Impact of a Model Used to Simulate Chronic Socio-Environmental Stressors Encountered during Spaceflight on Murine Intestinal Microbiota
Abstract
:1. Introduction
2. Results
2.1. Days of CUMS Exposure Do Not Induce a Major Stress Response
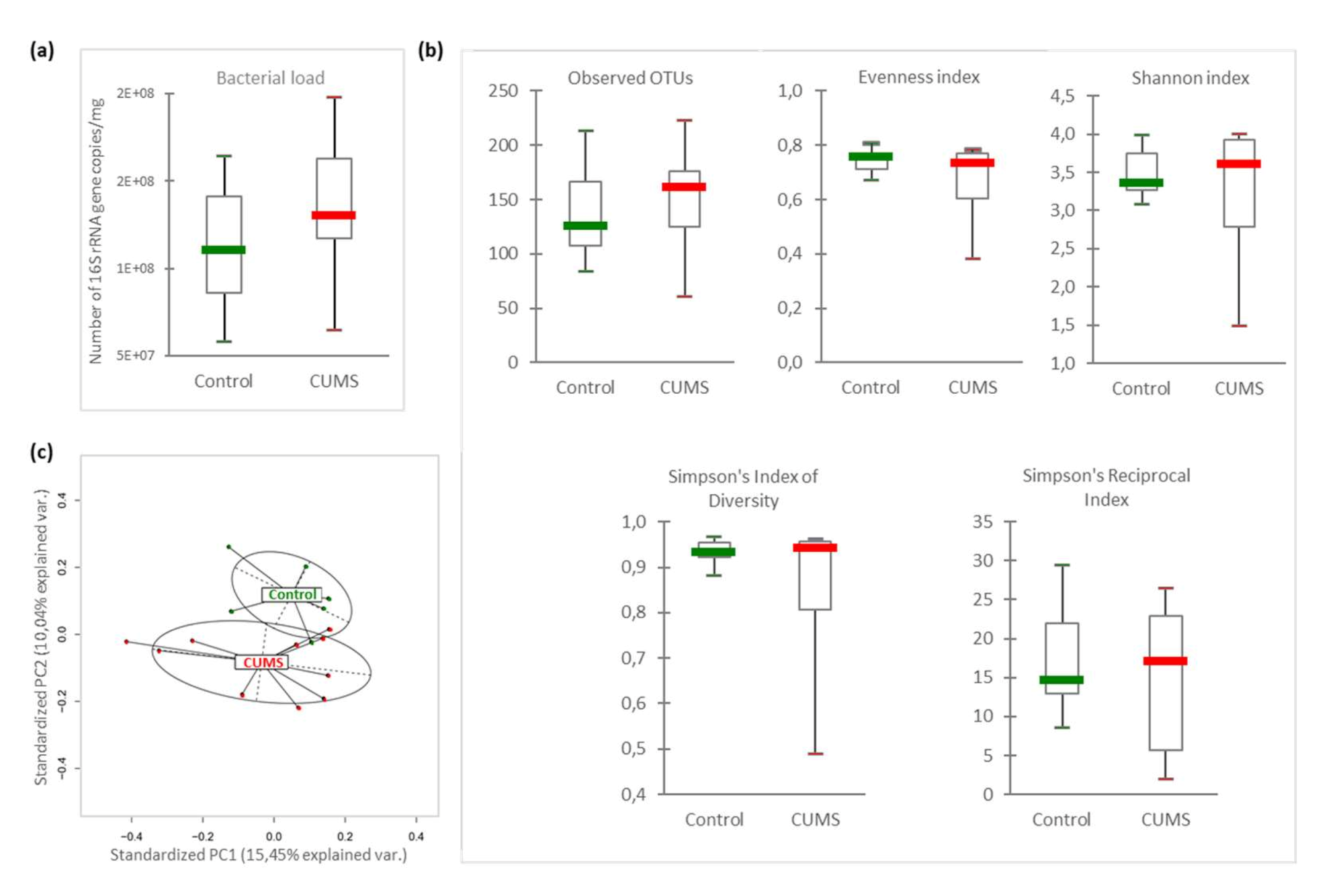
2.2. Intestinal Microbiome β-Diversity Is Significantly Modified by CUMS
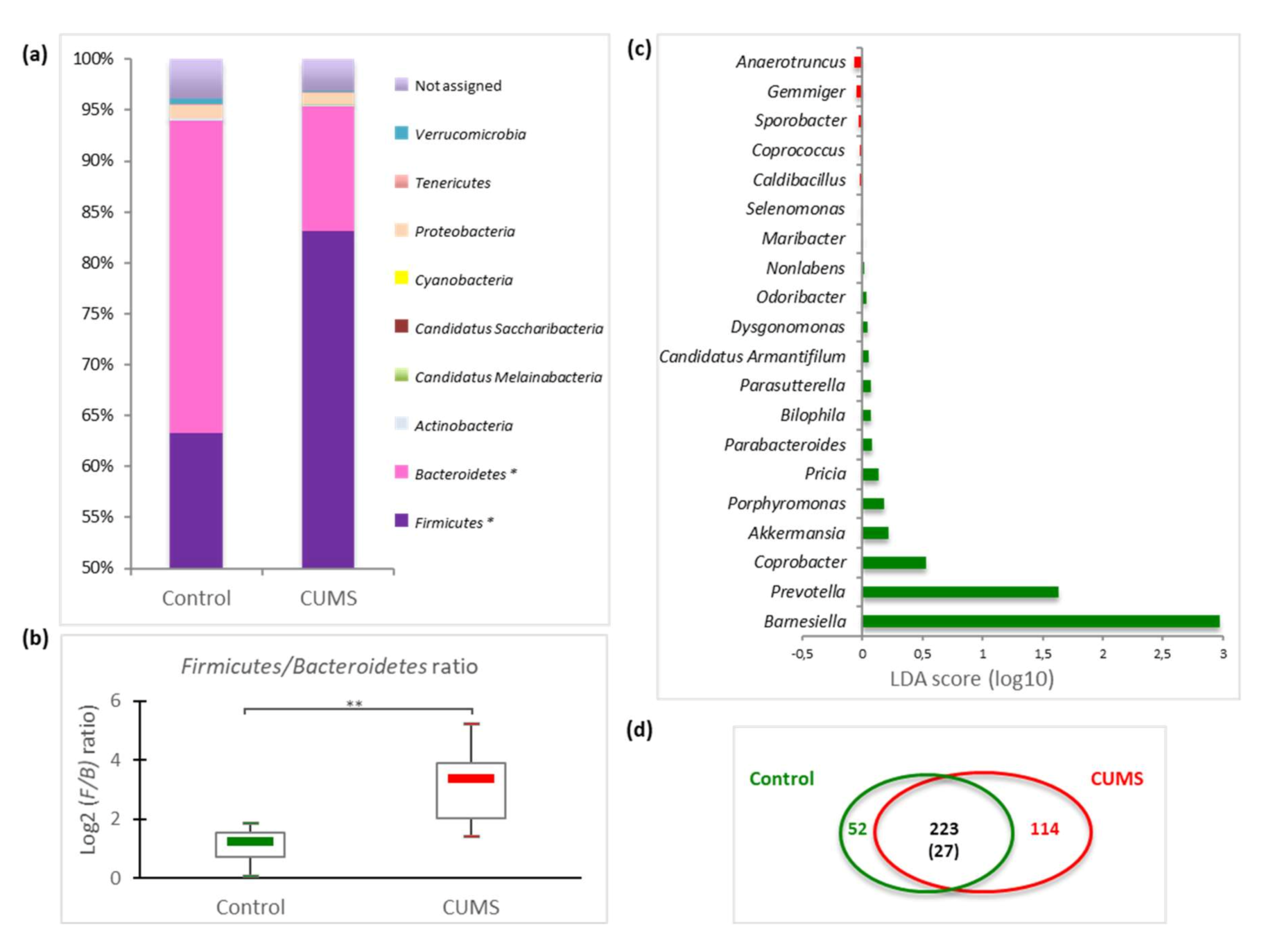
2.3. Impact on Caecal Microbiome Composition
3. Discussion
4. Materials and Methods
4.1. Experimental Animals and Ethics Statement
4.2. Exposure to Chronic Unpredictable Mild Psychosocial and Environmental Stressors (CUMS Model)
4.3. Sample Collection
4.4. Corticosterone Quantification
4.5. DNA Isolation
4.6. Intracaecal Microbiota Sequencing
4.7. Amplicon Sequencing Data Analysis
4.8. Intracaecal Bacterial Load Quantification
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CMS | chronic mild stress |
| CUMS | chronic unpredictable mild stress |
| GM | gut microbiota |
| HPA | hypothalamic-pituitary-adrenal |
| IBD | intestinal bowel disease |
| LDA | linear discriminant analysis |
| OTUs | operational taxonomic units |
| PCA | principal component analysis |
| TCR | T cell receptor |
References
- Lavelle, A.; Hoffmann, T.W.; Pham, H.-P.; Langella, P.; Guédon, E.; Sokol, H. Baseline microbiota composition modulates antibiotic-mediated effects on the gut microbiota and host. Microbiome 2019, 7, 111. [Google Scholar] [CrossRef] [PubMed]
- Rea, K.; Dinan, T.G.; Cryan, J.F. Gut Microbiota: A Perspective for Psychiatrists. Neuropsychobiology 2020, 79, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Alauzet, C.; Cunat, L.; Wack, M.; Lozniewski, A.; Busby, H.; Agrinier, N.; Cailliez-Grimal, C.; Frippiat, J.-P. Hypergravity disrupts murine intestinal microbiota. Sci. Rep. 2019, 9, 9410. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.T.; Coe, C.L. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev. Psychobiol. 1999, 35, 146–155. [Google Scholar] [CrossRef]
- Galley, J.D.; Mackos, A.R.; Varaljay, V.A.; Bailey, M.T. Stressor exposure has prolonged effects on colonic microbial community structure in Citrobacter rodentium-challenged mice. Sci. Rep. 2017, 7, 45012. [Google Scholar] [CrossRef] [Green Version]
- Geng, S.; Yang, L.; Cheng, F.; Zhang, Z.; Li, J.; Liu, W.; Li, Y.; Chen, Y.; Bao, Y.; Chen, L.; et al. Gut Microbiota Are Associated With Psychological Stress-Induced Defections in Intestinal and Blood–Brain Barriers. Front. Microbiol. 2020, 10, 3067. [Google Scholar] [CrossRef]
- Lv, L.-X.; Fang, D.-Q.; Shi, D.; Chen, D.-Y.; Yan, R.; Zhu, Y.-X.; Chen, Y.-F.; Shao, L.; Guo, F.-F.; Wu, W.-R.; et al. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis: Gut microbiome in PBC patients. Environ. Microbiol. 2016, 18, 2272–2286. [Google Scholar] [CrossRef]
- Michels, N.; Van de Wiele, T.; Fouhy, F.; O’Mahony, S.; Clarke, G.; Keane, J. Gut microbiome patterns depending on children’s psychosocial stress: Reports versus biomarkers. Brain. Behav. Immun. 2019, 80, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Qu, W.; Liu, S.; Zhang, W.; Zhu, H.; Tao, Q.; Wang, H.; Yan, H. Impact of traditional Chinese medicine treatment on chronic unpredictable mild stress-induced depression-like behaviors: Intestinal microbiota and gut microbiome function. Food Funct. 2019, 10, 5886–5897. [Google Scholar] [CrossRef]
- Dinan, T.G.; Cryan, J.F. The Microbiome-Gut-Brain Axis in Health and Disease. Gastroenterol. Clin. North. Am. 2017, 46, 77–89. [Google Scholar] [CrossRef] [Green Version]
- Jianguo, L.; Xueyang, J.; Cui, W.; Changxin, W.; Xuemei, Q. Altered gut metabolome contributes to depression-like behaviors in rats exposed to chronic unpredictable mild stress. Transl. Psychiatry 2019, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Mackner, L.M.; Hatzakis, E.; Allen, J.M.; Davies, R.H.; Kim, S.C.; Maltz, R.M.; Bailey, M.T. Fecal microbiota and metabolites are distinct in a pilot study of pediatric Crohn’s disease patients with higher levels of perceived stress. Psychoneuroendocrinology 2020, 111, 104469. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, L.; Xie, R.; Wang, B.; Jiang, K.; Cao, H. Stress Triggers Flare of Inflammatory Bowel Disease in Children and Adults. Front. Pediatrics 2019, 7, 432. [Google Scholar] [CrossRef] [PubMed]
- Frankiensztajn, L.M.; Elliott, E.; Koren, O. The microbiota and the hypothalamus-pituitary-adrenocortical (HPA) axis, implications for anxiety and stress disorders. Curr. Opin. Neurobiol. 2020, 62, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, H.; Cao, Y.; Wang, C.; Zhao, C.; Wang, H.; Cui, G.; Wang, M.; Pan, Y.; Shi, Y.; et al. Fluoxetine ameliorates dysbiosis in a depression model induced by chronic unpredicted mild stress in mice. Int. J. Med. Sci. 2019, 16, 1260–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, M.T.; Dowd, S.E.; Galley, J.D.; Hufnagle, A.R.; Allen, R.G.; Lyte, M. Exposure to a social stressor alters the structure of the intestinal microbiota: Implications for stressor-induced immunomodulation. Brain. Behav. Immun. 2011, 25, 397–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyte, M.; Vulchanova, L.; Brown, D.R. Stress at the intestinal surface: Catecholamines and mucosa-bacteria interactions. Cell Tissue Res. 2011, 343, 23–32. [Google Scholar] [CrossRef]
- Boyanova, L. Stress hormone epinephrine (adrenaline) and norepinephrine (noradrenaline) effects on the anaerobic bacteria. Anaerobe 2017, 44, 13–19. [Google Scholar] [CrossRef]
- Zhao, H.; Jiang, X.; Chu, W. Shifts in the gut microbiota of mice in response to dexamethasone administration. Int. Microbiol. 2020, 23, 565–573. [Google Scholar] [CrossRef]
- Frippiat, J.-P.; Crucian, B.E.; De Quervain, D.J.; Grimm, D.; Montano, N.; Praun, S.; Roozendaal, B.; Schelling, G.; Thiel, M.; Ullrich, O.; et al. Towards human exploration of space: The THESEUS review series on immunology research priorities. NPJ Microgravity 2016, 2, 16040. [Google Scholar] [CrossRef] [Green Version]
- Crucian, B.E.; Choukèr, A.; Simpson, R.J.; Mehta, S.; Marshall, G.; Smith, S.M.; Zwart, S.R.; Heer, M.; Ponomarev, S.; Whitmire, A.; et al. Immune System Dysregulation During Spaceflight: Potential Countermeasures for Deep Space Exploration Missions. Front. Immunol. 2018, 9, 1437. [Google Scholar] [CrossRef] [PubMed]
- Makedonas, G.; Mehta, S.; Choukèr, A.; Simpson, R.J.; Marshall, G.; Orange, J.S.; Aunon-Chancellor, S.; Smith, S.M.; Zwart, S.R.; Stowe, R.P.; et al. Specific Immunologic Countermeasure Protocol for Deep-Space Exploration Missions. Front. Immunol. 2019, 10, 2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voorhies, A.A.; Mark Ott, C.; Mehta, S.; Pierson, D.L.; Crucian, B.E.; Feiveson, A.; Oubre, C.M.; Torralba, M.; Moncera, K.; Zhang, Y.; et al. Study of the impact of long-duration space missions at the International Space Station on the astronaut microbiome. Sci. Rep. 2019, 9, 9911. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P. Impact of space flight on bacterial virulence and antibiotic susceptibility. Infect. Drug Resist. 2015, 8, 249–262. [Google Scholar] [CrossRef] [Green Version]
- Saei, A.A.; Barzegari, A. The microbiome: The forgotten organ of the astronaut’s body—Probiotics beyond terrestrial limits. Future Microbiol. 2012, 7, 1037–1046. [Google Scholar] [CrossRef]
- Gaignier, F.; Legrand-Frossi, C.; Stragier, E.; Mathiot, J.; Merlin, J.-L.; Cohen-Salmon, C.; Lanfumey, L.; Frippiat, J.-P. A Model of Chronic Exposure to Unpredictable Mild Socio-Environmental Stressors Replicates Some Spaceflight-Induced Immunological Changes. Front. Physiol. 2018, 9, 514. [Google Scholar] [CrossRef] [Green Version]
- Blanchard, E.B.; Lackner, J.M.; Jaccard, J.; Rowell, D.; Carosella, A.M.; Powell, C.; Sanders, K.; Krasner, S.; Kuhn, E. The role of stress in symptom exacerbation among IBS patients. J. Psychosom. Res. 2008, 64, 119–128. [Google Scholar] [CrossRef]
- Wei, L.; Li, Y.; Tang, W.; Sun, Q.; Chen, L.; Wang, X.; Liu, Q.; Yu, S.; Yu, S.; Liu, C.; et al. Chronic Unpredictable Mild Stress in Rats Induces Colonic Inflammation. Front. Physiol. 2019, 10, 1228. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Wang, Y.; He, J.; Li, P.; Jin, R.; Wang, K.; Xu, X.; Hao, J.; Zhang, Y.; Liu, H.; et al. Intestinal microbiota contributes to colonic epithelial changes in simulated microgravity mouse model. FASEB J. 2017, 31, 3695–3709. [Google Scholar] [CrossRef] [Green Version]
- Antoniuk, S.; Bijata, M.; Ponimaskin, E.; Wlodarczyk, J. Chronic unpredictable mild stress for modeling depression in rodents: Meta-analysis of model reliability. Neurosci. Biobehav. Rev. 2019, 99, 101–116. [Google Scholar] [CrossRef]
- Lv, W.; Wu, X.; Chen, W.; Li, Y.; Zhang, G.; Chao, L.; Zhou, J.; Guo, A.; Liu, C.; Guo, S. The Gut Microbiome Modulates the Changes in Liver Metabolism and in Inflammatory Processes in the Brain of Chronic Unpredictable Mild Stress Rats. Oxid. Med. Cell. Longev. 2019, 2019, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Siopi, E.; Chevalier, G.; Katsimpardi, L.; Saha, S.; Bigot, M.; Moigneu, C.; Eberl, G.; Lledo, P.-M. Changes in Gut Microbiota by Chronic Stress Impair the Efficacy of Fluoxetine. Cell Rep. 2020, 30, 3682–3690. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Sengupta, P. Men and mice: Relating their ages. Life Sci. 2016, 152, 244–248. [Google Scholar] [CrossRef]
- Bailey, M.T.; Dowd, S.E.; Parry, N.M.A.; Galley, J.D.; Schauer, D.B.; Lyte, M. Stressor Exposure Disrupts Commensal Microbial Populations in the Intestines and Leads to Increased Colonization by Citrobacter rodentium. Infect. Immun. 2010, 78, 1509–1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rincel, M.; Aubert, P.; Chevalier, J.; Grohard, P.-A.; Basso, L.; Monchaux de Oliveira, C.; Helbling, J.C.; Lévy, É.; Chevalier, G.; Leboyer, M.; et al. Multi-hit early life adversity affects gut microbiota, brain and behavior in a sex-dependent manner. Brain. Behav. Immun. 2019, 80, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.A.; Chassard, C.; Hennet, T. Selective proliferation of intestinal Barnesiella under fucosyllactose supplementation in mice. Br. J. Nutr. 2014, 111, 1602–1610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daillère, R.; Vétizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.M.; Flament, C.; Lepage, P.; Roberti, M.P.; et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef] [Green Version]
- Ubeda, C.; Bucci, V.; Caballero, S.; Djukovic, A.; Toussaint, N.C.; Equinda, M.; Lipuma, L.; Ling, L.; Gobourne, A.; No, D.; et al. Intestinal Microbiota Containing Barnesiella Species Cures Vancomycin-Resistant Enterococcus faecium Colonization. Infect. Immun. 2013, 81, 965–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guéguinou, N.; Bojados, M.; Jamon, M.; Derradji, H.; Baatout, S.; Tschirhart, E.; Frippiat, J.-P.; Legrand-Frossi, C. Stress response and humoral immune system alterations related to chronic hypergravity in mice. Psychoneuroendocrinology 2012, 37, 137–147. [Google Scholar] [CrossRef]
- Silberman, D.M.; Wald, M.; Genaro, A.M. Effects of chronic mild stress on lymphocyte proliferative response. Participation of serum thyroid hormones and corticosterone. Int. Immunopharmacol. 2002, 2, 487–497. [Google Scholar] [CrossRef]
- Fonte, C.; Kaminski, S.; Vanet, A.; Lanfumey, L.; Cohen-Salmon, C.; Ghislin, S.; Frippiat, J.-P. Socioenvironmental stressors encountered during spaceflight partially affect the murine TCR-β repertoire and increase its self-reactivity. FASEB J. 2018, 33, 896–908. [Google Scholar] [CrossRef] [PubMed]
- Ghislin, S.; Ouzren-Zarhloul, N.; Kaminski, S.; Frippiat, J.-P. Hypergravity exposure during gestation modifies the TCRβ repertoire of newborn mice. Sci. Rep. 2015, 5, 9318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glaser, R.; Kiecolt-Glaser, J.K. Stress-induced immune dysfunction: Implications for health. Nat. Rev. Immunol. 2005, 5, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Pardon, M.-C.; Gérardin, P.; Joubert, C.; Pérez-Diaz, F.; Cohen-Salmon, C. Influence of prepartum chronic ultramild stress on maternal pup care behavior in mice. Biol. Psychiatry 2000, 47, 858–863. [Google Scholar] [CrossRef]
- Ferrand, J.; Patron, K.; Legrand-Frossi, C.; Frippiat, J.-P.; Merlin, C.; Alauzet, C.; Lozniewski, A. Comparison of seven methods for extraction of bacterial DNA from fecal and cecal samples of mice. J. Microbiol. Methods 2014, 105, 180–185. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.-J.; Tappu, R. MEGAN Community Edition—Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [Green Version]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2001, 26, 32–46. [Google Scholar]
- Anderson, M.J.; Ellingsen, K.E.; McArdle, B.H. Multivariate dispersion as a measure of beta diversity. Ecol. Lett. 2006, 9, 683–693. [Google Scholar] [CrossRef] [PubMed]
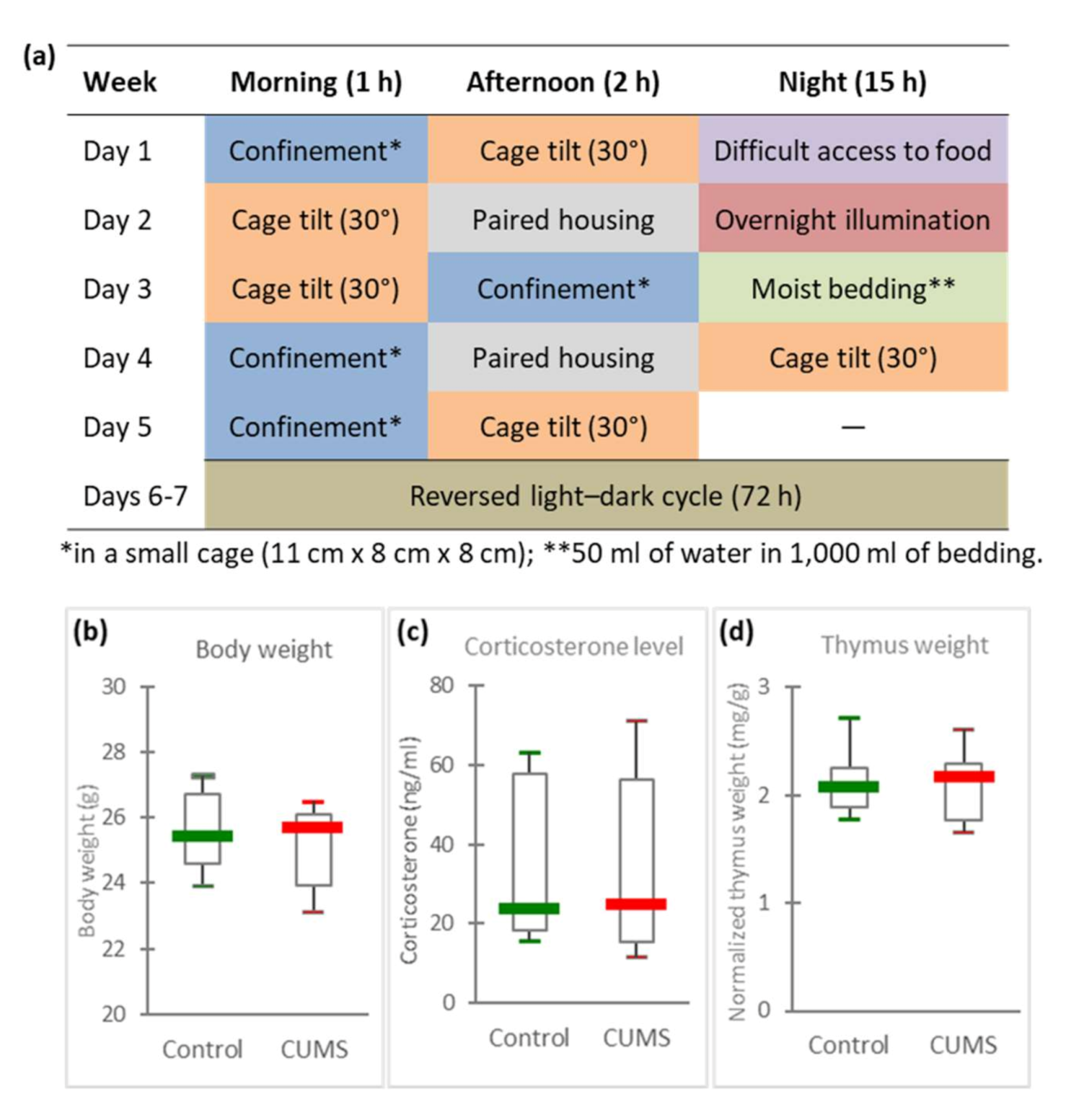
| Socio-Environmental Stressors Applied in the CUMS Model (Details in Figure 1a) | Socio-Environmental Stressors Encountered during Spaceflights | Limitations of CUMS Model |
|---|---|---|
| Mice confined in a small cage during 1 or 2 h. | Confinement throughout the mission. | Mice confined during short periods (ethical point of view) /astronauts confined for several months in the ISS. |
| Isolation of mice (sociable animals) during the whole CUMS procedure. | Isolation from friends and family. | |
| Reversed light/dark cycle during week-end. | Disrupted circadian rhythm. | Astronauts observe 16 sunrises and sunsets during a 24 h period. |
| Pair housing during 2 h. | Crew tension and other interpersonal issues. | Pair housing is of a limited duration. |
| Cage tilt for 1, 2 or 15 h. | Perturbation of spatial references. | Cage tilt is of a limited duration. |
| Period with difficult access to food, without a reduction in the daily food ration. | Lower dietary intake despite enough available food, changes in eating habits and rituals. | |
| Period in a soiled cage (mice do not like wet litter). | Uncomfortable living conditions. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alauzet, C.; Cunat, L.; Wack, M.; Lanfumey, L.; Legrand-Frossi, C.; Lozniewski, A.; Agrinier, N.; Cailliez-Grimal, C.; Frippiat, J.-P. Impact of a Model Used to Simulate Chronic Socio-Environmental Stressors Encountered during Spaceflight on Murine Intestinal Microbiota. Int. J. Mol. Sci. 2020, 21, 7863. https://doi.org/10.3390/ijms21217863
Alauzet C, Cunat L, Wack M, Lanfumey L, Legrand-Frossi C, Lozniewski A, Agrinier N, Cailliez-Grimal C, Frippiat J-P. Impact of a Model Used to Simulate Chronic Socio-Environmental Stressors Encountered during Spaceflight on Murine Intestinal Microbiota. International Journal of Molecular Sciences. 2020; 21(21):7863. https://doi.org/10.3390/ijms21217863
Chicago/Turabian StyleAlauzet, Corentine, Lisiane Cunat, Maxime Wack, Laurence Lanfumey, Christine Legrand-Frossi, Alain Lozniewski, Nelly Agrinier, Catherine Cailliez-Grimal, and Jean-Pol Frippiat. 2020. "Impact of a Model Used to Simulate Chronic Socio-Environmental Stressors Encountered during Spaceflight on Murine Intestinal Microbiota" International Journal of Molecular Sciences 21, no. 21: 7863. https://doi.org/10.3390/ijms21217863
APA StyleAlauzet, C., Cunat, L., Wack, M., Lanfumey, L., Legrand-Frossi, C., Lozniewski, A., Agrinier, N., Cailliez-Grimal, C., & Frippiat, J.-P. (2020). Impact of a Model Used to Simulate Chronic Socio-Environmental Stressors Encountered during Spaceflight on Murine Intestinal Microbiota. International Journal of Molecular Sciences, 21(21), 7863. https://doi.org/10.3390/ijms21217863





