Current Status of Circulating Tumor DNA Liquid Biopsy in Pancreatic Cancer
Abstract
1. Pancreatic Cancer: The Need for a Novel Diagnostic
2. cfDNA and ctDNA in PDAC
A Brief Mention of Exosomes
3. Liquid Biopsy Techniques in PDAC
4. Current Clinical Utility of cfDNA
4.1. ctDNA as a Screening Test
4.2. ctDNA as a Diagnostic and Prognostic Test
4.3. ctDNA in Assessing Resectability of Primary Tumor
4.4. Surveillance
4.5. Clinical Utility Of Exosomes
5. Ongoing Clinical Trials and the Future of Cfdna in Pdac
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PDAC | Pancreatic Ductal Adenocarcinoma |
| USPSTF | United States Preventative Service Task Force |
| OS | Overall Survival |
| EUS | Endoscopic Ultrasound |
| cfDNA | Cell Free Deoxyribonucleic Acid |
| ctDNA | Cell Free Tumor Deoxyribonucleic Acid |
| CTCs | Circulating Tumor Cells |
| PCR | Polymerase Chain Reaction |
| NGS | Next Generation Sequencing |
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- SEER Cancer Statistics Review, 1975–2017. Available online: https://seer.cancer.gov/csr/1975_2017/index.html (accessed on 13 July 2020).
- Ilic, M.; Ilic, I. Epidemiology of pancreatic cancer. World J. Gastroenterol. 2016, 22, 9694–9705. [Google Scholar] [CrossRef]
- Chang, J.S.; Chen, L.; Shan, Y.; Chu, P.; Tsai, C.; Tsai, H. The incidence and survival of pancreatic cancer by histology, including rare subtypes: A nation-wide cancer registry-based study from Taiwan. Cancer Med. 2018, 7, 5775–5788. [Google Scholar] [CrossRef]
- Huang, L.; Jansen, L.; Balavarca, Y.; Babaei, M.; van der Geest, L.; Lemmens, V.; Van Eycken, L.; De Schutter, H.; Johannesen, T.B.; Primic-Žakelj, M.; et al. Stratified survival of resected and overall pancreatic cancer patients in Europe and the USA in the early twenty-first century: A large, international population-based study. BMC Med. 2018, 16. [Google Scholar] [CrossRef] [PubMed]
- Shaib, Y.; Davila, J.; Naumann, C.; El-Serag, H. The Impact of Curative Intent Surgery on the Survival of Pancreatic Cancer Patients: A U.S. Population-Based Study. Off. J. Am. Coll. Gastroenterol. ACG 2007, 102, 1377–1382. [Google Scholar] [CrossRef]
- Mokdad, A.A.; Minter, R.M.; Zhu, H.; Augustine, M.M.; Porembka, M.R.; Wang, S.C.; Yopp, A.C.; Mansour, J.C.; Choti, M.A.; Polanco, P.M. Neoadjuvant Therapy Followed by Resection Versus Upfront Resection for Resectable Pancreatic Cancer: A Propensity Score Matched Analysis. J. Clin. Oncol. 2017, 35, 515–522. [Google Scholar] [CrossRef]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Gobbi, P.G.; Bergonzi, M.; Comelli, M.; Villano, L.; Pozzoli, D.; Vanoli, A.; Dionigi, P. The prognostic role of time to diagnosis and presenting symptoms in patients with pancreatic cancer. Cancer Epidemiol. 2013, 37, 186–190. [Google Scholar] [CrossRef]
- Force, U.P.S.T.; Owens, D.K.; Davidson, K.W.; Krist, A.H.; Barry, M.J.; Cabana, M.; Caughey, A.B.; Curry, S.J.; Doubeni, C.A.; Epling, J.W.; et al. Screening for Pancreatic Cancer: US Preventive Services Task Force Reaffirmation Recommendation Statement. JAMA 2019, 322, 438–444. [Google Scholar] [CrossRef]
- McAllister, F.; Montiel, M.F.; Uberoi, G.S.; Uberoi, A.S.; Maitra, A.; Bhutani, M.S. Current Status and Future Directions for Screening Patients at High Risk for Pancreatic Cancer. Gastroenterol. Hepatol. (N. Y.) 2017, 13, 268–275. [Google Scholar]
- Kitano, M.; Yoshida, T.; Itonaga, M.; Tamura, T.; Hatamaru, K.; Yamashita, Y. Impact of endoscopic ultrasonography on diagnosis of pancreatic cancer. J. Gastroenterol. 2019, 54, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Zhang, Y.; Chen, Q.; Sun, B.; Deng, Z.; Shan, H.; Dou, L.; Wang, J.; Li, Y.; Yang, X.; et al. Analysis of Fine-Needle Biopsy vs Fine-Needle Aspiration in Diagnosis of Pancreatic and Abdominal Masses: A Prospective, Multicenter, Randomized Controlled Trial. Clin. Gastroenterol. Hepatol. 2018, 16, 1314–1321. [Google Scholar] [CrossRef] [PubMed]
- Wittel, U.A.; Lubgan, D.; Ghadimi, M.; Belyaev, O.; Uhl, W.; Bechstein, W.O.; Grützmann, R.; Hohenberger, W.M.; Schmid, A.; Jacobasch, L.; et al. Consensus in determining the resectability of locally progressed pancreatic ductal adenocarcinoma—Results of the Conko-007 multicenter trial. BMC Cancer 2019, 19, 979. [Google Scholar] [CrossRef] [PubMed]
- Neoptolemos, J.P.; Stocken, D.D.; Friess, H.; Bassi, C.; Dunn, J.A.; Hickey, H.; Beger, H.; Fernandez-Cruz, L.; Dervenis, C.; Lacaine, F.; et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N. Engl. J. Med. 2004, 350, 1200–1210. [Google Scholar] [CrossRef]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouché, O.; Guimbaud, R.; Bécouarn, Y.; Adenis, A.; Raoul, J.-L.; Gourgou-Bourgade, S.; de la Fouchardière, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef]
- Ferrone, C.R.; Marchegiani, G.; Hong, T.S.; Ryan, D.P.; Deshpande, V.; McDonnell, E.I.; Sabbatino, F.; Santos, D.D.; Allen, J.N.; Blaszkowsky, L.S.; et al. Radiological and surgical implications of neoadjuvant treatment with FOLFIRINOX for locally advanced and borderline resectable pancreatic cancer. Ann. Surg. 2015, 261, 12–17. [Google Scholar] [CrossRef]
- Marrugo-Ramírez, J.; Mir, M.; Samitier, J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int. J. Mol. Sci. 2018, 19, 2877. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Vymetalkova, V.; Cervena, K.; Bartu, L.; Vodicka, P. Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. Int. J. Mol. Sci. 2018, 19, 3356. [Google Scholar] [CrossRef]
- Yadav, D.K.; Bai, X.; Yadav, R.K.; Singh, A.; Li, G.; Ma, T.; Chen, W.; Liang, T. Liquid biopsy in pancreatic cancer: The beginning of a new era. Oncotarget 2018, 9, 26900–26933. [Google Scholar] [CrossRef]
- Jacobson, R.A.; Munding, E.; Hayden, D.M.; Levy, M.; Kuzel, T.M.; Pappas, S.G.; Masood, A. Evolving Clinical Utility of Liquid Biopsy in Gastrointestinal Cancers. Cancers 2019, 11, 1164. [Google Scholar] [CrossRef] [PubMed]
- De Wit, S.; van Dalum, G.; Terstappen, L.W.M.M. Detection of Circulating Tumor Cells. Scientifica (Cairo) 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.; Okamura, R.; Fanta, P.; Patel, C.; Lanman, R.B.; Raymond, V.M.; Kato, S.; Kurzrock, R. Clinical correlates of blood-derived circulating tumor DNA in pancreatic cancer. J. Hematol. Oncol. 2019, 12, 130. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Circulating Tumor Cells: Liquid Biopsy of Cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Mandel, P.; Metais, P. Les acides nucleiques du plasma sanguin chez l’ homme. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Bendich, A.; Wilczok, T.; Borenfreund, E. Circulating DNA as a Possible Factor in Oncogenesis. Science 1965, 148, 374–376. [Google Scholar] [CrossRef] [PubMed]
- Samandari, M.; Julia, M.G.; Rice, A.; Chronopoulos, A.; Armando, E. Liquid biopsies for management of pancreatic cancer. Transl. Res. 2018, 201, 98–127. [Google Scholar] [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. JCO 2018, 36, 1631–1641. [Google Scholar] [CrossRef]
- Kinugasa, H.; Nouso, K.; Miyahara, K.; Morimoto, Y.; Dohi, C.; Tsutsumi, K.; Kato, H.; Matsubara, T.; Okada, H.; Yamamoto, K. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer 2015, 121, 2271–2280. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017, 32, 185–203. [Google Scholar] [CrossRef]
- San Lucas, F.A.; Allenson, K.; Bernard, V.; Castillo, J.; Kim, D.U.; Ellis, K.; Ehli, E.A.; Davies, G.E.; Petersen, J.L.; Li, D.; et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann. Oncol. 2016, 27, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Allenson, K.; Castillo, J.; San Lucas, F.A.; Scelo, G.; Kim, D.U.; Bernard, V.; Davis, G.; Kumar, T.; Katz, M.; Overman, M.J.; et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 2017, 28, 741–747. [Google Scholar] [CrossRef]
- Erb, U.; Zoller, M. Progress and potential of exosome analysis for early pancreatic cancer detection. Expert Rev. Mol. Diagn. 2016, 16, 757–767. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Wang, M.; McElyea, S.D.; Sherman, S.; House, M.; Korc, M. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 2017, 393, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Exosomes Facilitate Therapeutic Targeting of Oncogenic KRAS in Pancreatic Cancer | Nature. Available online: https://www.nature.com/articles/nature22341?__hstc=12316075.07430159d50a3c91e72c280a7921bf0d.1525305600142.1525305600143.1525305600144.1&__hssc=12316075.1.1525305600145&__hsfp=1773666937 (accessed on 12 September 2020).
- Diefenbach, R.J.; Lee, J.H.; Kefford, R.F.; Rizos, H. Evaluation of commercial kits for purification of circulating free DNA. Cancer Genet 2018, 228–229, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.P.; Hong, T.S.; Bardeesy, N. Pancreatic Adenocarcinoma. Available online: http://www.nejm.org/doi/10.1056/NEJMra1404198 (accessed on 15 September 2020).
- Takai, E.; Totoki, Y.; Nakamura, H.; Kato, M.; Shibata, T.; Yachida, S. Clinical Utility of Circulating Tumor DNA for Molecular Assessment and Precision Medicine in Pancreatic Cancer. Adv. Exp. Med. Biol. 2016, 924, 13–17. [Google Scholar] [CrossRef]
- Witkiewicz, A.K.; McMillan, E.A.; Balaji, U.; Baek, G.; Lin, W.-C.; Mansour, J.; Mollaee, M.; Wagner, K.-U.; Koduru, P.; Yopp, A.; et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015, 6, 6744. [Google Scholar] [CrossRef]
- Denis, J.A.; Guillerm, E.; Coulet, F.; Larsen, A.K.; Lacorte, J.-M. The Role of BEAMing and Digital PCR for Multiplexed Analysis in Molecular Oncology in the Era of Next-Generation Sequencing. Mol. Diagn. Ther. 2017, 21, 587–600. [Google Scholar] [CrossRef]
- Bennett, C.W.; Berchem, G.; Kim, Y.J.; El-Khoury, V. Cell-free DNA and next-generation sequencing in the service of personalized medicine for lung cancer. Oncotarget 2016, 7, 71013–71035. [Google Scholar] [CrossRef]
- Kyrochristos, I.D.; Ziogas, D.E.; Glantzounis, G.K.; Roukos, D.H. Prediction of pancreatic cancer risk and therapeutic response with next-generation sequencing. Biomark. Med. 2017, 12, 5–8. [Google Scholar] [CrossRef]
- Zill, O.A.; Greene, C.; Sebisanovic, D.; Siew, L.M.; Leng, J.; Vu, M.; Hendifar, A.E.; Wang, Z.; Atreya, C.E.; Kelley, R.K.; et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer Discov. 2015, 5, 1040–1048. [Google Scholar] [CrossRef]
- Park, G.; Park, J.K.; Son, D.S.; Shin, S.H.; Kim, Y.J.; Jeon, H.J.; Lee, J.; Park, W.Y.; Lee, K.H.; Park, D. Utility of targeted deep sequencing for detecting circulating tumor DNA in pancreatic cancer patients. Sci. Rep. 2018, 8, 11631. [Google Scholar] [CrossRef]
- Liang, W.S.; Craig, D.W.; Carpten, J.; Borad, M.J.; Demeure, M.J.; Weiss, G.J.; Izatt, T.; Sinari, S.; Christoforides, A.; Aldrich, J.; et al. Genome-wide characterization of pancreatic adenocarcinoma patients using next generation sequencing. PLoS ONE 2012, 7, e43192. [Google Scholar] [CrossRef]
- Sausen, M.; Phallen, J.; Adleff, V.; Jones, S.; Leary, R.J.; Barrett, M.T.; Anagnostou, V.; Parpart-Li, S.; Murphy, D.; Kay Li, Q.; et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 2015, 6, 7686. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Naranjo, J.C.; Wu, H.-J.; Ugaz, V.M. Microfluidics for exosome isolation and analysis: Enabling liquid biopsy for personalized medicine. Lab Chip 2017, 17, 3558–3577. [Google Scholar] [CrossRef]
- Ludwig, N.; Whiteside, T.L.; Reichert, T.E. Challenges in Exosome Isolation and Analysis in Health and Disease. Int. J. Mol. Sci. 2019, 20, 4684. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, H.; Chen, N.; Hao, J.; Jin, H.; Ma, X. Diagnostic value of various liquid biopsy methods for pancreatic cancer: A systematic review and meta-analysis. Medicine (Baltimore) 2020, 99, e18581. [Google Scholar] [CrossRef]
- Sefrioui, D.; Blanchard, F.; Toure, E.; Basile, P.; Beaussire, L.; Dolfus, C.; Perdrix, A.; Paresy, M.; Antonietti, M.; Iwanicki-Caron, I.; et al. Diagnostic value of CA19.9, circulating tumour DNA and circulating tumour cells in patients with solid pancreatic tumours. Br. J. Cancer 2017, 117, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.W.; Schwerdel, D.; Costa, I.G.; Hackert, T.; Strobel, O.; Lam, S.; Barth, T.F.; Schröppel, B.; Meining, A.; Büchler, M.W.; et al. Detection of Hot-Spot Mutations in Circulating Cell-Free DNA From Patients With Intraductal Papillary Mucinous Neoplasms of the Pancreas. Gastroenterology 2016, 151, 267–270. [Google Scholar] [CrossRef]
- Liggett, T.; Melnikov, A.; Yi, Q.L.; Replogle, C.; Brand, R.; Kaul, K.; Talamonti, M.; Abrams, R.A.; Levenson, V. Differential methylation of cell-free circulating DNA among patients with pancreatic cancer versus chronic pancreatitis. Cancer 2010, 116, 1674–1680. [Google Scholar] [CrossRef] [PubMed]
- Castells, A.; Puig, P.; Móra, J.; Boadas, J.; Boix, L.; Urgell, E.; Solé, M.; Capellà, G.; Lluís, F.; Fernández-Cruz, L.; et al. K-ras mutations in DNA extracted from the plasma of patients with pancreatic carcinoma: Diagnostic utility and prognostic significance. J. Clin. Oncol. 1999, 17, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Eissa, M.A.L.; Lerner, L.; Abdelfatah, E.; Shankar, N.; Canner, J.K.; Hasan, N.M.; Yaghoobi, V.; Huang, B.; Kerner, Z.; Takaesu, F.; et al. Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin. Epigenet. 2019, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Maire, F.; Micard, S.; Hammel, P.; Voitot, H.; Lévy, P.; Cugnenc, P.-H.; Ruszniewski, P.; Puig, P.L. Differential diagnosis between chronic pancreatitis and pancreatic cancer: Value of the detection of KRAS2 mutations in circulating DNA. Br. J. Cancer 2002, 87, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Javed, A.A.; Thoburn, C.; Wong, F.; Tie, J.; Gibbs, P.; Schmidt, C.M.; Yip-Schneider, M.T.; Allen, P.J.; Schattner, M.; et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 10202–10207. [Google Scholar] [CrossRef]
- Campos-Carrillo, A.; Weitzel, J.N.; Sahoo, P.; Rockne, R.; Mokhnatkin, J.V.; Murtaza, M.; Gray, S.W.; Goetz, L.; Goel, A.; Schork, N.; et al. Circulating tumor DNA as an early cancer detection tool. Pharmacol. Ther. 2020, 207, 107458. [Google Scholar] [CrossRef]
- Kato, S.; Honda, K. Use of Biomarkers and Imaging for Early Detection of Pancreatic Cancer. Cancers 2020, 12, 1965. [Google Scholar] [CrossRef]
- Henriksen, S.D.; Madsen, P.H.; Larsen, A.C.; Johansen, M.B.; Drewes, A.M.; Pedersen, I.S.; Krarup, H.; Thorlacius-Ussing, O. Cell-free DNA promoter hypermethylation in plasma as a diagnostic marker for pancreatic adenocarcinoma. Clin. Epigenet. 2016, 8, 117. [Google Scholar] [CrossRef]
- Shen, S.Y.; Singhania, R.; Fehringer, G.; Chakravarthy, A.; Roehrl, M.H.A.; Chadwick, D.; Zuzarte, P.C.; Borgida, A.; Wang, T.T.; Li, T.; et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018, 563, 579–583. [Google Scholar] [CrossRef]
- Poruk, K.E.; Gay, D.Z.; Brown, K.; Mulvihill, J.D.; Boucher, K.M.; Scaife, C.L.; Firpo, M.A.; Mulvihill, S.J. The Clinical Utility of CA 19-9 in Pancreatic Adenocarcinoma: Diagnostic and Prognostic Updates. Curr. Mol. Med. 2013, 13, 340–351. [Google Scholar]
- Kim, M.K.; Woo, S.M.; Park, B.; Yoon, K.-A.; Kim, Y.-H.; Joo, J.; Lee, W.J.; Han, S.-S.; Park, S.-J.; Kong, S.-Y. Prognostic Implications of Multiplex Detection of KRAS Mutations in Cell-Free DNA from Patients with Pancreatic Ductal Adenocarcinoma. Clin. Chem. 2018, 64, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gupta, S.; Pandey, R.M.; Chauhan, S.S.; Saraya, A. High levels of cell-free circulating nucleic acids in pancreatic cancer are associated with vascular encasement, metastasis and poor survival. Cancer Investig. 2015, 33, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Buscail, E.; Maulat, C.; Muscari, F.; Chiche, L.; Cordelier, P.; Dabernat, S.; Alix-Panabieres, C.; Buscail, L. Liquid Biopsy Approach for Pancreatic Ductal Adenocarcinoma. Cancers 2019, 11, 852. [Google Scholar] [CrossRef] [PubMed]
- Earl, J.; Garcia-Nieto, S.; Martinez-Avila, J.C.; Montans, J.; Sanjuanbenito, A.; Rodríguez-Garrote, M.; Lisa, E.; Mendía, E.; Lobo, E.; Malats, N.; et al. Circulating tumor cells (CTC) and KRAS mutant circulating free DNA (cfDNA) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer 2015, 15, 797. [Google Scholar] [CrossRef] [PubMed]
- Ako, S.; Nouso, K.; Kinugasa, H.; Dohi, C.; Matushita, H.; Mizukawa, S.; Muro, S.; Akimoto, Y.; Uchida, D.; Tomoda, T.; et al. Utility of serum DNA as a marker for KRAS mutations in pancreatic cancer tissue. Pancreatology 2017, 17, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Hadano, N.; Murakami, Y.; Uemura, K.; Hashimoto, Y.; Kondo, N.; Nakagawa, N.; Sueda, T.; Hiyama, E. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br. J. Cancer 2016, 115, 59–65. [Google Scholar] [CrossRef]
- Vietsch, E.E.; van Eijck, C.H.; Wellstein, A. Circulating DNA and Micro-RNA in Patients with Pancreatic Cancer. Pancreat. Disord. Ther. 2015, 5. [Google Scholar] [CrossRef]
- Pishvaian, M.J.; Joseph Bender, R.; Matrisian, L.M.; Rahib, L.; Hendifar, A.; Hoos, W.A.; Mikhail, S.; Chung, V.; Picozzi, V.; Heartwell, C.; et al. A pilot study evaluating concordance between blood-based and patient-matched tumor molecular testing within pancreatic cancer patients participating in the Know Your Tumor (KYT) initiative. Oncotarget 2016, 8, 83446–83456. [Google Scholar] [CrossRef]
- Shu, Y.; Wu, X.; Tong, X.; Wang, X.; Chang, Z.; Mao, Y.; Chen, X.; Sun, J.; Wang, Z.; Hong, Z.; et al. Circulating Tumor DNA Mutation Profiling by Targeted Next Generation Sequencing Provides Guidance for Personalized Treatments in Multiple Cancer Types. Sci. Rep. 2017, 7, 583. [Google Scholar] [CrossRef]
- Bitterman, D.S.; Price, K.S.; Van Seventer, E.E.; Clark, J.W.; Allen, J.N.; Blaszkowsky, L.S.; Ryan, D.P.; Eyler, C.E.; Wo, J.Y.-L.; Hong, T.S.; et al. Noninvasive comprehensive genomic profiling from plasma ctDNA in pancreatic cancer patients. JCO 2020, 38, 753. [Google Scholar] [CrossRef]
- Cheng, H.; Liu, C.; Jiang, J.; Luo, G.; Lu, Y.; Jin, K.; Guo, M.; Zhang, Z.; Xu, J.; Liu, L.; et al. Analysis of ctDNA to predict prognosis and monitor treatment responses in metastatic pancreatic cancer patients. Int. J. Cancer 2017, 140, 2344–2350. [Google Scholar] [CrossRef] [PubMed]
- Glazer, E.S.; Rashid, O.M.; Klapman, J.B.; Harris, C.L.; Hodul, P.J.; Pimiento, J.M.; Malafa, M.P. Endoscopic ultrasonography complements computed tomography in predicting portal or superior mesenteric vein resection in patients with borderline resectable pancreatic carcinoma. Pancreatology 2017, 17, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Sinn, M.; Bahra, M.; Liersch, T.; Gellert, K.; Messmann, H.; Bechstein, W.; Waldschmidt, D.; Jacobasch, L.; Wilhelm, M.; Rau, B.M.; et al. CONKO-005: Adjuvant Chemotherapy With Gemcitabine Plus Erlotinib Versus Gemcitabine Alone in Patients After R0 Resection of Pancreatic Cancer: A Multicenter Randomized Phase III Trial. J. Clin. Oncol. 2017, 35, 3330–3337. [Google Scholar] [CrossRef] [PubMed]
- Oettle, H.; Neuhaus, P.; Hochhaus, A.; Hartmann, J.T.; Gellert, K.; Ridwelski, K.; Niedergethmann, M.; Zulke, C.; Fahlke, J.; Arning, M.B.; et al. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: The CONKO-001 randomized trial. JAMA 2013, 310, 1473–1481. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, T.; Uesaka, K.; Kanemoto, H.; Mizuno, T.; Sasaki, K.; Furukawa, H.; Matsunaga, K.; Maeda, A. Serum CA19-9 is a significant predictor among preoperative parameters for early recurrence after resection of pancreatic adenocarcinoma. J. Gastrointest. Surg. 2012, 16, 977–985. [Google Scholar] [CrossRef]
- Fong, Z.V.; Alvino, D.M.L.; Fernandez-Del Castillo, C.; Mehtsun, W.T.; Pergolini, I.; Warshaw, A.L.; Chang, D.C.; Lillemoe, K.D.; Ferrone, C.R. Reappraisal of Staging Laparoscopy for Patients with Pancreatic Adenocarcinoma: A Contemporary Analysis of 1001 Patients. Ann. Surg. Oncol. 2017, 24, 3203–3211. [Google Scholar] [CrossRef]
- Adamo, P.; Cowley, C.M.; Neal, C.P.; Mistry, V.; Page, K.; Dennison, A.R.; Isherwood, J.; Hastings, R.; Luo, J.; Moore, D.A.; et al. Profiling tumour heterogeneity through circulating tumour DNA in patients with pancreatic cancer. Oncotarget 2017, 8, 87221–87233. [Google Scholar] [CrossRef]
- Brychta, N.; Krahn, T.; von Ahsen, O. Detection of KRAS Mutations in Circulating Tumor DNA by Digital PCR in Early Stages of Pancreatic Cancer. Clin. Chem. 2016, 62, 1482–1491. [Google Scholar] [CrossRef] [PubMed]
- Almoguera, C.; Shibata, D.; Forrester, K.; Martin, J.; Arnheim, N.; Perucho, M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 1988, 53, 549–554. [Google Scholar] [CrossRef]
- Takai, E.; Totoki, Y.; Nakamura, H.; Morizane, C.; Nara, S.; Hama, N.; Suzuki, M.; Furukawa, E.; Kato, M.; Hayashi, H.; et al. Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci. Rep. 2015, 5, 18425. [Google Scholar] [CrossRef]
- Nishio, K.; Kimura, K.; Amano, R.; Yamazoe, S.; Ohrira, G.; Nakata, B.; Hirakawa, K.; Ohira, M. Preoperative predictors for early recurrence of resectable pancreatic cancer. World J. Surg. Oncol. 2017, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Lipton, L.; Cohen, J.; Tie, J.; Javed, A.A.; Li, L.; Goldstein, D.; Burge, M.; Cooray, P.; Nagrial, A.; et al. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann. Oncol. 2019, 30, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- McDuff, S.; Parikh, A.R.; Hazar-Rethinam, M.; Zheng, H.; Van Seventer, E.; Nadres, B.; Ryan, D.P.; Weekes, C.D.; Clark, J.W.; Fernandez-del Castillo, C.; et al. Using circulating tumor DNA (ctDNA) to predict surgical outcome after neoadjuvant chemoradiation for locally advanced pancreatic cancer (LAPC). JCO 2018, 36, 272. [Google Scholar] [CrossRef]
- Lee, J.-S.; Rhee, T.-M.; Pietrasz, D.; Bachet, J.-B.; Laurent-Puig, P.; Kong, S.-Y.; Takai, E.; Yachida, S.; Shibata, T.; Lee, J.W.; et al. Circulating tumor DNA as a prognostic indicator in resectable pancreatic ductal adenocarcinoma: A systematic review and meta-analysis. Sci. Rep. 2019, 9, 16971. [Google Scholar] [CrossRef] [PubMed]
- Pietrasz, D.; Pécuchet, N.; Garlan, F.; Didelot, A.; Dubreuil, O.; Doat, S.; Imbert-Bismut, F.; Karoui, M.; Vaillant, J.-C.; Taly, V.; et al. Plasma Circulating Tumor DNA in Pancreatic Cancer Patients Is a Prognostic Marker. Clin. Cancer Res. 2017, 23, 116–123. [Google Scholar] [CrossRef]
- Nakano, Y.; Kitago, M.; Matsuda, S.; Nakamura, Y.; Fujita, Y.; Imai, S.; Shinoda, M.; Yagi, H.; Abe, Y.; Hibi, T.; et al. KRAS mutations in cell-free DNA from preoperative and postoperative sera as a pancreatic cancer marker: A retrospective study. Br. J. Cancer 2018, 118, 662–669. [Google Scholar] [CrossRef]
- Groot, V.P.; Mosier, S.; Javed, A.A.; Teinor, J.A.; Gemenetzis, G.; Ding, D.; Haley, L.M.; Yu, J.; Burkhart, R.A.; Hasanain, A.; et al. Circulating Tumor DNA as a Clinical Test in Resected Pancreatic Cancer. Clin. Cancer Res. 2019, 25, 4973–4984. [Google Scholar] [CrossRef]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.S.; et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef]
- Lee, B.; Cohen, J.; Lipton, L.R.; Tie, J.; Javed, A.A.; Li, L.; Goldstein, D.; Cooray, P.; Nagrial, A.; Burge, M.E.; et al. Potential role of circulating tumor DNA (ctDNA) in the early diagnosis and post-operative management of localised pancreatic cancer. JCO 2017, 35, 4101. [Google Scholar] [CrossRef]
- Azizian, A.; Rühlmann, F.; Krause, T.; Bernhardt, M.; Jo, P.; König, A.; Kleiß, M.; Leha, A.; Ghadimi, M.; Gaedcke, J. CA19-9 for detecting recurrence of pancreatic cancer. Sci. Rep. 2020, 10, 1332. [Google Scholar] [CrossRef]
- Däbritz, J.; Preston, R.; Hänfler, J.; Oettle, H. Follow-Up Study of K-ras Mutations in the Plasma of Patients With Pancreatic Cancer: Correlation With Clinical Features and Carbohydrate Antigen 19-9. Pancreas 2009, 38, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Del Re, M.; Vivaldi, C.; Rofi, E.; Vasile, E.; Miccoli, M.; Caparello, C.; d’Arienzo, P.D.; Fornaro, L.; Falcone, A.; Danesi, R. Early changes in plasma DNA levels of mutant KRAS as a sensitive marker of response to chemotherapy in pancreatic cancer. Sci. Rep. 2017, 7, 7931. [Google Scholar] [CrossRef] [PubMed]
- Bernard, V.; Kim, D.U.; San Lucas, F.A.; Castillo, J.; Allenson, K.; Mulu, F.C.; Stephens, B.M.; Huang, J.; Semaan, A.; Guerrero, P.A.; et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology 2019, 156, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.T.; Lim, D.H.; Jang, K.-T.; Lim, T.; Lee, J.; Choi, Y.-L.; Jang, H.-L.; Yi, J.H.; Baek, K.K.; Park, S.H.; et al. Impact of KRAS Mutations on Clinical Outcomes in Pancreatic Cancer Patients Treated with First-line Gemcitabine-Based Chemotherapy. Mol. Cancer Ther. 2011, 10, 1993–1999. [Google Scholar] [CrossRef] [PubMed]
- Semrad, T.; Barzi, A.; Lenz, H.-J.; Hutchins, I.M.; Kim, E.J.; Gong, I.-Y.; Tanaka, M.; Beckett, L.; Holland, W.; Burich, R.A.; et al. Pharmacodynamic separation of gemcitabine and erlotinib in locally advanced or metastatic pancreatic cancer: Therapeutic and biomarker results. Int. J. Clin. Oncol. 2015, 20, 518–524. [Google Scholar] [CrossRef]
- Nagai, M.; Sho, M.; Akahori, T.; Nakagawa, K.; Nakamura, K. Application of liquid biopsy for surgical management of pancreatic cancer. Ann. Gastroenterol. Surg. 2020, 4, 216–223. [Google Scholar] [CrossRef]
- Xu, Y.F.; Hannafon, B.N.; Zhao, Y.D.; Postier, R.G.; Ding, W.Q. Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer. Oncotarget 2017, 8, 77028–77040. [Google Scholar] [CrossRef]
- Yang, S.; Che, S.P.Y.; Kurywchak, P.; Tavormina, J.L.; Gansmo, L.B.; de Sampaio, P.C.; Tachezy, M.; Bockhorn, M.; Gebauer, F.; Haltom, A.R.; et al. Detection of mutant KRAS and TP53 DNA in circulating exosomes from healthy individuals and patients with pancreatic cancer. Cancer Biol. Ther. 2017, 18, 158–165. [Google Scholar] [CrossRef]
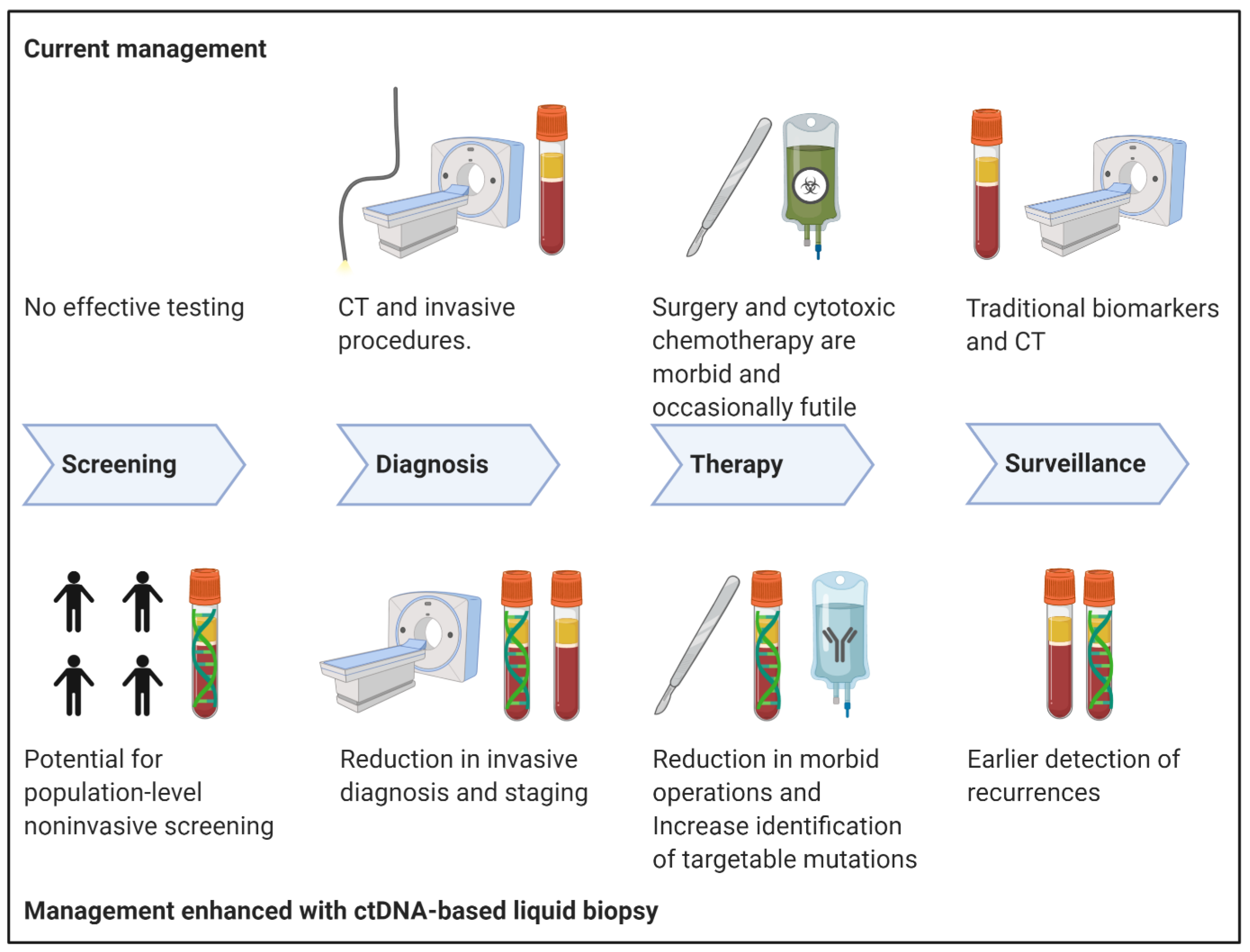
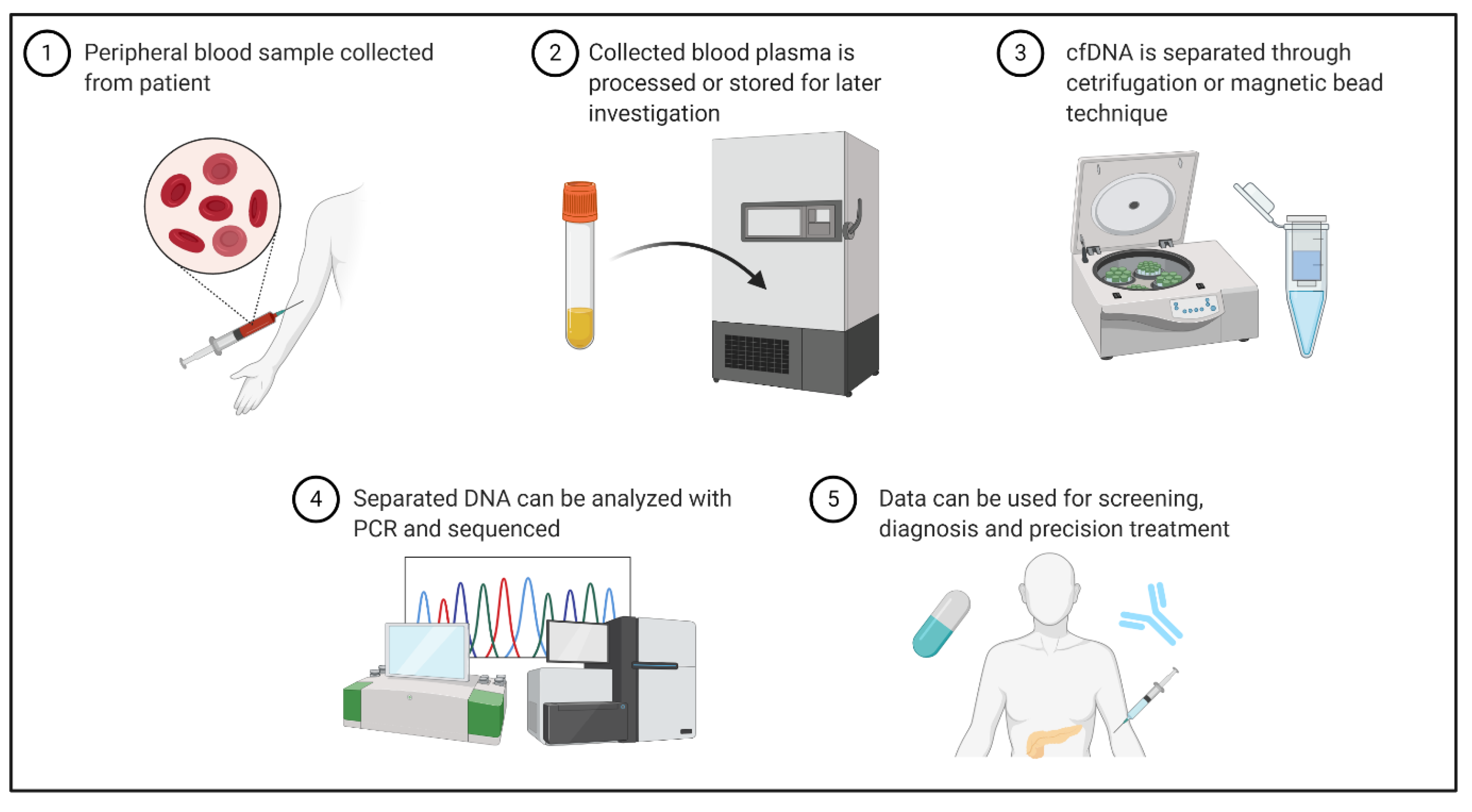
| Trial Number | Trial Name | ctDNA Liquid Biopsy Focus/Goals | Study Type | Estimated Completion | Recruitment Status |
|---|---|---|---|---|---|
| NCT02079363 | DNA Promoter Hypermethylation as a Blood Based Maker for Pancreatic Cancer | Assessment of hypermethylation as diagnostic, prognostic, and recurrence marker | Prospective Observational Cohort | January 2018 | Unknown |
| NCT03524677 | Mutation of K-RAS, CDKN2A, SMAD4 and TP53 in Pancreatic Cancer: Role of Liquid Biopsy in Preoperative Diagnosis | Assessment of four ctDNA mutations’ impact on preoperative staging and progression | Prospective Observational Cohort | January 2020 | Recruiting |
| NCT02934984 | Circulating Cell-free Tumor DNA (ctDNA) in Pancreatic Cancer | ctDNA as a tool for surveillance/screening for recurrence | Prospective Observational Cohort | January 2021 | Recruiting |
| NCT04246203 | Prognostic Role of Circulating Tumor DNA in Resectable Pancreatic Cancer (PROJECTION) | ctDNA as prognostic indicator in patients with radiographically resectable cancer | Prospective Observational Cohort | March 2025 | Not yet recruiting |
| NCT04241367 | Verification of Predictive Biomarkers for Pancreatic Cancer Treatment Using Multicenter Liquid Biopsy | Assessment of ctDNA KRAS mutation on outcomes in pancreas cancer. Will also assess ctDNA for specific gene targets | Prospective Observational Cohort | December 2025 | Recruiting |
| NCT04176952 | PRIMUS002: Looking at 2 Neo-adjuvant Treatment Regimens for Resectable and Borderline Resectable Pancreatic Cancer | ctDNA liquid biopsy as a means to stratify response to a chemotherapy regimen (secondary outcome) | Phase 2 Clinical Trial | December 2023 | Recruiting |
| NCT04484636 | PLATON - Platform for Analyzing Targetable Tumor Mutations (Pilot-study) (PLATON) | Assessing peripheral blood samples for targetable mutations in multiple gastrointestinal malignancies | Prospective Observational Cohort | June 2021 | Not yet recruiting |
| NCT03334708 | A Study of Blood Based Biomarkers for Pancreas Adenocarcinoma | Determination of sensitivity and specificity of ctDNA for the diagnosis of early stage pancreatic cancer | Prospective Observational Cohort | October 2021 | Recruiting |
| NCT03568630 | Blood Markers of Early Pancreas Cancer | Identification of blood markers of early pancreas cancer | Prospective Observational Cohort | July 2023 | Recruiting |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grunvald, M.W.; Jacobson, R.A.; Kuzel, T.M.; Pappas, S.G.; Masood, A. Current Status of Circulating Tumor DNA Liquid Biopsy in Pancreatic Cancer. Int. J. Mol. Sci. 2020, 21, 7651. https://doi.org/10.3390/ijms21207651
Grunvald MW, Jacobson RA, Kuzel TM, Pappas SG, Masood A. Current Status of Circulating Tumor DNA Liquid Biopsy in Pancreatic Cancer. International Journal of Molecular Sciences. 2020; 21(20):7651. https://doi.org/10.3390/ijms21207651
Chicago/Turabian StyleGrunvald, Miles W., Richard A. Jacobson, Timothy M. Kuzel, Sam G. Pappas, and Ashiq Masood. 2020. "Current Status of Circulating Tumor DNA Liquid Biopsy in Pancreatic Cancer" International Journal of Molecular Sciences 21, no. 20: 7651. https://doi.org/10.3390/ijms21207651
APA StyleGrunvald, M. W., Jacobson, R. A., Kuzel, T. M., Pappas, S. G., & Masood, A. (2020). Current Status of Circulating Tumor DNA Liquid Biopsy in Pancreatic Cancer. International Journal of Molecular Sciences, 21(20), 7651. https://doi.org/10.3390/ijms21207651





