A Novel HPLC-Based Method to Investigate on RNA after Fixation
Abstract
1. Introduction
2. Results
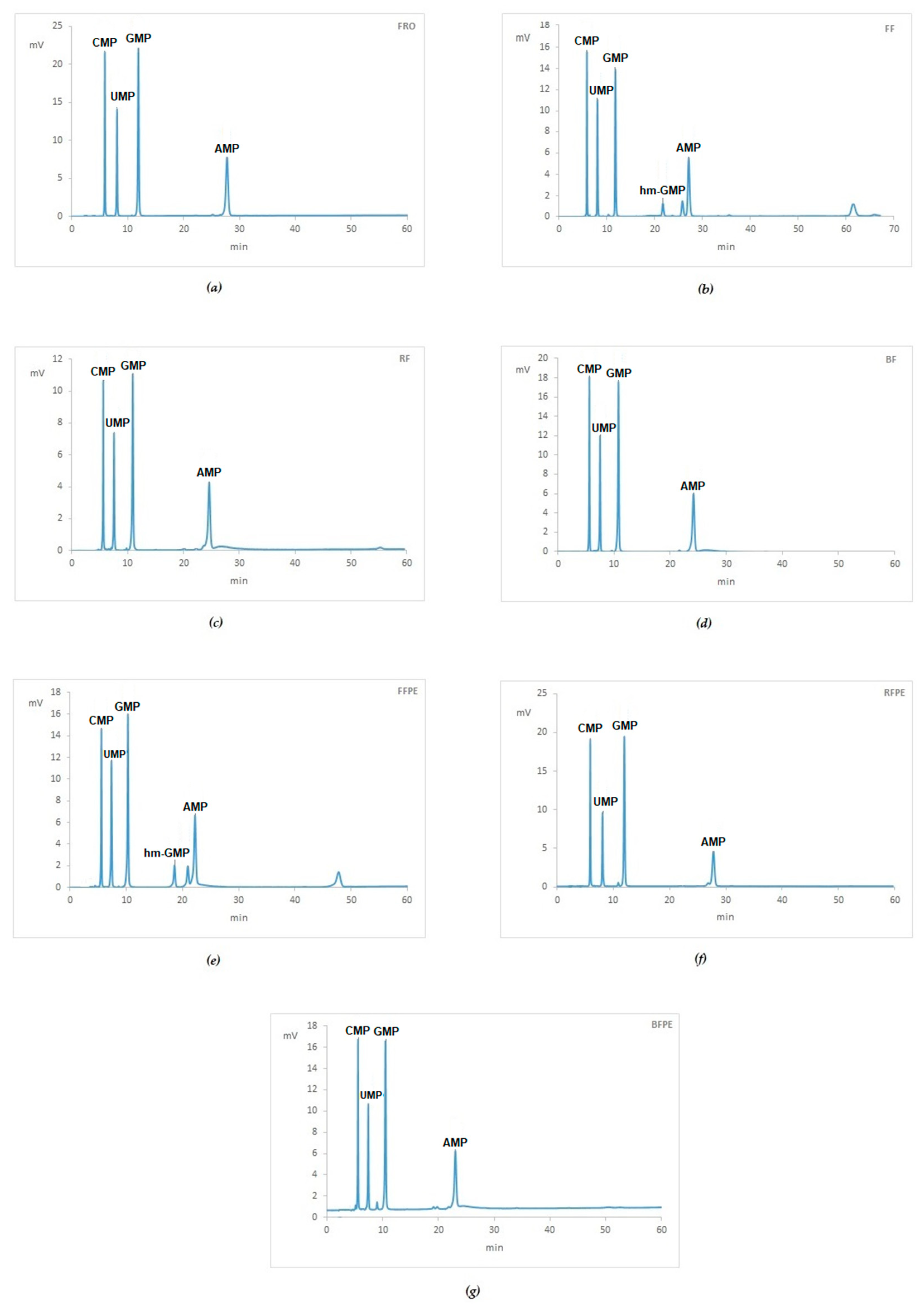
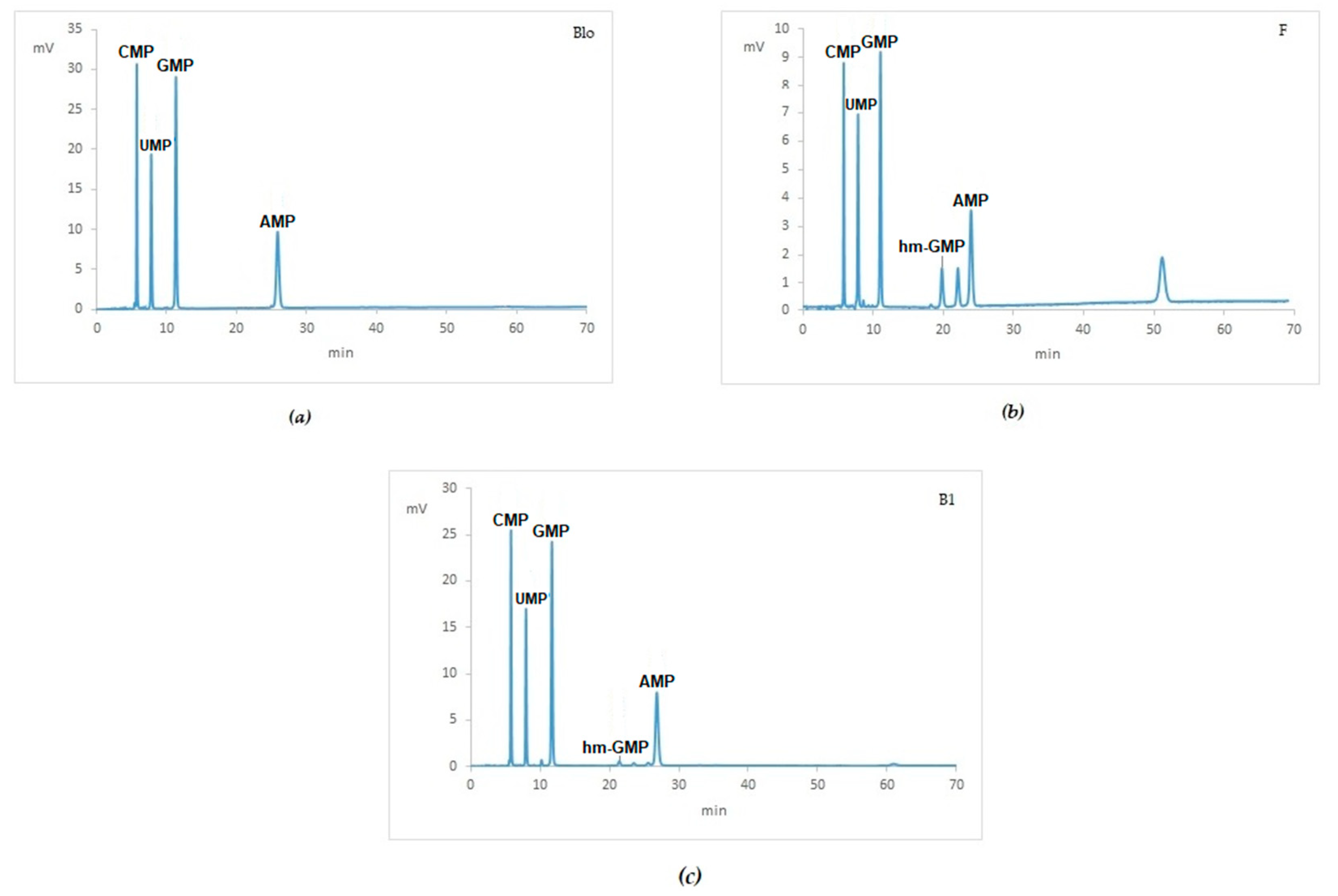
2.1. HPLC Analysis-Quantification of NMPs and Resolution of NMPs and hm-NMPs
2.2. RNA Digestion Set-Up
2.3. Fixatives Conditions in Mouse Livers
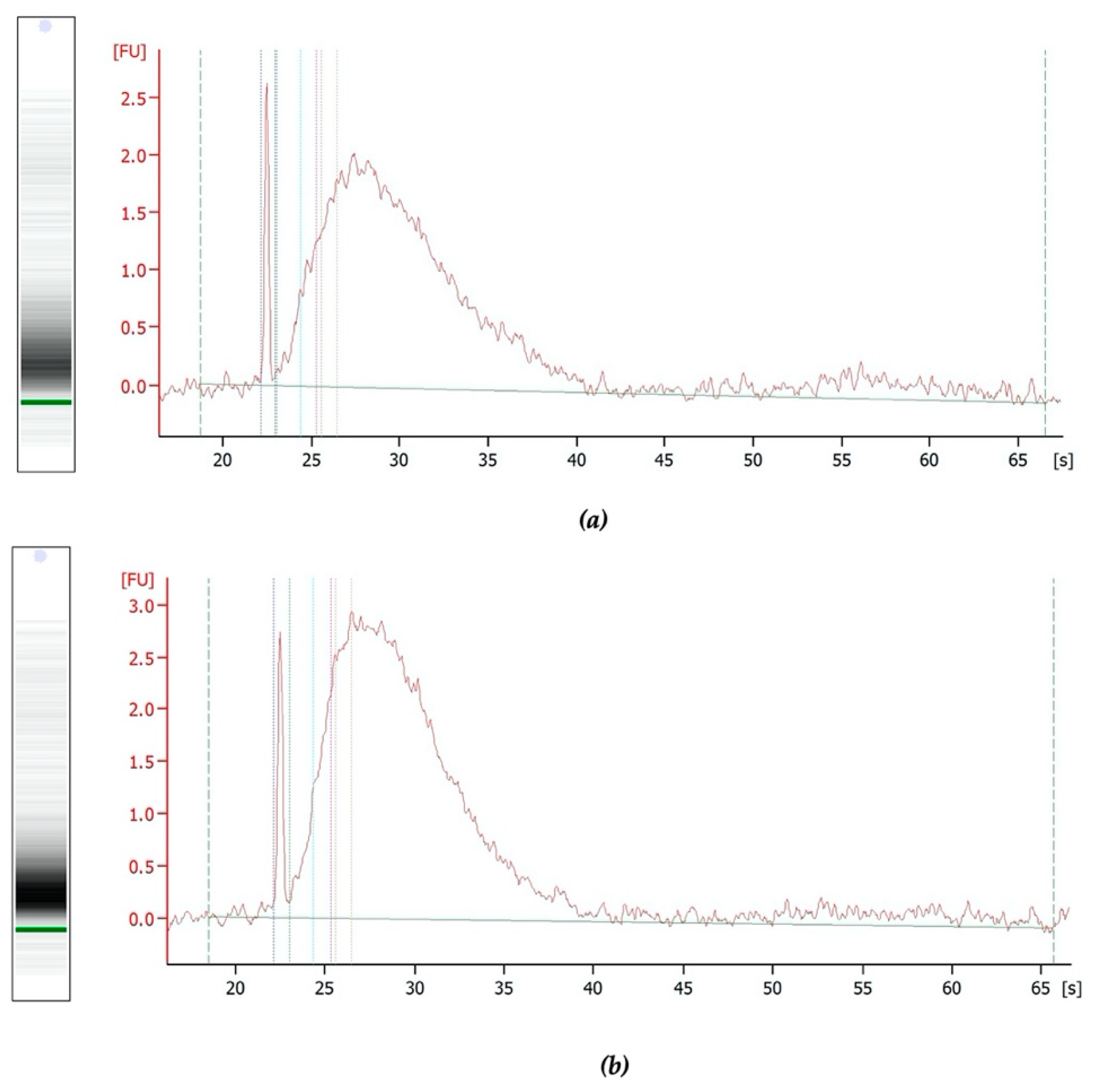
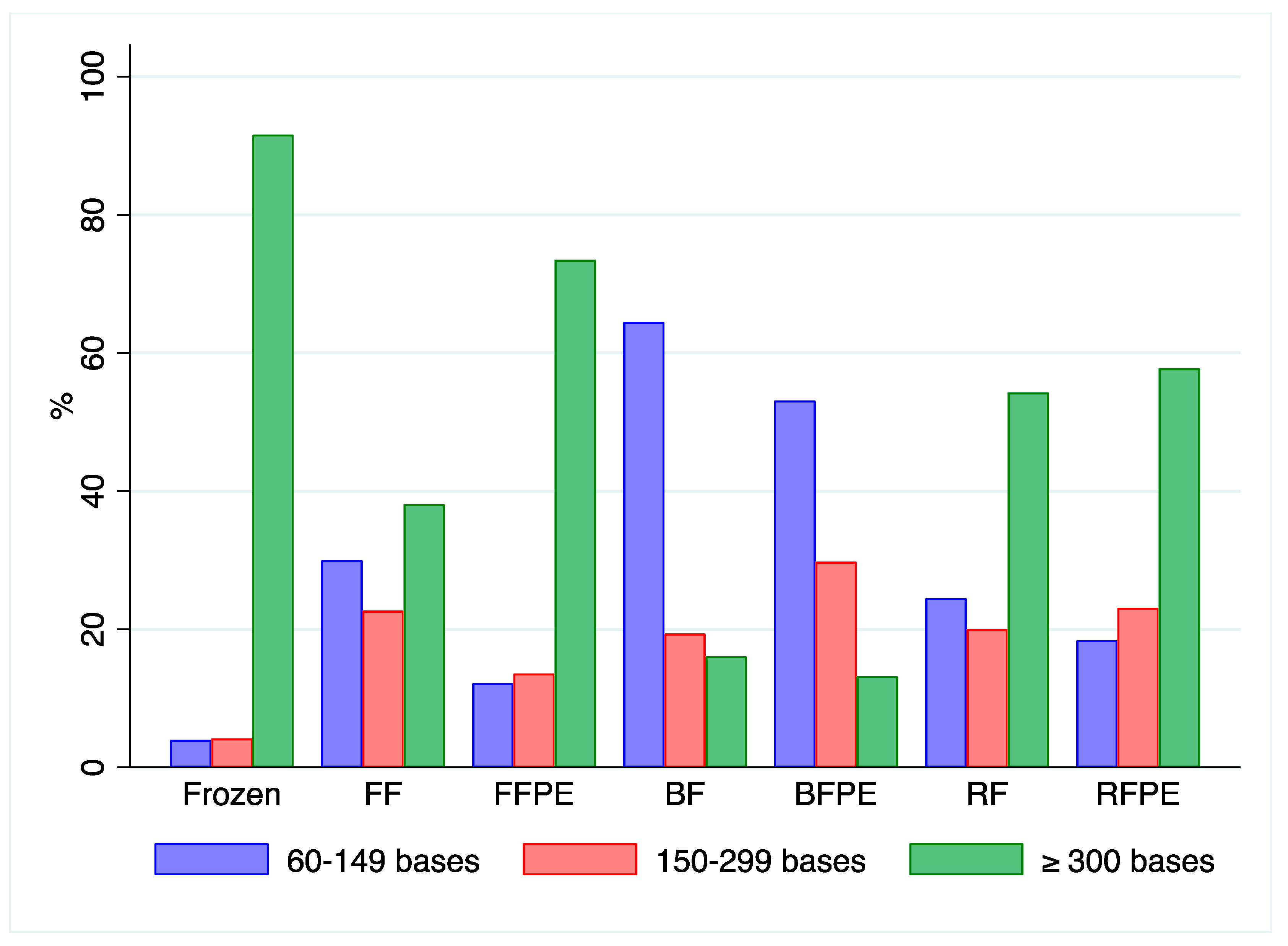
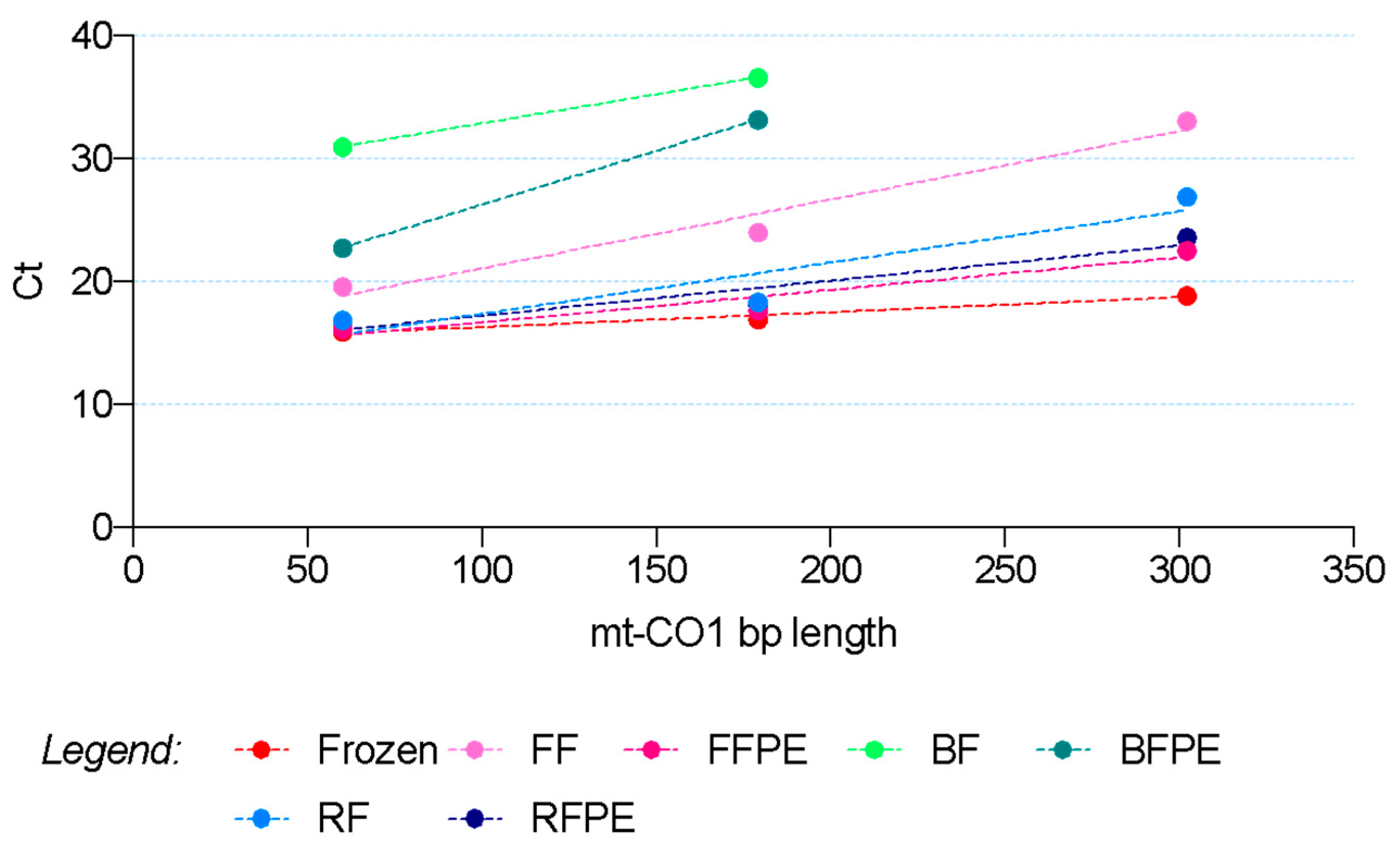
2.4. RNA Quantification and Integrity from Mouse Livers
2.5. Clinical Samples
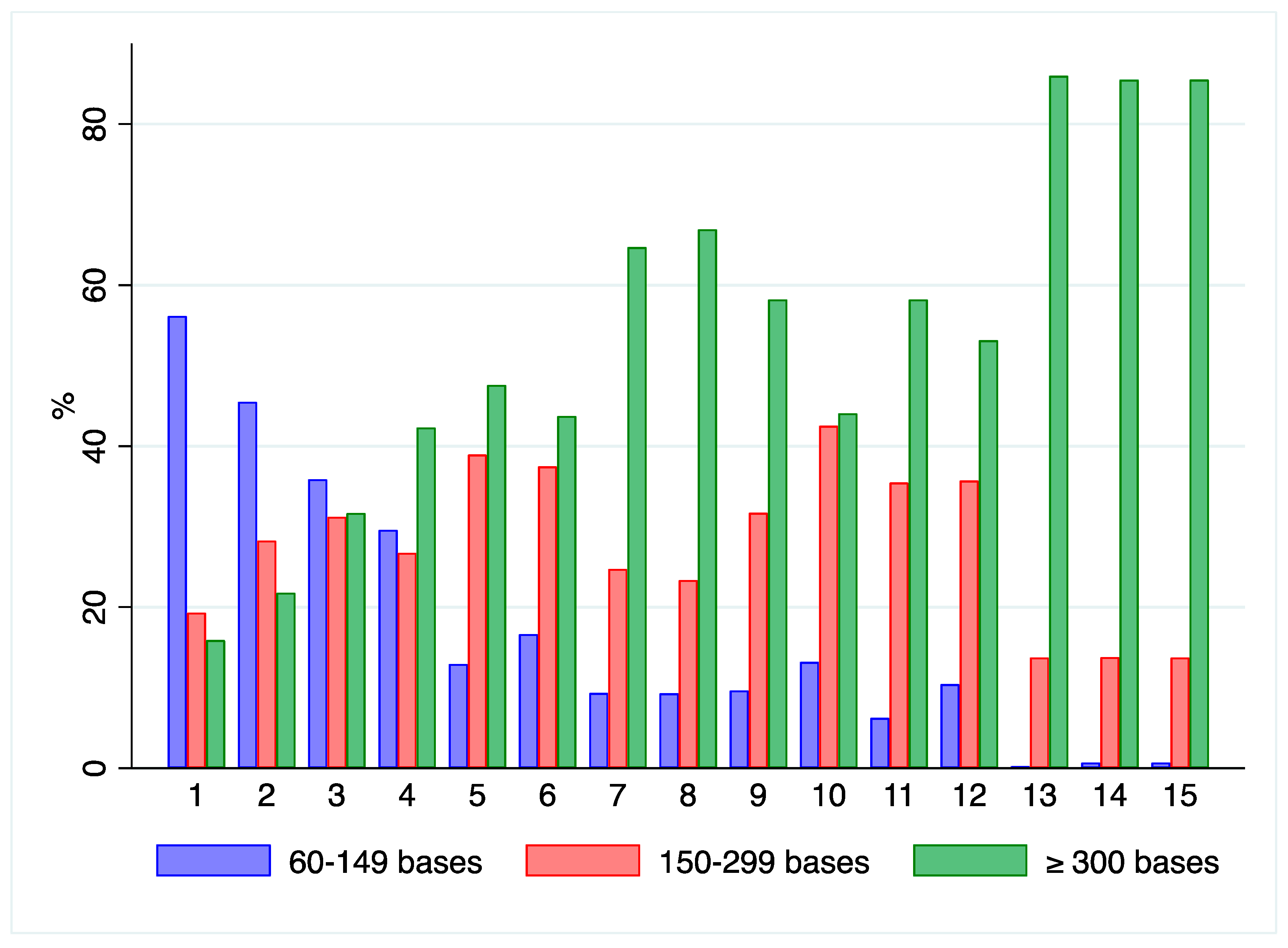
2.6. RNA Quantification and Integrity of Clinical Samples
3. Discussion
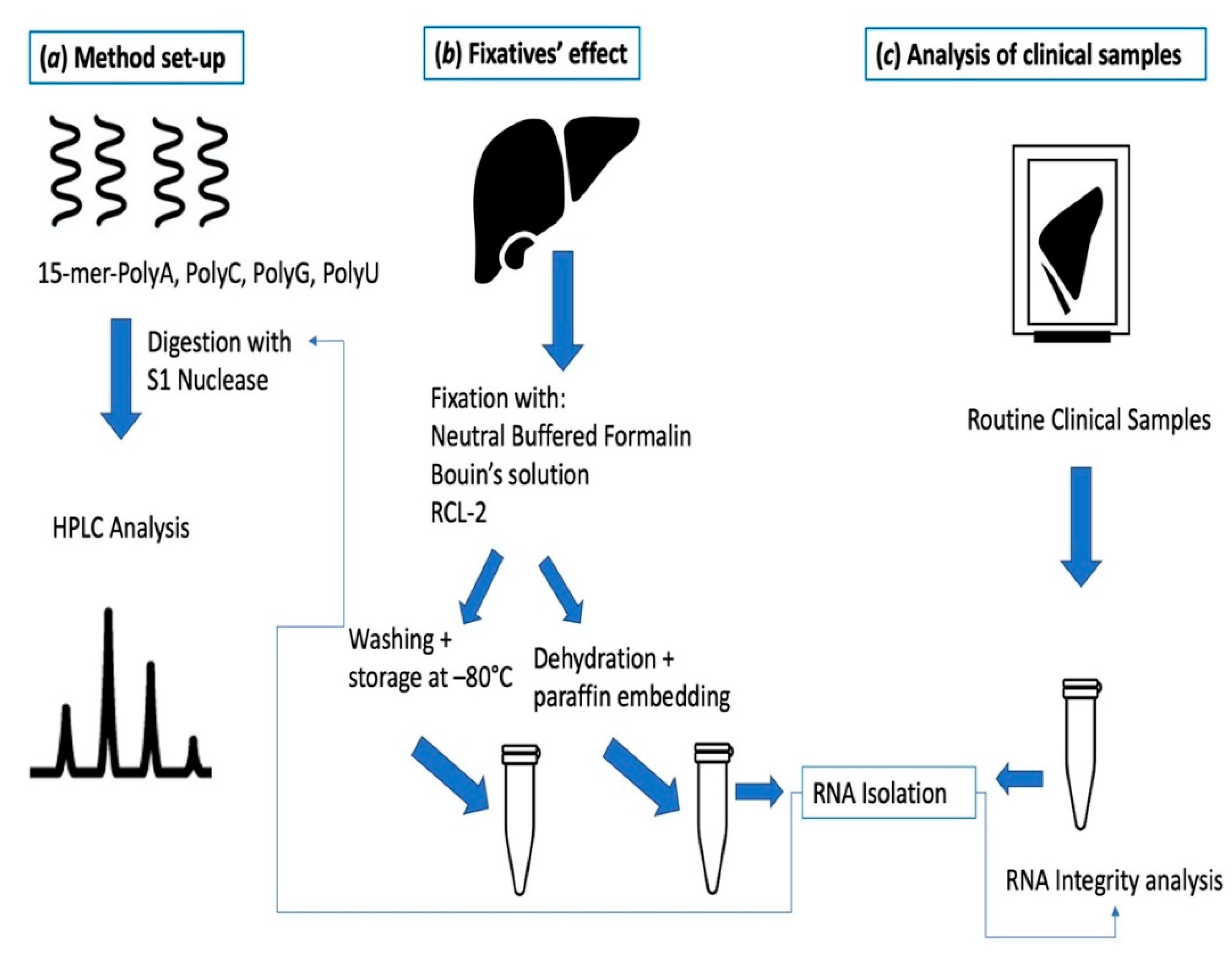
4. Materials and Methods
4.1. RNA Digestion
4.2. Fixation Procedure
4.3. HPLC Analysis
4.4. RNA Isolation
4.4.1. RNA Isolation from Fixed Specimens
4.4.2. RNA Isolation from Fixed and Paraffin-Embedded Specimens
4.4.3. RNA Isolation from Peripheral Blood
4.5. DNase Digestion
4.6. RNA Integrity
4.7. Reverse Transcription and Real-Time PCR Assay
4.8. ∆Amp Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BF | Bouin’s solution fixed |
| BFPE | Bouin’s solution fixed and paraffin-embedded |
| c-NMP | Cyclic-NMP |
| Ct | Threshold cycle |
| FF | Formalin-fixed |
| FFPE | Formalin-fixed and paraffin-embedded |
| Hm-NMP | Hydroxymethyl- nucleotide monophosphate |
| HPLC | High performance liquid chromatography |
| NMP | Nucleotide monophosphate |
| Mt-CO1 | Mitochondrially encoded cytochrome C oxidase I |
| RIN | RNA integrity number |
| RF | RCL2® fixed |
| RFPE | RCL2® fixed and paraffin-embedded |
| Rt-PCR | Reverse transcription-polymerase chain reaction |
Appendix A
Appendix A.1. Synthesis of Hydroxymethyl-AMP, -CMP, -UMP and –GMP
Appendix A.2. 1H-NMR Characterization of Hydroxymethyl-AMP, -CMP, -UMP and –GMP
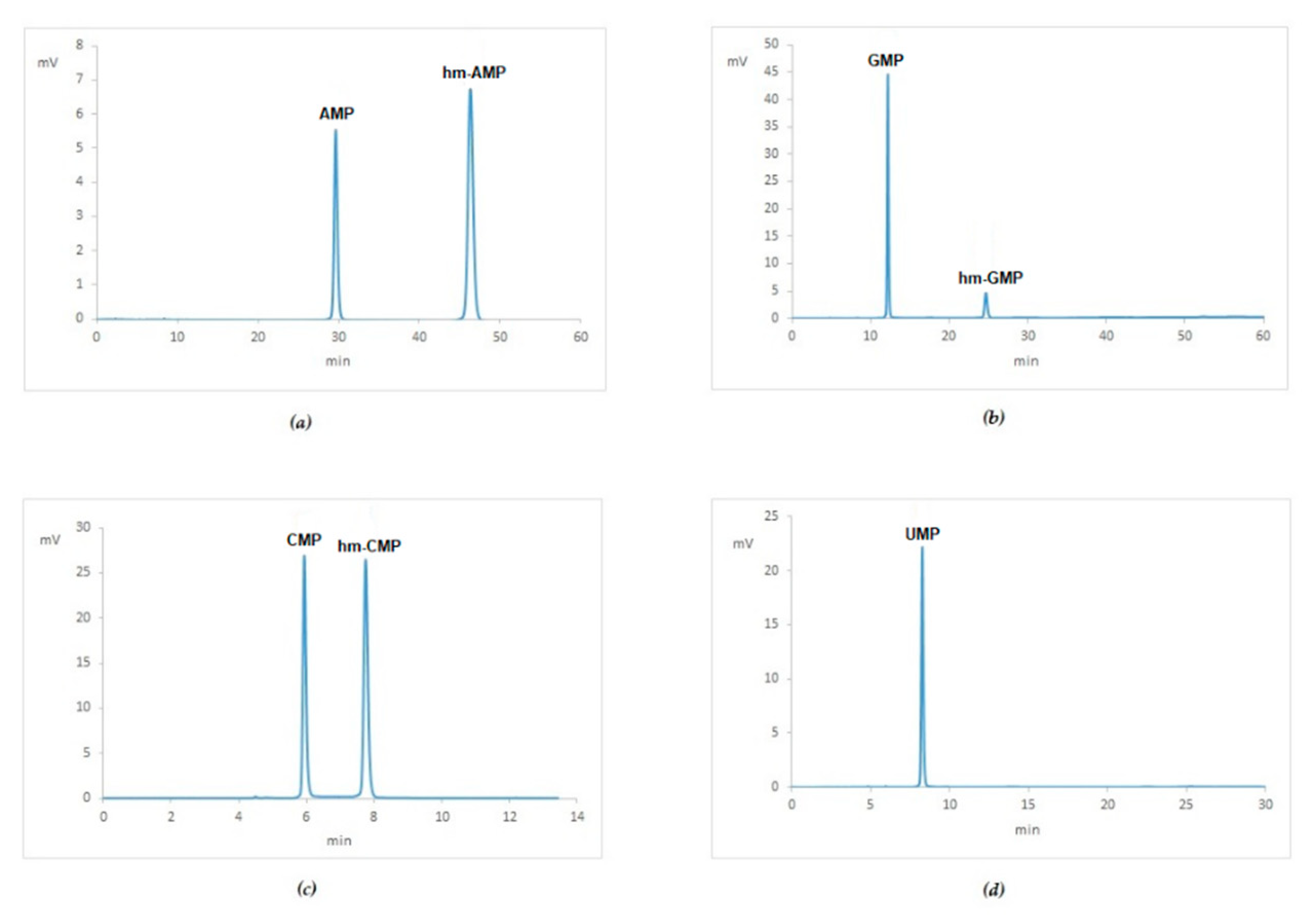
Appendix A.3. HPLC Characterization of Hydroxymethyl-AMP, -CMP, -UMP and –GMP
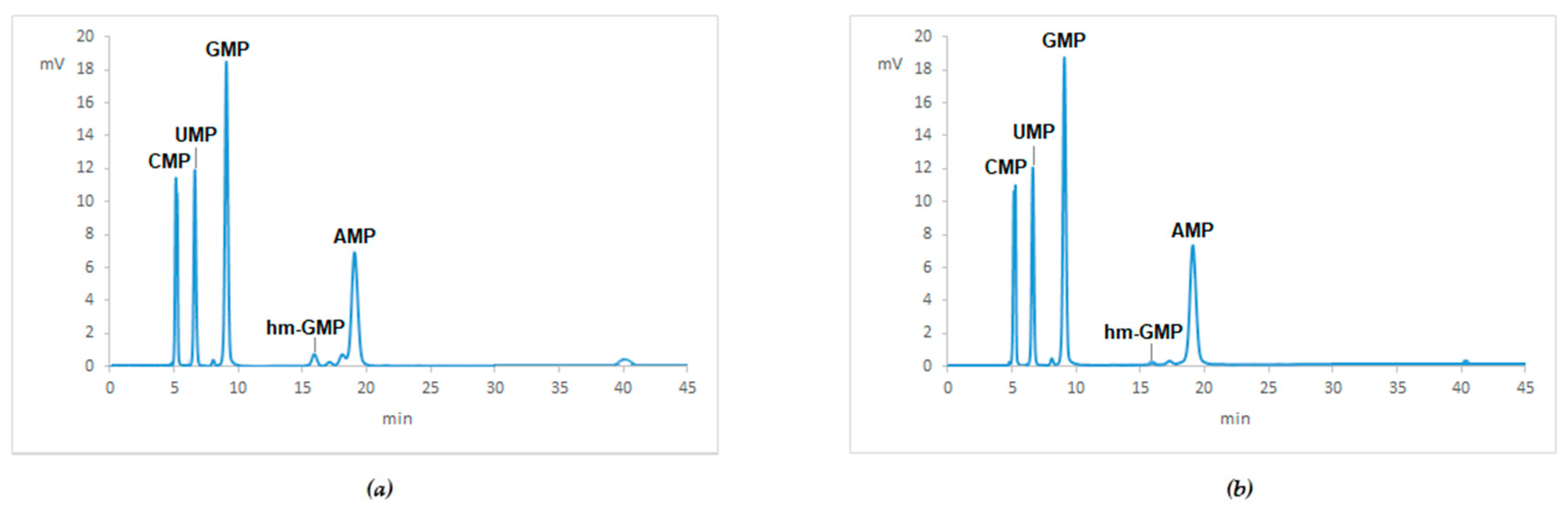
Appendix B
| Specimen | ∆Amp M-S (60–179) | ∆Amp L-S (302–60) |
|---|---|---|
| Frozen | 1.00 | 2.93 |
| FF | 4.43 | 13.48 |
| BF | 5.63 | N.A. 1 |
| RF | 1.48 | 10.05 |
| FFPE | 1.50 | 6.37 |
| BFPE | 10.41 | N.A. |
| RFPE | 1.44 | 6.87 |
Appendix C
References
- Bussolati, G.; Annaratone, L.; Medico, E.; D’Armento, G.; Sapino, A. Formalin fixation at low temperature better preserves nucleic acid integrity. PLoS ONE 2011, 6, e21043. [Google Scholar] [CrossRef] [PubMed]
- Azzalini, E.; De Martino, E.; Fattorini, P.; Canzonieri, V.; Stanta, G.; Bonin, S. Reliability of miRNA Analysis from Fixed and Paraffin-Embedded Tissues. Int. J. Mol. Sci. 2019, 20, 4819. [Google Scholar] [CrossRef] [PubMed]
- Delfour, C.; Roger, P.; Bret, C.; Berthe, M.L.; Rochaix, P.; Kalfa, N.; Raynaud, P.; Bibeau, F.; Maudelonde, T.; Boulle, N. RCL2, a new fixative, preserves morphology and nucleic acid integrity in paraffin-embedded breast carcinoma and microdissected breast tumor cells. J. Mol. Diagn. 2006, 8, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Dotti, I.; Bonin, S.; Basili, G.; Nardon, E.; Balani, A.; Siracusano, S.; Zanconati, F.; Palmisano, S.; De Manzini, N.; Stanta, G. Effects of formalin, methacarn, and fineFIX fixatives on RNA preservation. Diagn. Mol. Pathol. 2010, 19, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Viertler, C.; Groelz, D.; Gundisch, S.; Kashofer, K.; Reischauer, B.; Riegman, P.H.; Winther, R.; Wyrich, R.; Becker, K.F.; Oelmuller, U.; et al. A new technology for stabilization of biomolecules in tissues for combined histological and molecular analyses. J. Mol. Diagn. 2012, 14, 458–466. [Google Scholar] [CrossRef]
- Bonin, S.; Petrera, F.; Rosai, J.; Stanta, G. DNA and RNA obtained from Bouin’s fixed tissues. J. Clin. Pathol. 2005, 58, 313–316. [Google Scholar] [CrossRef]
- Masuda, N.; Ohnishi, T.; Kawamoto, S.; Monden, M.; Okubo, K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999, 27, 4436–4443. [Google Scholar] [CrossRef]
- AbouHaidar, M.G.; Ivanov, I.G. Non-enzymatic RNA hydrolysis promoted by the combined catalytic activity of buffers and magnesium ions. Z. Naturforsch. C 1999, 54, 542–548. [Google Scholar] [CrossRef]
- Crain, P.F. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990, 193, 782–790. [Google Scholar] [CrossRef]
- Dizdaroglu, M. Chemical determination of oxidative DNA damage by gas chromatography-mass spectrometry. Methods Enzymol. 1994, 234, 3–16. [Google Scholar] [CrossRef]
- Lipkin, D.; Talbert, P.T.; Cohn, M. The Mechanism of the Alkaline Hydrolysis of Ribonucleic Acids. J. Am. Chem. Soc. 1954, 76, 2871–2872. [Google Scholar] [CrossRef]
- Perret, D. HPLC of Small Molecules a Practical Approach; Lim, C.K., Ed.; Oxford IRL Press: Oxford, UK, 1986; pp. 221–225. [Google Scholar]
- Adams, R.L.P.; Knowler, J.; Leader, D. (Eds.) The Biochemistry of Nucleic Acids, 11th ed.; Chapman & Hall Ltd.: London, UK, 1992; ISBN 978-94-011-2290-0. [Google Scholar]
- ISO 20166-1:2018. Molecular In Vitro Diagnostic Examinations—Specifications for Pre-Examination Processes for Formalin-Fixed and Paraffin-Embedded (FFPE) Tissue—Part 1: Isolated RNA. 2018. Available online: https://www.iso.org/standard/67179.html (accessed on 12 October 2020).
- Bjorkman, J.; Svec, D.; Lott, E.; Kubista, M.; Sjoback, R. Differential amplicons (DeltaAmp)-a new molecular method to assess RNA integrity. Biomol. Detect. Quantif. 2016, 6, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.Y.; Cho, H.; Hewitt, S.M. The paraffin-embedded RNA metric (PERM) for RNA isolated from formalin-fixed, paraffin-embedded tissue. Biotechniques 2016, 60, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Wehmas, L.C.; Wood, C.E.; Chorley, B.N.; Yauk, C.L.; Nelson, G.M.; Hester, S.D. Enhanced Quality Metrics for Assessing RNA Derived From Archival Formalin-Fixed Paraffin-Embedded Tissue Samples. Toxicol. Sci. 2019, 170, 357–373. [Google Scholar] [CrossRef]
- Bjoerkman, J.; Kubista, M. Methods for Assessing RNA Quality. WO2013139860A1, 26 September 2013. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2013139860&tab=PCTDESCRIPTION (accessed on 12 October 2020).
- Srinivasan, M.; Sedmak, D.; Jewell, S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am. J. Pathol. 2002, 161, 1961–1971. [Google Scholar] [CrossRef]
- Evers, D.L.; Fowler, C.B.; Cunningham, B.R.; Mason, J.T.; O’Leary, T.J. The effect of formaldehyde fixation on RNA: Optimization of formaldehyde adduct removal. J. Mol. Diagn. 2011, 13, 282–288. [Google Scholar] [CrossRef]
- Wehmas, L.C.; Wood, C.E.; Gagne, R.; Williams, A.; Yauk, C.; Gosink, M.M.; Dalmas, D.; Hao, R.; O’Lone, R.; Hester, S. Demodifying RNA for Transcriptomic Analyses of Archival Formalin-Fixed Paraffin-Embedded Samples. Toxicol. Sci. 2018, 162, 535–547. [Google Scholar] [CrossRef]
- Beland, F.A.; Fullerton, N.F.; Heflich, R.H. Rapid isolation, hydrolysis and chromatography of formaldehyde-modified DNA. J. Chromatogr. 1984, 308, 121–131. [Google Scholar] [CrossRef]
- Fennell, T.R. Development of methods for measuring biological markers of formaldehyde exposure. Res. Rep. Health Eff. Inst. 1994, 67, 1–20. [Google Scholar]
- Karmakar, S.; Harcourt, E.M.; Hewings, D.S.; Scherer, F.; Lovejoy, A.F.; Kurtz, D.M.; Ehrenschwender, T.; Barandun, L.J.; Roost, C.; Alizadeh, A.A.; et al. Organocatalytic removal of formaldehyde adducts from RNA and DNA bases. Nat. Chem. 2015, 7, 752–758. [Google Scholar] [CrossRef]
- McGhee, J.D.; von Hippel, P.H. Formaldehyde as a probe of DNA structure. I. Reaction with exocyclic amino groups of DNA bases. Biochemistry 1975, 14, 1281–1296. [Google Scholar] [CrossRef] [PubMed]
- Rait, V.K.; Zhang, Q.; Fabris, D.; Mason, J.T.; O’Leary, T.J. Conversions of formaldehyde-modified 2′-deoxyadenosine 5′-monophosphate in conditions modeling formalin-fixed tissue dehydration. J. Histochem. Cytochem. 2006, 54, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Howat, W.J.; Wilson, B.A. Tissue fixation and the effect of molecular fixatives on downstream staining procedures. Methods 2014, 70, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Guillou, L.; Coindre, J.; Gallagher, G.; Terrier, P.; Gebhard, S.; de Saint Aubain Somerhausen, N.; Michels, J.; Jundt, G.; Vince, D.R.; Collin, F.; et al. Detection of the synovial sarcoma translocation t(X;18) (SYT;SSX) in paraffin-embedded tissues using reverse transcriptase-polymerase chain reaction: A reliable and powerful diagnostic tool for pathologists. A molecular analysis of 221 mesenchymal tumors fixed in different fixatives. Hum. Pathol. 2001, 32, 105–112. [Google Scholar] [PubMed]
- Bonin, S.; Stanta, G. Pre-analytics and tumor heterogeneity. N. Biotechnol. 2020, 55, 30–35. [Google Scholar] [CrossRef]
- Hester, S.D.; Bhat, V.; Chorley, B.N.; Carswell, G.; Jones, W.; Wehmas, L.C.; Wood, C.E. Editor’s Highlight: Dose-Response Analysis of RNA-Seq Profiles in Archival Formalin-Fixed Paraffin-Embedded Samples. Toxicol. Sci. 2016, 154, 202–213. [Google Scholar] [CrossRef]
- Kashofer, K.; Viertler, C.; Pichler, M.; Zatloukal, K. Quality control of RNA preservation and extraction from paraffin-embedded tissue: Implications for RT-PCR and microarray analysis. PLoS ONE 2013, 8, e70714. [Google Scholar] [CrossRef]
- Sanchez, I.; Betsou, F.; Culot, B.; Frasquilho, S.; McKay, S.C.; Pericleous, S.; Smith, C.; Thomas, G.; Mathieson, W. RNA and microRNA Stability in PAXgene-Fixed Paraffin-Embedded Tissue Blocks After Seven Years’ Storage. Am. J. Clin. Pathol. 2018, 149, 536–547. [Google Scholar] [CrossRef]
- Groelz, D.; Viertler, C.; Pabst, D.; Dettmann, N.; Zatloukal, K. Impact of storage conditions on the quality of nucleic acids in paraffin embedded tissues. PLoS ONE 2018, 13, e0203608. [Google Scholar] [CrossRef]
- Dotti, I.; Bonin, S. DNase Treatment of RNA. In Guidelines for Molecular Analysis in Archive Tissues; Stanta, G., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 87–90. ISBN 978-3-642-17890-0-18. [Google Scholar]
- Nardon, E.; Donada, M.; Bonin, S.; Dotti, I.; Stanta, G. Higher random oligo concentration improves reverse transcription yield of cDNA from bioptic tissues and quantitative RT-PCR reliability. Exp. Mol. Pathol. 2009, 87, 146–151. [Google Scholar]


| Variables | CMP | UMP | GMP | AMP |
|---|---|---|---|---|
| 130 ng/µL | 98 ± 1 1 | 99 ± 1 | 98 ± 1 | 97 ± 1 |
| 65 ng/µL | 98 ± 1 | 98 ± 1 | 98 ± 1 | 99 ± 1 |
| 39 ng/µL | 98 ± 1 | 99 ± 1 | 95 ± 1 | 98 ± 1 |
| 13 ng/µL | 97 ± 1 | 97 ± 1 | 93 ± 1 | 93 ± 1 |
| 1.3 ng/µL | 91 ± 4 | 98 ± 2 | 90 ± 3 | 92 ± 3 |
| LRE | 5.8 × 10−5 × −3.9 × 10−4 | 4.6 × 10−5 × +8.4 × 10−5 | 4.6 × 10−5 × −1.0 × 10−3 | 2.5 × 10−5 × −2.8 × 10−4 |
| R2 | >0.999 | >0.999 | >0.999 | >0.999 |
| LOQ (ng/µL) 10 (σ/s) 1 | 7.1 | 4.4 | 14.9 | 11.2 |
| LOD (ng/µL) 3.3 (σ/s) | 2.3 | 1.4 | 4.9 | 3.8 |
| Analytical Session | CMP | Hm-CMP | UMP | AMP | Hm-AMP | GMP | Hm-GMP | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | RT | n | RT | n | RT | n | RT | n | RT | n | RT | n | RT | |
| 1°—02/2019 | 45 | 6.0 (2.3) | 3 | 7.8 (0.4) | 42 | 8.2 (2.9) | 45 | 28.2 (7.3) | 3 | 46.3 (0.4) | 45 | 12.0 (4.1) | 24 | 22.1 (6.7) |
| 2°—04/2019 | 18 | 5.8 (0.8) | 0 | - | 18 | 7.9 (1.5) | 18 | 26.1 (4.1) | 0 | - | 17 | 11.5 (2.1) | 12 | 20.8 (3.4) |
| 3°—07/2019 | 24 | 5.8 (2.4) | 0 | - | 24 | 7.8 (3.8) | 24 | 25.9 (7.1) | 0 | - | 24 | 11.3 (5.6) | 6 | 21 (3.3) |
| 4°—02/2020 | 27 | 5.4 (4.0) | 6 | 6.5 (1.4) | 27 | 7.1 (2.6) | 24 | 21.9 (5.8) | 3 | 40.8 (0.3) | 27 | 10.1 (3.6) | 18 | 18.5 (7.0) |
| 5°—06/2020 | 17 | 5.3 (1.3) | 0 | - | 17 | 6.8 (2.5) | 19 | 20.35 (7.1) | 6 | 34.1 (0.3) | 19 | 9.52 (4.8) | 12 | 17.5 (5.7) |
| 6°—08/2020 | 4 | 5.2 (1.4) | 2 | 6.2 (0.9) | 4 | 6.6 (0.8) | 4 | 19.0 (0.6) | 2 | 28.3 (0.8) | 4 | 9.04 (0.9) | 4 | 16.05 (1.2) |
| Total | 135 | 5.7 (5.4) | 11 | 6.7 (9.4) | 132 | 7.6 (7.7) | 134 | 25.0 (14.1) | 14 | 36.7 (17.6) | 134 | 11.0 (9.8) | 79 | 20.2 (12.2) |
| Variables | CMP | UMP | GMP | AMP |
|---|---|---|---|---|
| PolyC | 93 ± 1 | |||
| PolyU | 91 ± 1 | |||
| PolyG | 89 ± 1 | |||
| PolyA | 92 ± 1 | |||
| NMP/cNMP | 8.6 ± 1 | 9.0 ± 1 | 10.7 ± 1 | 7.3 ± 1 |
| Sample 1 | CMP | UMP | GMP | AMP | TOT |
|---|---|---|---|---|---|
| Frozen | 21 ± 1% | 18 ± 1% | 35 ± 1% | 26 ± 1% | 100 ± 1% |
| FF | 21 ± 3% | 17 ± 2% | 26 ± 1% | 21 ± 3% | 85 ± 3% |
| RF | 21 ± 1% | 17 ± 1% | 38 ± 1% | 24 ± 1% | 99 ± 1% |
| BF | 21 ± 1% | 18 ± 1% | 37 ± 1% | 24 ± 1% | 100 ± 1% |
| FFPE | 15 ± 1% | 16 ± 1% | 28 ± 1% | 22 ± 1% | 80 ± 1% |
| RFPE | 21 ± 3% | 15 ± 1% | 32 ± 3% | 20 ± 1% | 88 ± 5% |
| BFPE | 21 ± 1% | 16 ± 1% | 38 ± 1% | 21 ± 2% | 96 ± 1% |
| Samples 1 | A260/A280 | A260/A230 | RIN |
|---|---|---|---|
| Frozen | 2.10 | 2.07 | 4.0 |
| FF liver | 1.95 | 2.14 | 2.1 |
| BF liver | 1.70 | 2.09 | N.A. 2 |
| RF liver | 2.09 | 2.12 | 2.2 |
| FFPE liver | 2.02 | 2.12 | 2.2 |
| BFPE liver | 1.93 | 2.14 | 2.4 |
| RFPE liver | 2.05 | 2.11 | 1.7 |
| Frozen | FF | BF | RF | FFPE | BFPE | RFPE | p1 | |
|---|---|---|---|---|---|---|---|---|
| Slope | 0.01 | 0.06 | 0.05 | 0.04 | 0.03 | 0.09 | 0.03 | 0.14 |
| Y-intercept | 15.08 | 15.50 | 28.14 | 13.20 | 14.04 | 17.50 | 14.40 | <0.0001 |
| R2 | 0.97 | 0.96 | 0.86 | 0.92 | 0.9 |
| Sample | Sample | Age | RIN | CMP | UMP | GMP | AMP | TOT |
|---|---|---|---|---|---|---|---|---|
| 1—HGSOC B1 | BFPE | 11 | 1.7 | 19% | 18% | 35% | 24% | 96% |
| 2—HGSOC B2 | BFPE | 10 | 2.4 | 19% | 17% | 34% | 24% | 94% |
| 3—HGSOC | FFPE | 9 | 2.3 | 12% | 15% | 23% | 18% | 63% |
| 4—Breast 1 | FFPE | 30 | 2.4 | 16% | 17% | 31% | 24% | 88% |
| 5—Breast 2 | FFPE | 28 | 2.4 | 22% | 17% | 33% | 25% | 97% |
| 6—Colon 1 | FFPE | 20 | N.A. 1 | 19% | 16% | 32% | 22% | 89% |
| 7—Colon 2 | FFPE | 18 | 2.0 | 22% | 17% | 33% | 23% | 95% |
| 8—Glioma | FFPE | 11 | 2.2 | 18% | 15% | 34% | 25% | 92% |
| 9—Melanoma | FFPE | 13 | 2.5 | 20% | 16% | 35% | 24% | 95% |
| 10—Pancreas | FFPE | 15 | 2.4 | 20% | 14% | 36% | 19% | 89% |
| 11—Prostate | FFPE | 18 | 2.3 | 19% | 17% | 35% | 26% | 97% |
| 12—Uterine cervix | FFPE | 28 | 2.2 | 6% | 11% | 10% | 4% | 31% |
| 13—Blood 1 | Na2EDTA | 0 | 9.1 | 20% | 17% | 33% | 30% | 100% |
| 14—Blood 2 | Na2EDTA | 0 | 8.8 | 20% | 17% | 33% | 30% | 100% |
| 15–Blood 3 | Na2EDTA | 0 | 8.6 | 20% | 18% | 33% | 29% | 100% |
| Standard RNA | Solution | - | 9.7 | 22% | 17% | 34% | 27% | 100% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fattorini, P.; Forzato, C.; Tierno, D.; De Martino, E.; Azzalini, E.; Canzonieri, V.; Stanta, G.; Bonin, S. A Novel HPLC-Based Method to Investigate on RNA after Fixation. Int. J. Mol. Sci. 2020, 21, 7540. https://doi.org/10.3390/ijms21207540
Fattorini P, Forzato C, Tierno D, De Martino E, Azzalini E, Canzonieri V, Stanta G, Bonin S. A Novel HPLC-Based Method to Investigate on RNA after Fixation. International Journal of Molecular Sciences. 2020; 21(20):7540. https://doi.org/10.3390/ijms21207540
Chicago/Turabian StyleFattorini, Paolo, Cristina Forzato, Domenico Tierno, Eleonora De Martino, Eros Azzalini, Vincenzo Canzonieri, Giorgio Stanta, and Serena Bonin. 2020. "A Novel HPLC-Based Method to Investigate on RNA after Fixation" International Journal of Molecular Sciences 21, no. 20: 7540. https://doi.org/10.3390/ijms21207540
APA StyleFattorini, P., Forzato, C., Tierno, D., De Martino, E., Azzalini, E., Canzonieri, V., Stanta, G., & Bonin, S. (2020). A Novel HPLC-Based Method to Investigate on RNA after Fixation. International Journal of Molecular Sciences, 21(20), 7540. https://doi.org/10.3390/ijms21207540







