoString | Control (n = 7) | 84 (70–89) | 2/5 | 14 (6–42) |
| PVL (n = 5) | 88 (85–95) | 2/3 | 28 (18–72) |
| Antibody | Isotype | Dilution (Time, Temperature) | Supplier |
|---|---|---|---|
| CD163 | Monoclonal Mouse IgG | 1:100 (1 h RT) | Bio-Rad, UK |
| CD68 (PG-M1) | Mouse, IgG3κ | 1:100 (1 h RT) | Dako, UK |
| CD74 | Polyclonal Rabbit IgG | 1:200 (1 h RT) | Sigma, UK |
| CD80/B7-1 | Monoclonal Mouse IgG1 | 1:20 (overnight 4 °C) | R&D Systems, UK |
| CD86/B7-2 | Polyclonal goat IgG | 1:20 (overnight 4 °C) | R&D Systems, UK |
| CX3CR1 | Monoclonal Mouse IgG1 | 1:25 (overnight 4 °C) | Biolegend, UK |
| IL-1β | Polyclonal Rabbit IgG | 1:200 (1 h RT) | Proteintech, UK |
| MHC-II (HLA-DR) | Mouse Monoclonal IgG | 1:20 (1 h RT) | Dako, UK |
| Gene Symbol | Gene Name | |
|---|---|---|
| Group A | CD317 (BST2) | Bone marrow stromal cell antigen 2 |
| Group B | CD39 (ENTPD1) | Ectonucleoside Triphosphate Diphosphohydrolase 1 |
| HLA-DRA | Major Histocompatibility Complex, Class II, DR Alpha | |
| CD86 | Cluster of differentiation 86 | |
| CD80 | Cluster of differentiation 80 | |
| CD274 | Cluster of differentiation 274 | |
| TIMD4 | T Cell Immunoglobulin and Mucin Domain Containing 4 | |
| CD11C (ITGAX) | Integrin Subunit Alpha X | |
| TREM2 | Triggering Receptor Expressed on Myeloid Cells 2 | |
| CCR5 | C-C chemokine receptor type 5 | |
| MRC1 | Mannose Receptor C-Type 1 |
| Function | Gene Symbol | Gene Name |
|---|---|---|
| Inflammation | AKT3 | V-akt murine thymoma viral oncogene homolog 3/RAC-gamma serine/threonine-protein kinase |
| CCL2 | C-C motif chemokine ligand 2 | |
| CCL3 | C-C motif chemokine ligand 3 | |
| CCL4 | C-C motif chemokine ligand 4 | |
| CCR5 | C-C motif chemokine receptor 5 (gene/pseudogene) | |
| COL6A3 | Collagen type VI alpha 3 chain | |
| CX3CR1 | C-X3-C motif chemokine receptor 1 | |
| IFNAR1 | Interferon alpha and beta receptor subunit 1 | |
| IKBKB | Inhibitor of nuclear factor kappa B kinase subunit beta | |
| IL1A | Interleukin 1 alpha | |
| IL1B | Interleukin 1 beta | |
| JUN | Jun proto-oncogene, AP-1 transcription factor subunit | |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | |
| Antigen processing and presentation | CD74 | CD74 molecule, HLA class II histocompatibility antigen gamma chain |
| HLA-DRA | Major histocompatibility complex, class II, DR alpha | |
| CD8A | CD8a molecule, T-cell surface glycoprotein CD8 alpha chain | |
| CD163 | CD163 molecule, Scavenger Receptor Cysteine-Rich Type 1 Protein M130 | |
| CD14 Monocytes | EPSTI1 | Epithelial stromal interaction protein 1 |
| FGL2 | Fibrinogen like 2 | |
| FPR1 | Formyl peptide receptor 1 | |
| IGSF6 | Immunoglobulin superfamily member 6 | |
| PHF21A | PHD finger protein 21A | |
| Inactivated Microglia | IRF7 | Interferon regulatory factor 7 |
| OAS1 | 2’-5’-oligoadenylate synthetase 1 | |
| RSAD2 | Radical S-adenosyl methionine domain containing 2 | |
| Activated Microglia | FCER1G | Fc fragment of IgE receptor Ig |
| MSR1 | Macrophage scavenger receptor 1 | |
| SLAMF8 | SLAM family member 8 | |
| CYBB | Cytochrome b-245, beta polypeptide | |
| Housekeeping Genes | ACTB | Actin cytoplasmic 1 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | |
| RPLP0 | 60S acidic ribosomal protein P0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fadul, M.M.; Heath, P.R.; Cooper-Knock, J.; Kurz, J.M.; Al-Azzawi, H.A.; Ali, Z.; Smith, T.; Matthews, F.E.; Brayne, C.; Wharton, S.B.; et al. Transcriptomic Analysis of Age-Associated Periventricular Lesions Reveals Dysregulation of the Immune Response. Int. J. Mol. Sci. 2020, 21, 7924. https://doi.org/10.3390/ijms21217924
Fadul MM, Heath PR, Cooper-Knock J, Kurz JM, Al-Azzawi HA, Ali Z, Smith T, Matthews FE, Brayne C, Wharton SB, et al. Transcriptomic Analysis of Age-Associated Periventricular Lesions Reveals Dysregulation of the Immune Response. International Journal of Molecular Sciences. 2020; 21(21):7924. https://doi.org/10.3390/ijms21217924
Chicago/Turabian StyleFadul, Motaz M., Paul R. Heath, Johnathan Cooper-Knock, Julian M. Kurz, Hayder A. Al-Azzawi, Zarki Ali, Taylor Smith, Fiona E. Matthews, Carol Brayne, Stephen B. Wharton, and et al. 2020. "Transcriptomic Analysis of Age-Associated Periventricular Lesions Reveals Dysregulation of the Immune Response" International Journal of Molecular Sciences 21, no. 21: 7924. https://doi.org/10.3390/ijms21217924
APA StyleFadul, M. M., Heath, P. R., Cooper-Knock, J., Kurz, J. M., Al-Azzawi, H. A., Ali, Z., Smith, T., Matthews, F. E., Brayne, C., Wharton, S. B., & Simpson, J. E., on behalf of the Cognitive Function and Ageing Neuropathology Study Group. (2020). Transcriptomic Analysis of Age-Associated Periventricular Lesions Reveals Dysregulation of the Immune Response. International Journal of Molecular Sciences, 21(21), 7924. https://doi.org/10.3390/ijms21217924





