In the Model Host Caenorhabditis elegans, Sphingosine-1-Phosphate-Mediated Signaling Increases Immunity toward Human Opportunistic Bacteria
Abstract
:1. Introduction
2. Results
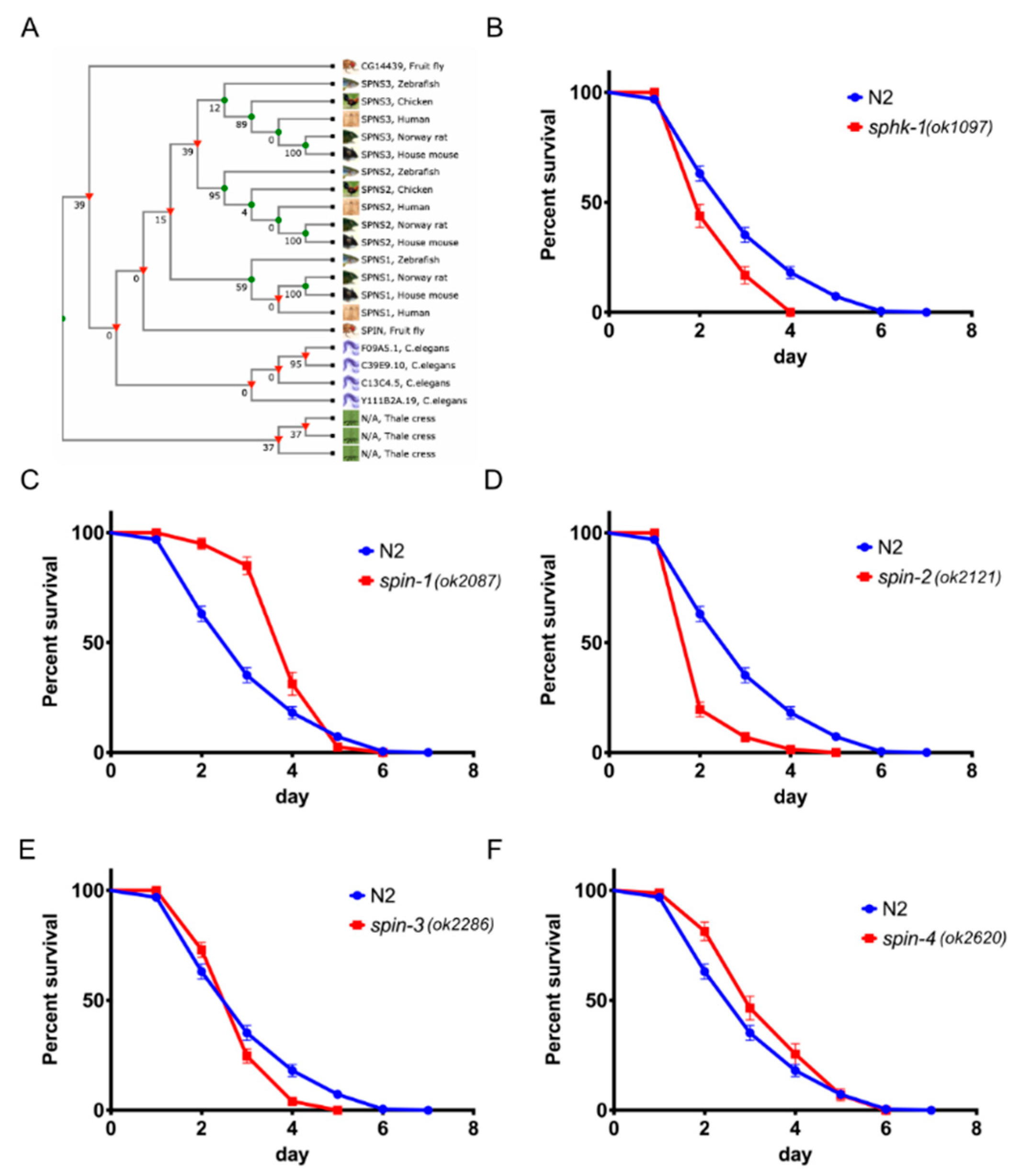
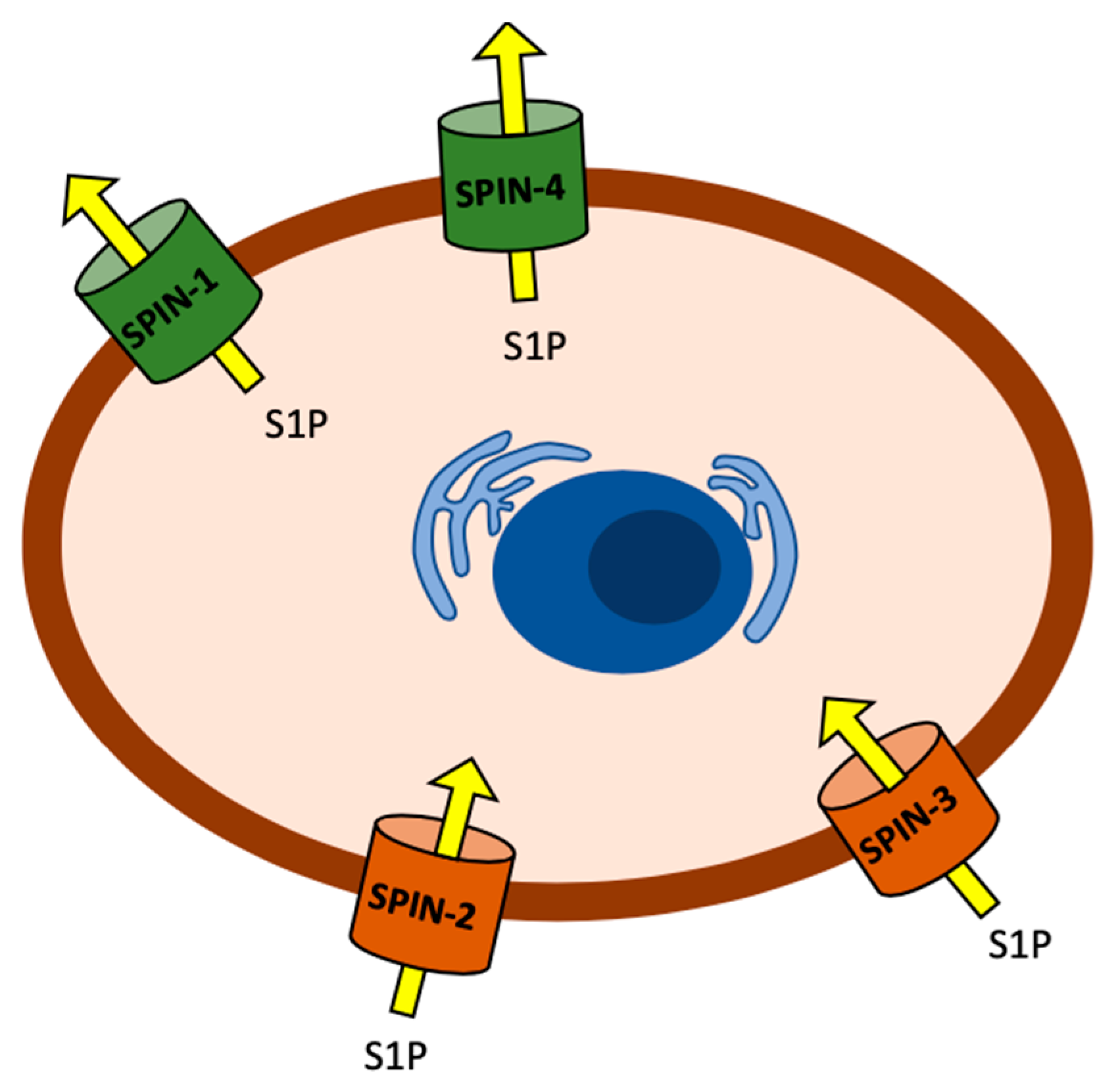
2.1. Sphingosine Kinase and S1P Transporters Affect the C. elegans Immune Response
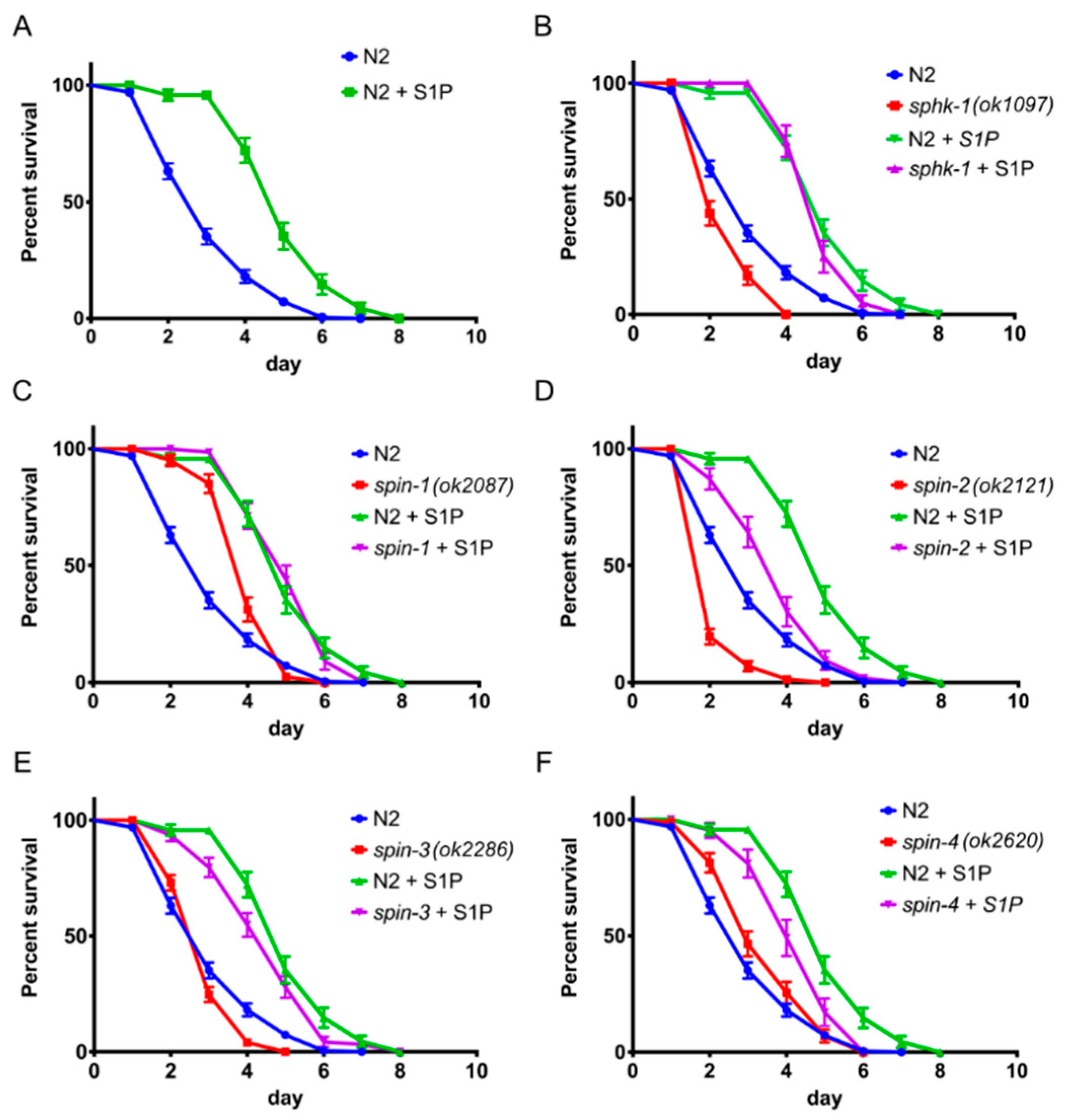
2.2. External Supplementation of S1P Extends Worm Survival
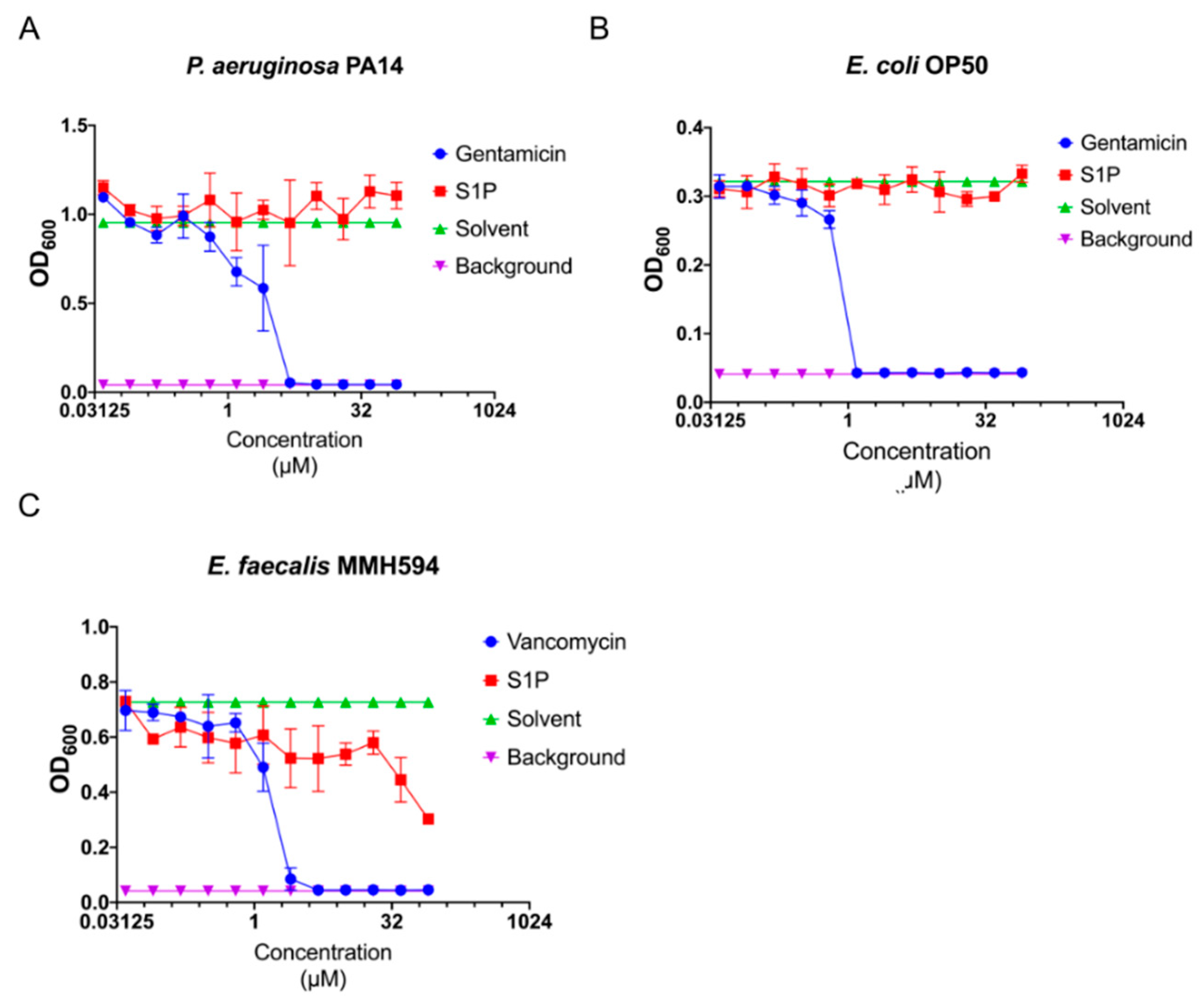
2.3. S1P Has No or Nominal Antimicrobial Activity
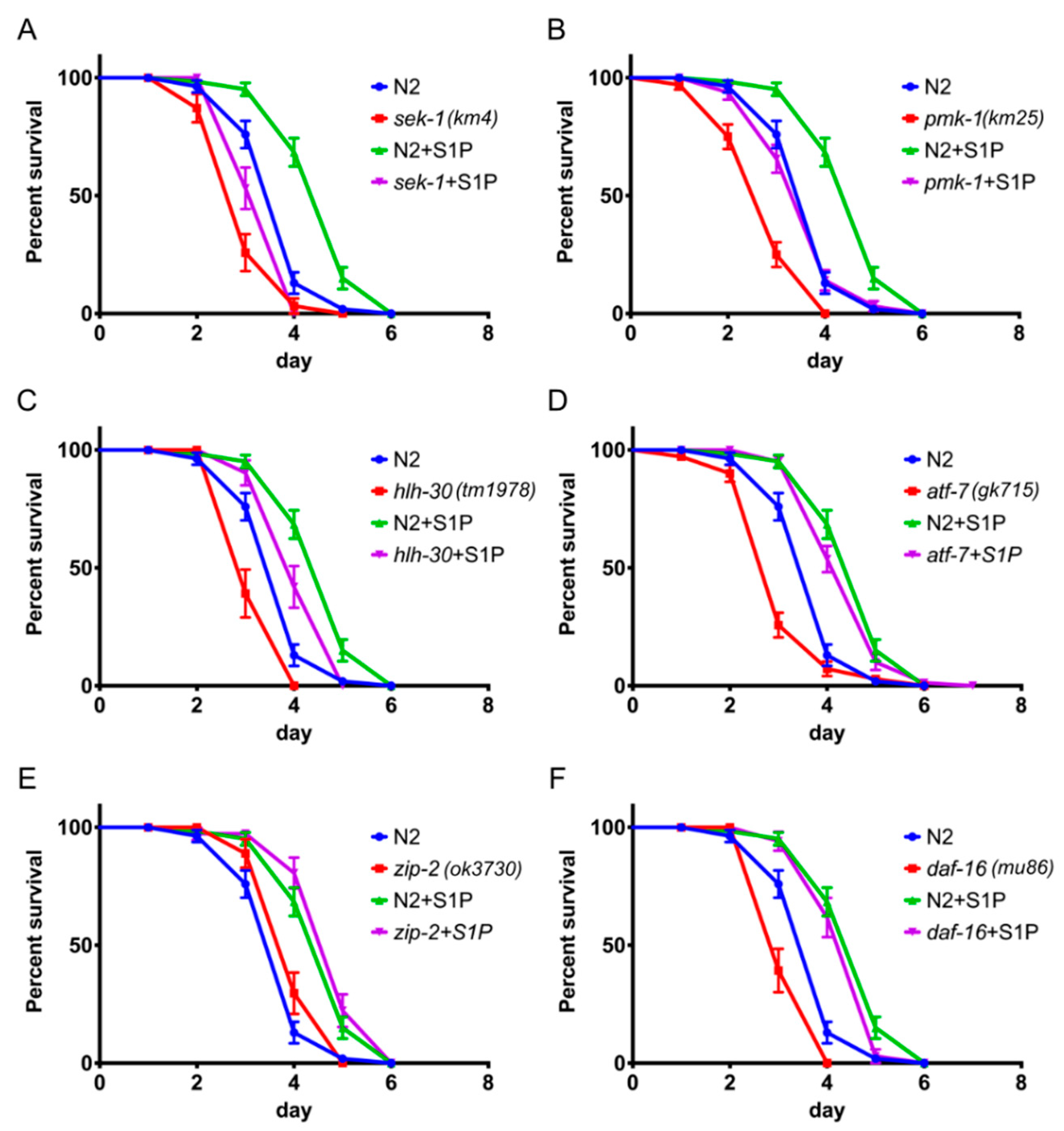
2.4. S1P Signaling Depends on p38 MAPK Pathway and the Transcription Factor hlh-30
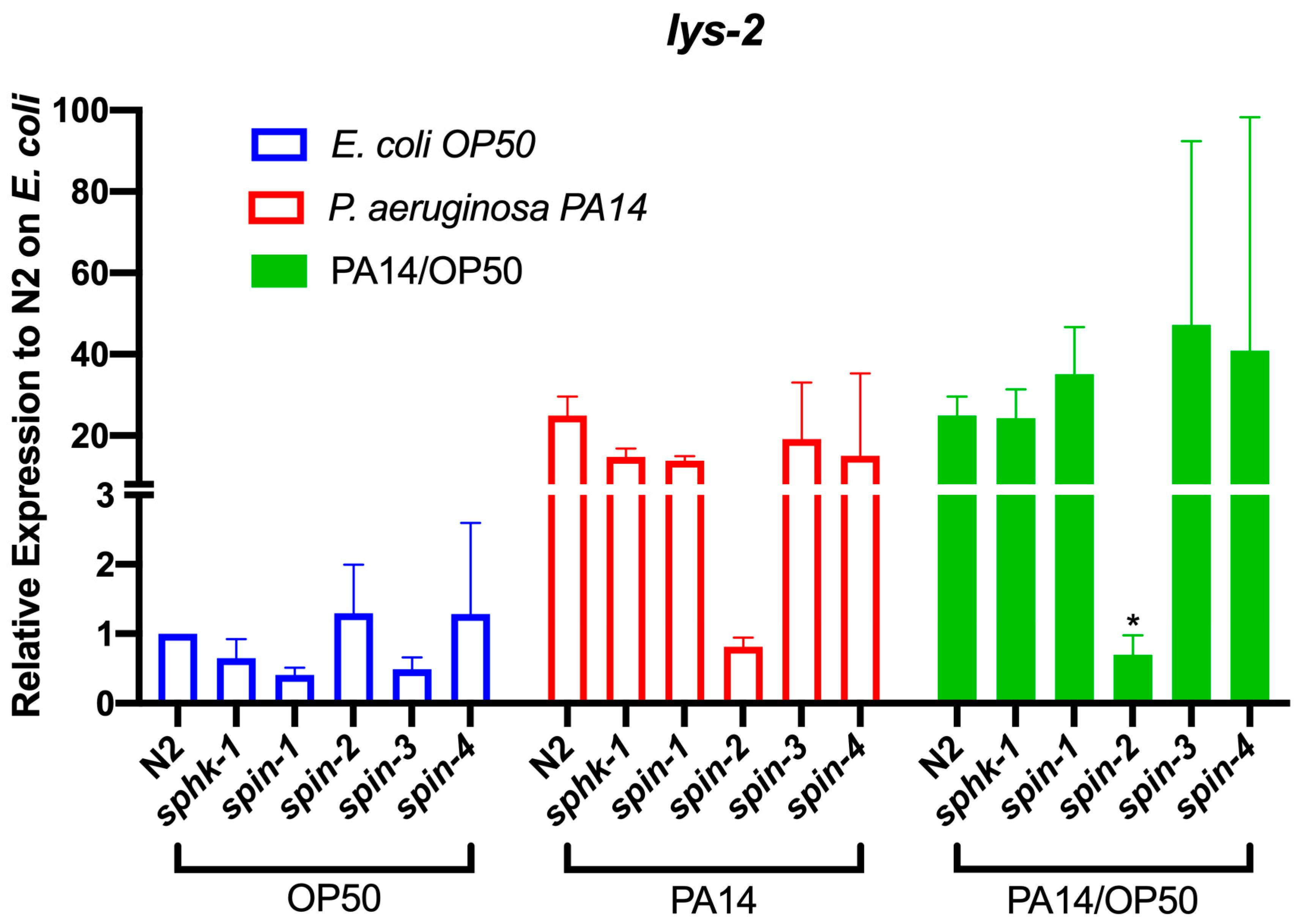
2.5. S1P Signaling Affects C. Elegans Immune Effect Gene Lys-2
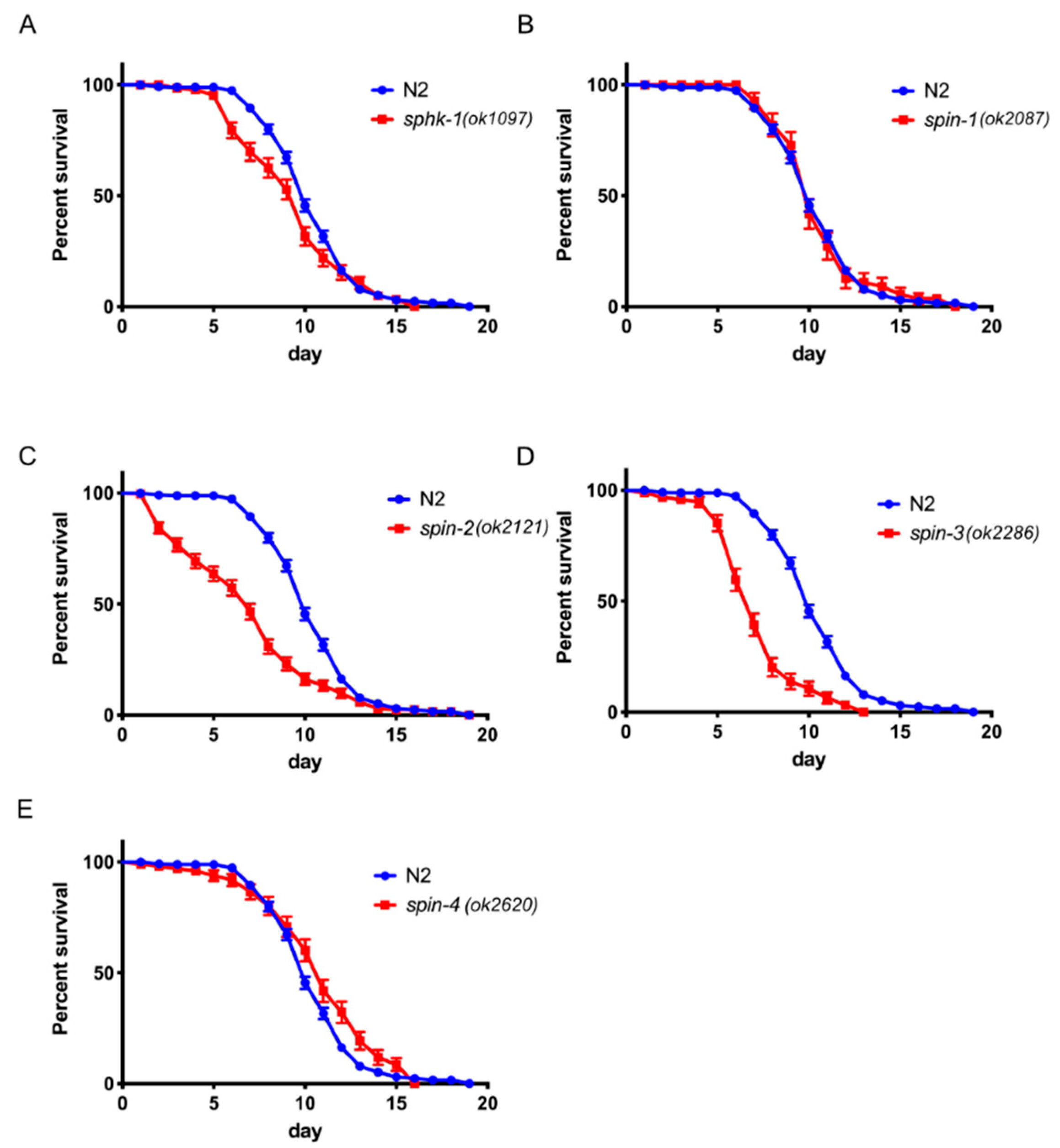
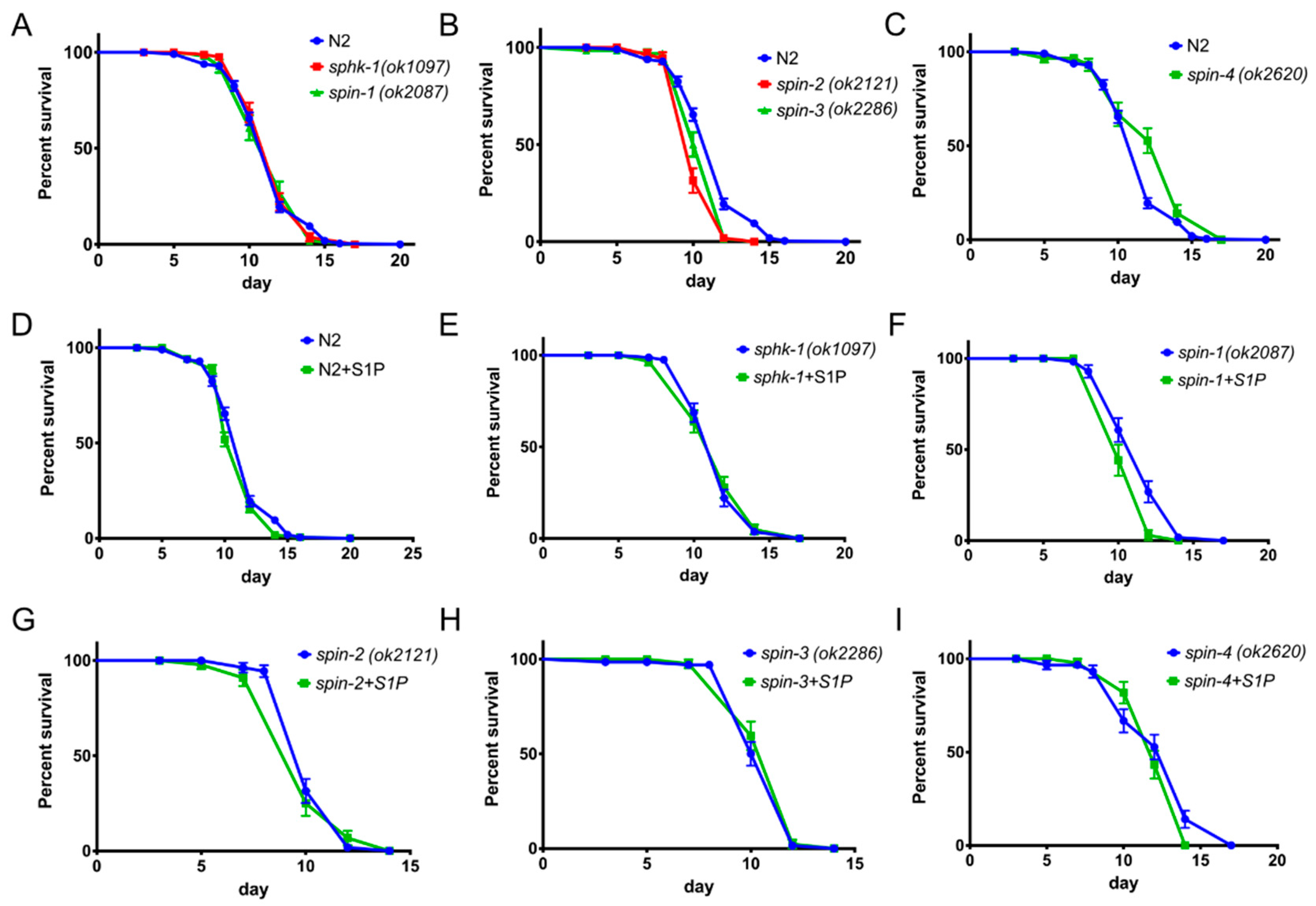
2.6. S1P Signaling Has a Modest Effect on C. Elegans Lifespan in a Normal Food Source
3. Discussion
4. Materials and Methods
4.1. Nematode Strains
4.2. Bacterial Strains and Maintenance
4.3. Reagents and Media Preparation
4.4. C. elegans Lifespan and Killing Assay
4.5. Measuring Minimum Inhibitory Concentration
4.6. Sequence Alignment
4.7. RNA Preparation, cDNA Synthesis, and Quantitative PCR
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| S1P | Sphingosine-1-phosphate |
| C. elegans | Caenorhabditis elegans |
| P. aeruginosa | Pseudomonas aeruginosa |
| E. faecalis | Enterococcus faecalis |
| UPRmt | Mitochondrial Unfolded Protein Response |
References
- Visvikis, O.; Ihuegbu, N.; Labed, S.A.; Luhachack, L.G.; Alves, A.-M.F.; Wollenberg, A.C.; Stuart, L.M.; Stormo, G.D.; Irazoqui, J.E. Innate Host Defense Requires TFEB-Mediated Transcription of Cytoprotective and Antimicrobial Genes. Immunity 2014, 40, 896–909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Troemel, E.R.; Chu, S.W.; Reinke, V.; Lee, S.S.; Ausubel, F.M.; Kim, D.H. p38 MAPK Regulates Expression of Immune Response Genes and Contributes to Longevity in C. elegans. PLoS Genet. 2006, 2, e183. [Google Scholar] [CrossRef] [PubMed]
- Shivers, R.P.; Pagano, D.J.; Kooistra, T.; Richardson, C.E.; Reddy, K.C.; Whitney, J.K.; Kamanzi, O.; Matsumoto, K.; Hisamoto, N.; Kim, D.H. Phosphorylation of the Conserved Transcription Factor ATF-7 by PMK-1 p38 MAPK Regulates Innate Immunity in Caenorhabditis elegans. PLoS Genet. 2010, 6, e1000892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irazoqui, J.E.; Urbach, J.M.; Ausubel, F.M. Evolution of host innate defence: Insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 2010, 10, 47–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engelmann, I.; Griffon, A.; Tichit, L.; Montañana-Sanchis, F.; Wang, G.; Reinke, V.; Waterston, R.H.; Hillier, L.W.; Ewbank, J.J. A Comprehensive Analysis of Gene Expression Changes Provoked by Bacterial and Fungal Infection in C. elegans. PLoS ONE 2011, 6, e19055-13. [Google Scholar] [CrossRef] [PubMed]
- Rosen, H.; Germana Sanna, M.; Gonzalez-Cabrera, P.J.; Roberts, E. The organization of the sphingosine 1-phosphate signaling system. Curr. Top. Microbiol. Immunol. 2014, 378, 1–21. [Google Scholar] [PubMed]
- Maceyka, M.; Spiegel, S. Sphingolipid metabolites in inflammatory disease. Nature 2014, 510, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Van Brocklyn, J.R.; Hobson, J.P.; Movafagh, S.; Zukowska-Grojec, Z.; Milstien, S.; Spiegel, S. Sphingosine 1-phosphate stimulates cell migration through a G(i)-coupled cell surface receptor. Potential involvement in angiogenesis. J. Biol. Chem. 1999, 274, 35343–35350. [Google Scholar] [CrossRef] [Green Version]
- Spiegel, S.; Milstien, S. The outs and the ins of sphingosine-1-phosphate in immunity. Nat. Rev. Immunol. 2011, 11, 403–415. [Google Scholar] [CrossRef]
- Pantoja, M.; Fischer, K.A.; Ieronimakis, N.; Reyes, M.; Ruohola-Baker, H. Genetic elevation of sphingosine 1-phosphate suppresses dystrophic muscle phenotypes in Drosophila. Development 2013, 140, 136–146. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, A.; Nishi, T.; Hisano, Y.; Fukui, H.; Yamaguchi, A.; Mochizuki, N. The Sphingolipid Transporter Spns2 Functions in Migration of Zebrafish Myocardial Precursors. Science 2009, 323, 524–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hisano, Y.; Kobayashi, N.; Yamaguchi, A.; Nishi, T. Mouse SPNS2 Functions as a Sphingosine-1-Phosphate Transporter in Vascular Endothelial Cells. PLoS ONE 2012, 7, e38941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaye, D.D.; Greenwald, I. OrthoList: A compendium of C. elegans genes with human orthologs. PLoS ONE 2011, 6, e20085. [Google Scholar] [CrossRef]
- Kim, W.; Underwood, R.S.; Greenwald, I.; Shaye, D.D. OrthoList 2: A New Comparative Genomic Analysis of Human and Caenorhabditis elegans Genes. Genetics 2018, 210, 445–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.P.; Sieburth, D. Localized sphingolipid signaling at presynaptic terminals is regulated by calcium influx and promotes recruitment of priming factors. J. Neurosci. 2012, 32, 17909–17920. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.P.; Brown, J.; Hark, B.; Nolan, A.; Servello, D.; Hrobuchak, H.; Staab, T.A. Loss of Sphingosine Kinase Alters Life History Traits and Locomotor Function in Caenorhabditis elegans. Front. Genet. 2017, 8, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.P.; Hu, Z.; Sieburth, D. Recruitment of sphingosine kinase to presynaptic terminals by a conserved muscarinic signaling pathway promotes neurotransmitter release. Genes Dev. 2012, 26, 1070–1085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Sieburth, D. Sphingosine Kinase Regulates Neuropeptide Secretion During the Oxidative Stress-Response through Intertissue Signaling. J. Neurosci. 2018, 38, 8160–8176. [Google Scholar] [CrossRef]
- Kim, S.; Sieburth, D. Sphingosine Kinase Activates the Mitochondrial Unfolded Protein Response and Is Targeted to Mitochondria by Stress. Cell Rep. 2018, 24, 2932–2945.e4. [Google Scholar] [CrossRef] [Green Version]
- Runkel, E.D.; Liu, S.; Baumeister, R.; Schulze, E. Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response. PLoS Genet. 2013, 9, e1003346. [Google Scholar] [CrossRef] [Green Version]
- Pellegrino, M.W.; Nargund, A.M.; Kirienko, N.V.; Gillis, R.; Fiorese, C.J.; Haynes, C.M. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 2014, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melber, A.; Haynes, C.M. UPR mt regulation and output: A stress response mediated by mitochondrial-nuclear communication. Cell Res. 2018, 28, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.; Bazopoulou, D.; Pujol, N.; Tavernarakis, N.; Ewbank, J.J. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007, 8, R194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The C. elegans Deletion Mutant Consortium Large-Scale Screening for Targeted Knockouts in the Caenorhabditis elegans Genome. G3 Genes Genomes Genet. 2012, 2, 1415–1425. [CrossRef] [Green Version]
- Kim, D.H. Signaling in the innate immune response. WormBook 2018, 2018, 1–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.H. Bacteria and the aging and longevity of Caenorhabditis elegans. Annu. Rev. Genet. 2013, 47, 233–246. [Google Scholar] [CrossRef] [Green Version]
- Takabe, K.; Paugh, S.W.; Milstien, S.; Spiegel, S. “Inside-Out” Signaling of Sphingosine-1-Phosphate: Therapeutic Targets. Pharmacol. Rev. 2008, 60, 181–195. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.-C. The Role of Sphingolipids on Innate Immunity to Intestinal Salmonella Infection. Int. J. Mol. Sci. 2017, 18, 1720. [Google Scholar] [CrossRef] [Green Version]
- Bargmann, C.I. Neurobiology of the Caenorhabditis elegans Genome. Science 1998, 282, 2028–2033. [Google Scholar] [CrossRef]
- Vassilatis, D.K.; Hohmann, J.G.; Zeng, H.; Li, F.; Ranchalis, J.E.; Mortrud, M.T.; Brown, A.; Rodriguez, S.S.; Weller, J.R.; Wright, A.C.; et al. The G protein-coupled receptor repertoires of human and mouse. Proc. Natl. Acad. Sci. USA 2003, 100, 4903–4908. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Kolesnick, R. Caenorhabditis elegans as a model to study sphingolipid signaling. Biol. Chem. 2015, 396, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.-E.; Lee, D.; Hwang, S.; Lee, Y.; Lee, J.; Seo, M.; Hwang, W.; Seo, K.; Hwang, A.B.; Artan, M.; et al. Mitochondrial chaperone HSP -60 regulates anti-bacterial immunity via p38 MAP kinase signaling. EMBO J. 2017, 36, 1046–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapira, M.; Hamlin, B.J.; Rong, J.; Chen, K.; Ronen, M.; Tan, M.-W. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 14086–14091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menuz, V.; Howell, K.S.; Gentina, S.; Epstein, S.; Riezman, I.; Fornallaz-Mulhauser, M.; Hengartner, M.O.; Gomez, M.; Riezman, H.; Martinou, J.-C. Protection of C. elegans from anoxia by HYL-2 ceramide synthase. Science 2009, 324, 381–384. [Google Scholar] [CrossRef] [Green Version]
- Bolz, D.D.; Tenor, J.L.; Aballay, A. A conserved PMK-1/p38 MAPK is required in caenorhabditis elegans tissue-specific immune response to Yersinia pestis infection. J. Biol. Chem. 2010, 285, 10832–10840. [Google Scholar] [CrossRef] [Green Version]
- Dierking, K.; Yang, W.; Schulenburg, H. Antimicrobial effectors in the nematode Caenorhabditis elegans: An outgroup to the Arthropoda. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150299. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, M.; Tillman, E.J.; Butty, V.L.; Levine, S.S.; Kim, D.H. Global transcriptional regulation of innate immunity by ATF-7 in C. elegans. PLoS Genet. 2019, 15, e1007830. [Google Scholar] [CrossRef] [Green Version]
- Simonsen, K.T.; Gallego, S.F.; Færgeman, N.J.; Kallipolitis, B.H. Strength in numbers: “Omics” studies of C. elegans innate immunity. Virulence 2012, 3, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Garigan, D.; Hsu, A.-L.; Fraser, A.G.; Kamath, R.S.; Ahringer, J.; Kenyon, C. Genetic analysis of tissue aging in Caenorhabditis elegans: A role for heat-shock factor and bacterial proliferation. Genetics 2002, 161, 1101–1112. [Google Scholar]
- Youngman, M.J.; Rogers, Z.N.; Kim, D.H. A Decline in p38 MAPK Signaling Underlies Immunosenescence in Caenorhabditis elegans. PLoS Genet. 2011, 7, e1002082. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Mylonakis, E. An Intestine-Derived Neuropeptide Controls Avoidance Behavior in Caenorhabditis elegans. Cell Rep. 2017, 20, 2501–2512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CLSI (Clinical and Laboratory Standards Institute) - Document CLSI M7-A7; CLSI; 2012 Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. Available online: https://clsi.org/standards/products/microbiology/documents/m07/ (accessed on 19 October 2020).
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Ding, J.S.O.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Boehnisch, C.; Wong, D.; Habig, M.; Isermann, K.; Michiels, N.K.; Roeder, T.; May, R.C.; Schulenburg, H. Protist-type lysozymes of the nematode Caenorhabditis elegans contribute to resistance against pathogenic Bacillus thuringiensis. PLoS ONE 2011, 6, e24619. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Liu, X.; Yu, H.; Yin, X.; Nie, S.-P.; Xie, M.-Y.; Chen, W.; Gong, J. Cell Signaling of Caenorhabditis elegans in Response to Enterotoxigenic Escherichia coli Infection and Lactobacillus zeae Protection. Front. Immunol. 2018, 9, 1745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di, R.; Zhang, H.; Lawton, M.A. Transcriptome Analysis of C. elegans Reveals Novel Targets for DON Cytotoxicity. Toxins (Basel) 2018, 10. [Google Scholar] [CrossRef]
- Block, D.H.S.; Twumasi-Boateng, K.; Kang, H.S.; Carlisle, J.A.; Hanganu, A.; Lai, T.Y.-J.; Shapira, M. The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult C. elegans. PLoS Genet. 2015, 11, e1005265. [Google Scholar] [CrossRef] [Green Version]
- Hoogewijs, D.; Houthoofd, K.; Matthijssens, F.; Vandesompele, J.; Vanfleteren, J.R. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 2008, 9, 9. [Google Scholar] [CrossRef] [Green Version]
| References | Sequence Name | Gene Name | Direction | Sequence |
|---|---|---|---|---|
| [44] | Y22F5A.5 | lys-2 | F | GCTGGATTGGGAATTGAGAC |
| Y22F5A.5 | lys-2 | R | GACGTTGGCAGTTGGATTG | |
| [23] | F35C5.6 | clec-63 | F | AGGAGCTGCTCTTCAAACCA |
| F35C5.6 | clec-63 | R | TCCAGGATGAGGAGATGGTG | |
| [45] | T07C4.4 | spp-1 | F | TGGACTATGCTGTTGCCGTT |
| T07C4.4 | spp-1 | R | ACGCCTTGTCTGGAGAATCC | |
| [46] | K08D8.5 | F | CCGGGAAGTCGAATGAACAT | |
| K08D8.5 | R | GATGCAACACCTGCCAAAGA | ||
| [47] | F08G5.6 | irg-4 | F | CACAATGATTTCAATGCGAGA |
| F08G5.6 | irg-4 | R | GTTTCGACCGAGAAATCGAG | |
| [47] | F55G11.2 | F | TGGTTCTCCAGACGTGTTCA | |
| F55G11.2 | R | CAGCCTTGCCTTTACTGACA | ||
| [48] | C54G10.3 | pmp-3 | F | GTTCCCGTGTTCATCACTCAT |
| C54G10.3 | pmp-3 | R | ACACCGTCGAGAAGCTGTAGA | |
| [48] | Y45F10D.4 | F | GTCGCTTCAAATCAGTTCAGC | |
| Y45F10D.4 | R | GTTCTTGTCAAGTGATCCGACA | ||
| [23] | F56D6.2 | clec-67 | F | CGGGCTGGGAATATATCAAT |
| F56D6.2 | clec-67 | R | CAATAGGTTGTGGCGTATGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.; Escobar, I.; Jang, Y.; Kim, W.; Ausubel, F.M.; Mylonakis, E. In the Model Host Caenorhabditis elegans, Sphingosine-1-Phosphate-Mediated Signaling Increases Immunity toward Human Opportunistic Bacteria. Int. J. Mol. Sci. 2020, 21, 7813. https://doi.org/10.3390/ijms21217813
Lee K, Escobar I, Jang Y, Kim W, Ausubel FM, Mylonakis E. In the Model Host Caenorhabditis elegans, Sphingosine-1-Phosphate-Mediated Signaling Increases Immunity toward Human Opportunistic Bacteria. International Journal of Molecular Sciences. 2020; 21(21):7813. https://doi.org/10.3390/ijms21217813
Chicago/Turabian StyleLee, Kiho, Iliana Escobar, Yeeun Jang, Wooseong Kim, Frederick M. Ausubel, and Eleftherios Mylonakis. 2020. "In the Model Host Caenorhabditis elegans, Sphingosine-1-Phosphate-Mediated Signaling Increases Immunity toward Human Opportunistic Bacteria" International Journal of Molecular Sciences 21, no. 21: 7813. https://doi.org/10.3390/ijms21217813





