Abstract
The success of seed germination and the successful establishment of seedlings across diverse environmental conditions depends on seed vigour, which is of both economic and ecologic importance. The smoke-derived exogenous compound karrikins (KARs) and the endogenous plant hormone strigolactone (SL) are two classes of butanolide-containing molecules that follow highly similar signalling pathways to control diverse biological activities in plants. Unravelling the precise mode-of-action of these two classes of molecules in model species has been a key research objective. However, the specific and dynamic expression of biomolecules upon stimulation by these signalling molecules remains largely unknown. Genomic and post-genomic profiling approaches have enabled mining and association studies across the vast genetic diversity and phenotypic plasticity. Here, we review the background of smoke-assisted germination and vigour and the current knowledge of how plants perceive KAR and SL signalling and initiate the crosstalk with the germination-associated hormone pathways. The recent advancement of ‘multi-omics’ applications are discussed in the context of KAR signalling and with relevance to their adoption for superior agronomic trait development. The remaining challenges and future opportunities for integrating multi-omics datasets associated with their application in KAR-dependent seed germination and abiotic stress tolerance are also discussed.
1. Introduction
Dormancy breakdown and seedling establishments are critical steps of the plant life cycle and are simultaneously controlled by multiple external cues, for instance light, temperature, minerals, water, and nutrients. Smoke produced from the burning or charring of plant materials can stimulate seed germination and improve seed vigour, i.e., rapid and uniform emergence in a range of field conditions; and provide ways to mitigate and overcome abiotic stresses in the field [1,2,3,4]. The clearing of ground cover by wildfires assists the light and water to enter the soil more easily and creates an opportunity for seed germination and the establishment for many species whose germination commences upon sensing smoke or generated heat.
Chemical stimulants present in the char and smoke from fires promote seed germination and seedling establishment. Chemicals such as nitrogen oxides (NOx) and cyanohydrins, when present in the smoke or aqueous smoke solutions, have shown a stimulating effect on germination [5,6]. Among the thousands of compounds present in the smoke, the butanolide moiety containing molecule 3-methyl-2H-furo-[2,3-c]pyran-2-one and its derivatives, commonly known as karrikin (KAR) [7,8], has shown the highest potential to promote seed germination and assist seedlings with abiotic stress. KARs have been shown to interact with plant phytohormones and currently unknown transcription factor (TF)-like proteins to process endogenous signals for dormancy breakdown and seedling vigour process [9,10,11]. To understand the smoke-assisted regulatory pathways in model and non-model species, large scale genomic screening studies along with targeted experiments have been conducted [12,13,14,15]. These studies were often designed to generate and analyse each dataset in isolation—ignoring the possible interactions between the underlying biomolecules—resulting in findings independent of one another. The investigation and integration of multi-omics datasets are essential to fully understand the complex nature of this enigma. In this review, we first outline the current knowledge about seed dormancy, germination, and vigour; and explain how these stages can be affected with the application of smokes produced from burning plant materials. Then, the interconnection between KARs, strigolactones (SLs) and other phytohormones will be discussed in terms of their role in seed germination and abiotic stress management. Finally, the application of proteomics and multi-omics strategies will be discussed for agronomically important crop species to demonstrate how different ‘omics’ data can be investigated to explore the crosstalk between different pathways and abiotic stress tolerance in crop species. As an example, a protein–protein interaction map will be built using proteins related to KAR and SL-associated Gene Ontology (GO) terms in Arabidopsis thaliana. Then, gene expression patterns of homologous genes associated with KAR and SL-metabolism or signalling GO terms in wheat will be analysed using the transcriptome data of different plant development stages and abiotic and biotic stress conditions, which lay the foundation for future experiments to explore their application potential in crops.
2. Seed Dormancy/Germination and Environmental Factors that Affect Seed Germination
2.1. Seed Dormancy, Germination, and Vigour
Seed dormancy is a static state where the seed can halt germination until favourable conditions are present. This state of dormancy is maintained in various ways across the plant kingdom to ensure the establishment of a new plant generation [16,17]. The dormant seed is unable to germinate within a given period under the effect of any abiotic factors such as temperature, light, water, etc. The seed dormancy mechanisms are interconnected with the plant genetic systems and the abiotic environment, both required to initiate the germination process. Dormancy is not just defined as the characteristics of the seed that is unable to germinate but also as an inherent trait of the seed that determines the conditions required for germination [18,19]. Seed dormancy can be classified into five sub-classes: physiological, morphological, morphophysiological, physical and combinatorial dormancy [20].
Seed germination is a complex process where the seed undergoes several changes including: recovery from maturation drying; starting metabolic operations; completing critical cellular events that allow the embryo to emerge; and preparing for seedling growth [21,22,23]. This process spans from water uptake (imbibition) by the seeds to the appearance of the embryo, throughout the surrounding structures. Germination begins with the imbibition of water by the dry seed (Phase I) until the matrices and cell contents are adequately hydrated. Then, the seeds uptake a limited amount of water (Phase II), followed by the increase re-uptake of water (Phase III) eventually completing the germination cycle. Overall, the mature seed must rapidly shift from a desiccated state to a germination-dependent development program for seedling growth [22].
Seed vigour is an integral seed feature that determines its potential for rapid uniform germination success and subsequent development across various environmental conditions. Seed germination and seedling emergence is controlled by a network of molecular mechanisms that are cumulatively defined as seed vigour. Experiments using ‘omics-based’ approaches have found multiple factors including the quality of messenger RNAs stored during the seed maturation stage, proteostasis, DNA integrity, cell metabolism and connection with hormonal pathways, to all play a key role in seed vigour [22]. The loss of seed vigour is related to the reduction of the ability of seeds to carry out all the physiological functions during germination. Physiological ageing causes the reduction of seed vigour, which includes changes in cell membrane integrity, enzyme activity and protein synthesis. Seed vigour is not a single factor but a concept associated with multiple factors: the rate and uniformity of seed germination and seedling growth; ability to receive the environmental signals of favourable conditions; storage conditions; and performance during the storage conditions, in particular to retain their ability to germinate [24]. Proteomics and bioinformatic studies have found that the activation of pathways such as sulphur amino acid, lipid and starch metabolism, protein synthesis (increase the abundance of translation initiation factors) and abscisic acid (ABA) signalling pathways correlate with increased seed vigour [25]. Although studies have tried to establish a seed vigour mechanism, the interaction between genetic, DNA repair mechanisms, storage conditions and environmental factors with these biochemical changes are not fully understood.
2.2. Environmental Factors that Affect Seed Germination
Seeds start to breakdown the dormancy state and activate the germination pathways upon receiving cues from surrounding environments. The surrounding environment and the physical soil characteristics help to establish their microclimate conditions. Various mechanisms are involved in the breakdown of dormancy. For instance, after ripening or chilling, dormancy breakdown depends upon whether the seeds are dry or wet. Once dormancy starts to breakdown, the seeds respond to the cues from the microenvironment and sense the overall environment. Light, temperature, soil moisture, nutrients and chemical cues, i.e., smoke, determine the rate and extent of germination of non-dormant seeds [26]. Comprehensive reviews have been published on how various biotic and abiotic factors influence the seed germination process [16,18,21,22,23,24]. The role of chemical cues, i.e., the smoke-assisted germination process will be discussed in later sections.
3. Role of Smoke, Smoke Generation, and Components
3.1. Role of Smoke in Germination
Wildfire plays a crucial role in ecosystems to facilitate the germination process for many plant species around the world. Plants have evolved with the fire-dependent reproductive strategies over time to build natural ecosystems. Smoke generated from burning plant materials enhances seed germination in over 1200 phylogenetically diverse plant species [27,28,29,30]. Studies have shown the impact of smoke originating from wildfire to restore the vegetation of fire-prone native species in the Mediterranean region [31]; Australia [32]; South Africa [33]; California [34]; and, the Mediterranean basin [35]. Additionally, its positive impact on the germination of cereal crops [2,36]; weed species [1]; horticultural crops [14]; and medicinal plants [37] has also been confirmed. The smoke-dependent germination process improves germination success across phylogenetically diverse plant groups including gymnosperms; angiosperms [33]; annuals; bi-annuals; short-lived species; and, perennial herbs [38], in different weather regions around the globe from tropical to the fire-prone Mediterranean region, disturbed habitats, representing a broad array of fire-prone environments [30,32]. Smoke has been used as an effective mediator to release dormancy in the laboratory [39] and field conditions [14]. Altogether, the germination response to smoke is not just associated with plant traits such as habitat requirements, seed bank, type, life form, and seed morphology, but also the chemical stimuli that alleviate the dormancy state.
Seeds receive natural cues from the environment that help them to respond to the physical (temperature and light) and chemical (smoke, gas, nutrients) conditions as germination cues are associated with fire [29,30]. For example, the heat produced from the fire can help to breakdown or desiccate the seed coat [40] or may reduce the expression of dormancy controlling hormone abscisic acid (ABA)-related genes during post-germination events [41]. Hard-coated seeds with a prominent waxy cuticle and dense palisade layer of sclereids enforce dormancy by making the seed coat impermeable to water. Brief heat shock between 80 and 120 °C is enough to cause the seed to imbibe water by loosening cells or by denaturing germination inhibitors. For some species, heat shock alone may work, but some heat-induced species also require light and/or cold stratification [9,42]. Heat and the chemical constituents generated from the fire events may also stimulate the embryo growth by inducing somatic embryogenesis during seed germination [39]. The application of smoke has been shown to activate the KAR receptors in Arabidopsis [43], presenting the question as to whether smoke can be applied to crop species to develop drought-tolerant plants.
3.2. Smoke Generation and Application
Smoke water produced by the combustion of vegetation has been widely used to increase germination and seedling vigour due to its convenient mode of application. The chemical constituents present in the smoke readily dissolve in water and the dilution of this smoke water has been shown to markedly improve germination rates [1,7,44] and abiotic stress tolerance [3,45,46]. In this technique, smoke was generated in a drum, and using compressed air, was bubbled through distilled water [33]. A wide range of plant materials has been used to generate aqueous smoke extracts using this technique [1,29,30,47]. Studies have shown that all plant materials produce similar chemical compositions and can be applied in diluted forms ranging from 1:10 to 1:100, v/v [48] or 1000–2000 ppm [2] to enhance germination events. However, a recent study has shown that the dilution ratio of smoke water and distilled water 1:2500 (v/v) from the sub-dilution 25:75 (v/v) of smoke water was the best for the germination of lettuce seeds in the dark, achieving a 91% germination rate against the water control that had only 7–10% germination [47]. Although these dilution ranges have been found to be effective to promote the germination events, the response varies widely across plant species.
3.3. Active Components in Smoke
Chemical compounds present in the smoke stimulate germination and enhance seedling vigour in several agricultural, horticultural and wild plant species [24,30]. Smoke contains thousands of compounds including organic acids, organic bases, alcohols, aldehydes, esters, alkyl aryl ethers, furan and pyran derivatives, ketones and diketones, lactones, phenolic derivatives, guaiacol derivatives, syringol derivatives, hydrocarbons, and nitrogenated derivatives [29,32,49,50,51]. De Lange and Boucher (1990) demonstrated that active component(s) of gas phase smoke are water-soluble and that a smoke-saturated solution is effective in improving seed germination [33]. Since then bioactive compounds having either stimulatory or inhibitory activity on seed germination have been identified in smoke [47,52]. Both organic and inorganic chemicals originating or released from smoke help stimulate the germination of seeds. Nitric oxide was discovered as an active component in the smoke that can stimulate the maximum level of germination in the California chaparral annual Emmenanthe penduliflora [53]. Dormant seeds from this species germinate fully upon brief exposure to smoke or vapor generated from smoke-treated sand or paper, and the germination capacity can be comparable with 500 ppm NO2 [53]. Chaparral wildfires generate enough NO2 from the combustion of organic matter or from postfire nitrification, which helps trigger germination events in a process independent from imbibition. Notably, all pyroendemics (seedling germination and successful seedling recruitment for plant varieties that depend on postfire environments) in chaparral showed a significant response to this gas and thus it appears that other chemicals may also act as germination stimulants in smoke and charred wood [54]. In addition, other nitrogenous compounds generated from combustion include cyanohydrin (glyceronitrile), which can be slowly converted to cyanide and stimulate seed germination in a wide range of fire-responsive species from different continents [6].
Of the thousands of different compounds in smoke, some of them act as enhancers while some inhibit the germination process [55,56]. Dose-dependent responses of smoke compounds have also been demonstrated [49]. Studies have been conducted in recent decades to identify the active constituents from smoke [28,53,57]. A compound was purified as 3-methyl-2H-furo-[2,3-c] pyran-2-one from the burning of cellulose- and plant-derived smoke material by using bioassay-guided fractionation [7,8]. Later, this compound was named as karrikinolide (or KAR1 or KAR) after the Noongar aboriginal word for smoke “Karrik” [50]. KARs are produced by the pyrolysis of cellulose and sugars—thought to be deposited on the soil surface during a fire, and eventually absorbed by the seeds buried in the soil over the time after being dissolved by rain. To date, six different isoforms of the KAR family (KAR1–KAR6) have been reported; all of them contain a five-membered butanolide ring and a six-membered pyran ring [27,50]. The main difference among the KAR family members is the number and location of methyl group(s) [50]. KAR1 is the most abundant and bioactive within smoke [50]. Notably, KARs share structural similarities with plant hormone SLs—both belong to the butanolide class and trigger germination but originate from different sources [10].
The effect of KARs is not limited to the fire-prone plant species but improves seedling vigour, root development and abiotic stress tolerance in many crop species [35,42,43]. In Arabidopsis, KARs have been found to enhance germination and seedling responses to light [58]. Variable responses to KARs in terms of germination are observed to be dependent on plant species, their origin (geographical location) and/or seed maturity stage. For example, germination is delayed upon KAR exposure in soybean by increasing ABA synthesis and impairing GA biogenesis [59]. Additionally, the germination of other species such as Capsella bursapastoris, Bromus sterilis and Alopecurus myosuroide are also delayed by KARs [9]. The ubiquitous nature of KAR-induced germination success indicates that it is an ecologically significant germination cue [58]. However, interestingly, the activity of KAR is also dependent on light intensity, the duration that seeds spend in dormancy and the surrounding ecological conditions [60].
KAR1 has the highest water solubility, low volatility and is the most abundant compound among the other subtypes in smoke residue [51]. Studies have reported that KAR1 can be produced by burning pure xylose, glucose, or cellulose, and it has been proposed that KAR1 is derived from a pyranose sugar [6]. KAR1 produced during combustion condenses and is deposited as the smoke cools and remains close to the fire-affected areas [4]. Karrikin is deposited along with the nutrients present in the ash. The removal of litter containing allelopathic chemicals and changes in light quality or accessibility affects the rate of germination in the fire-affected sites, but not in the unburned areas in the rainy months [27]. Overall, the seeds’ response to KARs can provide an inherent capacity for plants co-located with annual fire events to enhance their ability and potency in the post-fire landscape.
5. Co-Activation of KAR and Plant Innate-Hormonal Pathways
Plant hormones play key roles in the integration of diverse environmental cues with signalling networks and pathways in plants. KAR is considered a plant hormone, though it is not produced in planta, due to its diverse function in seed germination and plant vigour [4]. Gibberellic acid (GA) helps to breakdown dormancy, while ABA and indole-3-acetic acid (IAA) promote dormancy and delay seed germination [20,21]. Recent studies have shown that KARs can regulate seed germination and the seedling establishment process by alleviating shade avoidance through triggering the plant phytochrome signalling pathways such as ABA, GA and IAA [9,79]. For example, ABA antagonistically regulates the KARs’ effects during germination and thus KARs requires the activation of the GA biosynthetic pathways to complete the germination process in Arabidopsis [80]. In Arabidopsis, KARs can promote the expression of GA biosynthesis genes GA3ox1 and GA3ox2 during germination [58]. Both ABA and IAA can synergistically control seed dormancy; however, the mechanism of how these two hormones work together to establish the dormant state and the interrelationship between GA and IAA is still unknown. Of interest, KARs can suppress the expression of the auxin-responsive protein IAA1 in Arabidopsis during germination [11,81]. IAA regulates the shade avoidance response in plants by controlling hypocotyl elongations along with GA and brassinosteroid [9,82]. Given that KARs suppress IAA1 expression, it may thus be possible that the biosynthesis and transport of IAA during seedling establishment may be inhibited by KARs. However, the precise mechanism of how KARs dominate seedling establishment by the regulation of IAA biosynthesis is unclear. Overall, KARs and IAA interact with ABA during germination, but how the KARs interact with the ABA pathways by regulating the IAA signalling pathway remains unknown.
Hormone receptors in plants perform two major roles: signal perception and signal transfer to downstream molecules. The hormone binding sites of the receptors generally contain hydrophobic pockets with polar patches to trigger specific interactions with the polar groups of the hormones. These hormones are allosteric inducers that induce conformational changes in receptors upon hormone perception to switch on or facilitate the transfer of signals to downstream effector proteins. Hormone-dependent proteolysis of cullin-RING ubiquitin ligase (CRL) complexes play a key role in the signalling pathways of major plant hormones such as IAA, jasmonic acid (JA), GA, SL, KAR, ABA and ethylene [79]. The auxin and JA receptors are F-box proteins with leucine-rich repeat (LRR) domains that recognise the substrate proteins for ubiquitinylation in a hormone-dependent manner. Notably, the GA, SL, and KAR receptors are members of the α/β hydrolase superfamily and act as F-box protein-bound adaptor proteins that recognise substrate proteins for ubiquitylation in a hormone-dependent manner [83]. Receptor GID1 activates upon binding to GA exposing the hydrophobic surfaces of the catalytic His residue of the Ser–His–Asp catalytic triad with a Val/Ile residue that can interact with DELLA proteins [70]. In contrast to GID1, the SL and KAR receptors possess a conserved catalytic triad system. These catalytic residues are essential for the action of SL and KAR receptors, and the SL receptor obviously exhibits catalytic activity with respect to SL hydrolysis, which is essential for SL function [84]. Although the genetic homology-based approach has been used to elucidate the hormonal KAR signalling pathways’ activation through KAI2 and SL signalling, the KAR-induced KAR–KAI2–D3/SMAX1 interactions and MAX2-dependent proteolysis of SMAX1 remain biochemically untested [10]. Additionally, how the SMXL/D53 proteins regulate the germination and growth upon hormone signal perception is currently unknown.
MAX2-dependent KAR signalling pathways have a role in seed germination and development. The MAX2 mutants have shown extended dormancy, epinastic leaves and long hypocotyls under various photomorphogenesis conditions [11,85]. Studies have reported the connection between MAX2 and plant phytohormones, i.e., IAA biosynthesis pathways [9,75]. In Arabidopsis, the expression pattern of MAX genes from SL pathways can modify IAA transportation, and thereby regulate vascular tissue formation and regeneration [86]. This result suggests that IAA and SL can control their levels and distribution in a thorough MAX2 pathway by creating a dynamic feedback loop required for the coordinated control of auxiliary branching. Additional studies have shown that MAX2 can suppress IAA transport through the inhibition of PIN genes responsible for IAA transportation [87]. Furthermore, the ABA and GA biosynthesis pathways are also co-regulated by the MAX2-dependent signalling pathway [88], indicating a shared pathway activation mechanism between the phytohormones and KARs to regulate seed germination and photomorphogenesis. Although the crosstalk between MAX2 and phytohormones has been investigated, the relationship between KAI2 receptors and other photomorphogenesis factors is yet to be understood.
6. Application of Proteomics to Understand Smoke Water-Assisted Germination and Abiotic Tolerance in Crops
The advancement of mass spectrometry-based proteomics has enabled the routine measurements of thousands of proteins in terms of their abundance, localisation, interactions, and modifications while connecting the large-scale data to gene expression regulation and function [89,90]. Targeted and untargeted proteomics experiments have been widely used to investigate crop responses to the abiotic stress [89,90,91,92]. Proteomics studies have shown that plant-derived smoke water solutions can improve the abiotic stress tolerance capacity in soybean [3], chickpea [93] and rice [36]. Zhong et al. have shown that treatment with plant-derived smoke can enhance soybean plant growth under flooding stress [94]. A combination of proteomics and metabolomics revealed that smoke water helps to alter arginine metabolism and regulates ornithine synthesis/ubiquitin–proteasome pathways to provide early stage soybean plant development under stress conditions [94]. Interestingly, a recent study has shown that the SL and KAR signalling pathways activate the polyubiquitination and degradation of SMXL2 to regulate hypocotyl elongations in a D14 and KAI2-dependent manner in Arabidopsis [95]. Another proteomics-based study on flooding-affected soybean plants has shown that the treatment with smoke water can increase the accumulation of cell wall-related proteins, sucrose/starch metabolism and glycolysis [3]. Likewise, the application of smoke water has shown to increase the weight and length of flood-stressed soybean root by activating the ascorbate/glutathione pathways [46]. Using proteomics, smoke water-treated chickpea plants showed a perturbation of proteins such as glyceraldehyde-3-phosphate dehydrogenase and fructose-bisphosphate aldolase to support glycolysis pathways in order to enhance seed germination and plant growth rates [93]. Overall, proteomics-based studies have shown great potential to understand the smoke water-assisted germination pathways, which can lead to future research opportunities for agricultural crop improvement.
7. Applying Multi-Omics Strategies to Identify Karrikin Receptor Proteins and Establish Their Pathways in Agronomically Important Crops
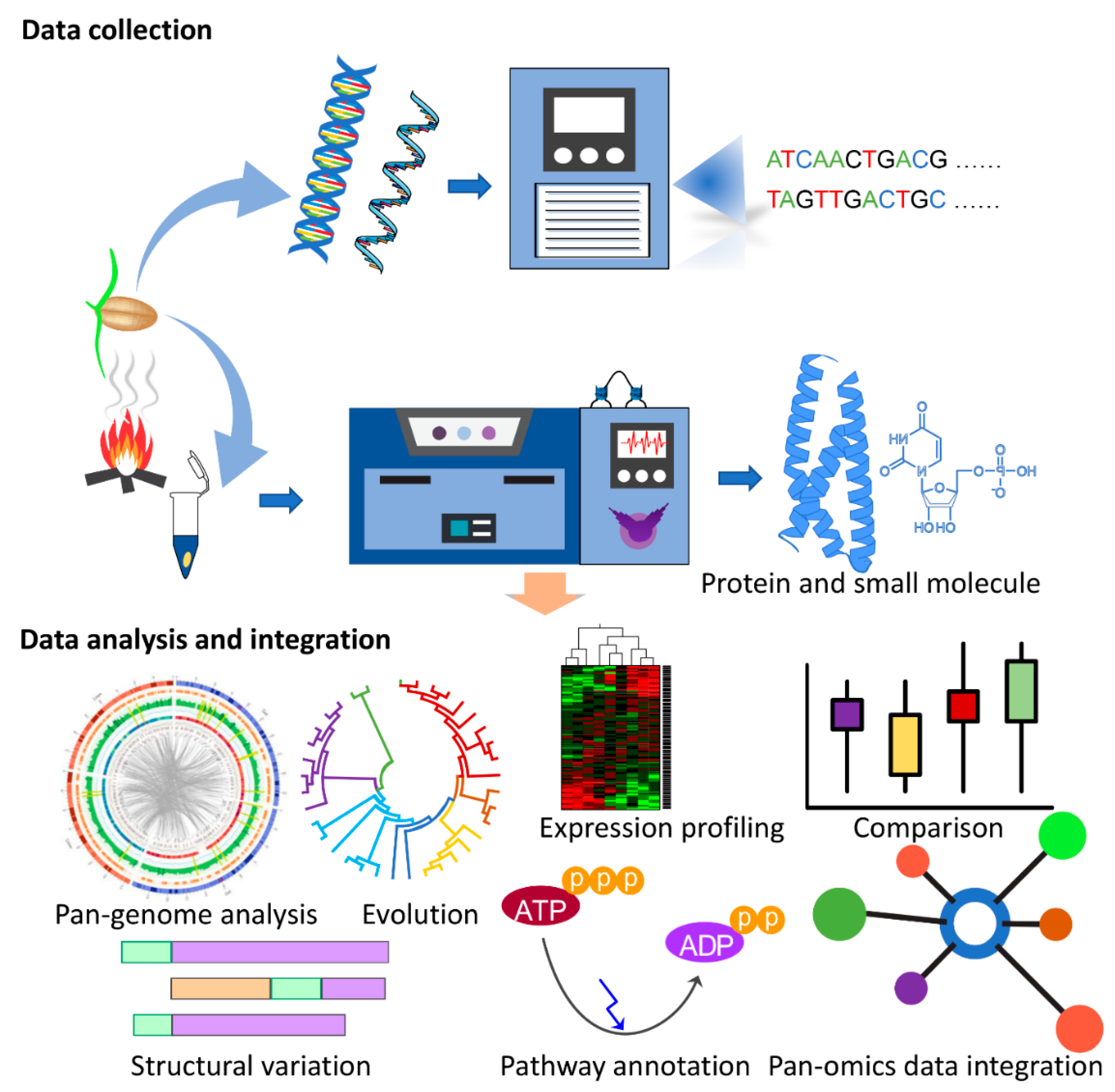
Decoding the genetic basis of agronomic traits is critical for crop improvement. The in-depth knowledge of the underlying genetic variation offers resources for marker development to link genetic variation with agronomic traits via quantitative trait loci (QTL) and genome-wide association studies (GWAS) studies. The availability of reference genome sequencing data from various crop species and their comparison with other species through the comparative pan-genome approach has enabled the understanding of the underlying molecular genetics such as gene function and pathways for biotic and abiotic stress resistance [96,97]. Advancements in next generation sequencing and data processing techniques have improved the understanding of genetic diversity, making it feasible to look beyond the knowledge generated from a model species and explore the evolution of signalling pathways in the specific ecological context of crop breeding programs. Studies have applied comparative genomic approaches to select agronomic traits such as grain yield and quality for rice [98,99], photoperiod sensitivity in maize [100] and grain size and shape in wheat [101]. Despite the wide application of genome sequencing and pan-genome analysis for improving cereal traits, the diversity and role of KAR and SL pathways remain unexplored with regard to their role in shade avoidance, the regulation of germination, seed vigour and plant development across cereal species such as wheat, maize, rice and barley. Notably, many signalling-related experiments were performed on model species such as Arabidopsis. To translate this knowledge from model species to agronomically important crops, the integration of genomics resources with QTL, GWAS, molecular marker and integrated-omics studies can be beneficial and represents a promising strategy for understanding critical insights into the regulation of KAR and SL pathways. The unprecedented availability of reference and pangenome assemblies will facilitate further interdisciplinary research and generate resources to study the combinatorial differences in gene content between crops, supporting the functional characterisation of genes, their evolutionary history and function and facilitate the design of future ‘omics-guided synthetic biology-based experiments to validate their functional importance in crops (Figure 1).
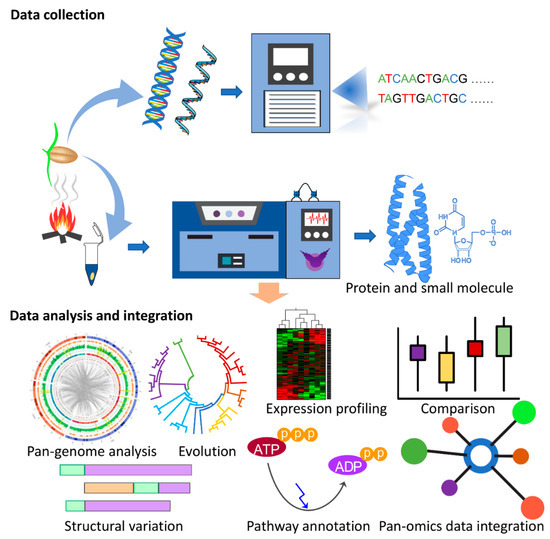
Figure 1.
Multi-omics experimental strategy for decoding karrikin (KAR) signalling pathways in crops. The first step is to collect omics data and then analyse and integrate the data in combination to identify the key regulatory mechanisms in plants.
Of the multi-omics approaches, one avenue is to build mutant- or species-specific genomic resources through DNA and RNA sequencing technologies and then use proteomics-guided techniques to validate the gene model and understand the extent of modifications or perturbations at the system level. These proteogenomic models have been developed and widely applied for personalised medical treatment development [102]. Adapting this methodology to plant science also has the potential to answer the remaining questions in KAR signalling that genomic or other ‘omics’ strategies alone have been unable to answer. This will include the investigation of post-translational modification that is critical to signalling and the impact of environmental changes. Specific attention should also be paid towards transcription factors (TFs) as the targeted and dynamic expression of genes, proteins and metabolite biosynthesis and signalling are independently or cooperatively dependent on these in planta [103,104]. To elucidate how TFs influence the KAR-dependent signalling pathways for crop species, additional studies are required to investigate the interaction among TFs and the quantitative responses between DNA, RNA, proteins and the small molecules.
Besides traditional marker selection methods used for breeding, data-driven analyses have been used to establish and identify the molecular strengths (targeting the genome, transcriptome and proteome) of plants, providing a pathway to upregulate their defence-related genes [105] or enzymes [106]. For example, the application of jasmonic acid can induce insect resistance in rice [105]. Likewise, a transmembrane protein BIL4 that regulates cell elongation was found to be affected by the brassinosteroid synthesis inhibitor brasinazole [107]. In a similar way, upon identifying and validating the molecular targets for KAR-dependent seed vigour and/or plant abiotic stress-tolerant pathways for crops, molecular strengthening techniques can be developed by applying exogenous substances that can control or enhance plant growth and development.
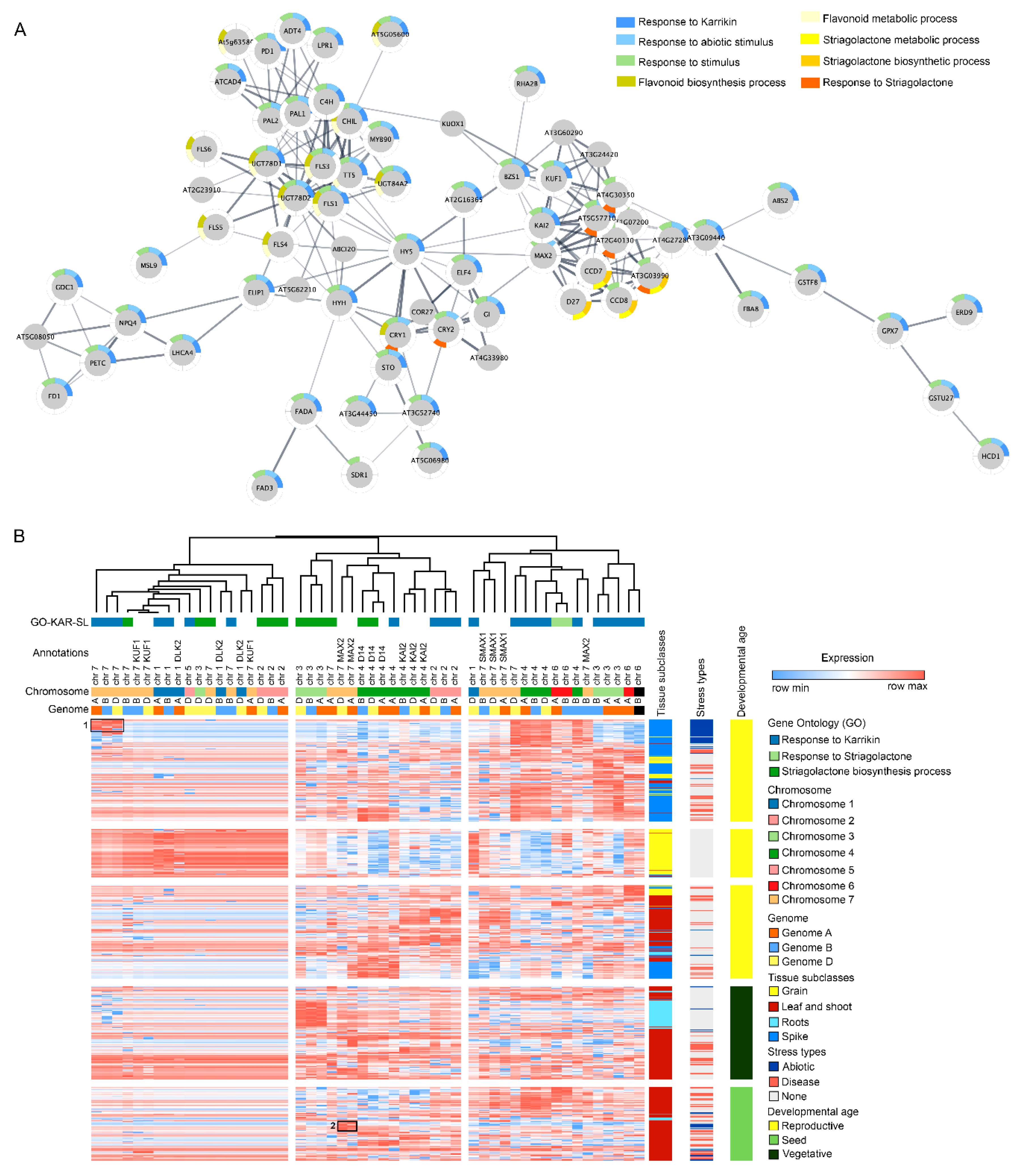
To map the tentative orthologous receptor genes for KARs and SLs identified in A. thaliana, a genome-wide search was performed using the published genome of agronomically important crops like wheat, barley, rice, maize and sorghum. Gene ontology (GO) terms associated with response to karrikin (GO:0080167), response to strigolactone (GO:190234); strigolactone metabolic process (GO:1901600) and strigolactone biosynthetic process (GO:1901601) were identified from A. thaliana and searched against publicly available databases for the aforementioned genomes (Table S1). In Arabidopsis, 133 genes have been associated with KAR response, many of these are present in multiple protein isoforms (Figure 2A). In wheat, 22 genes representing all seven chromosome groups are associated with response to karrikin. In barley, there are 13 genes associated with the GO term. Additionally, 22 genes have been identified in rice, 20 in sorghum and 23 in maize.
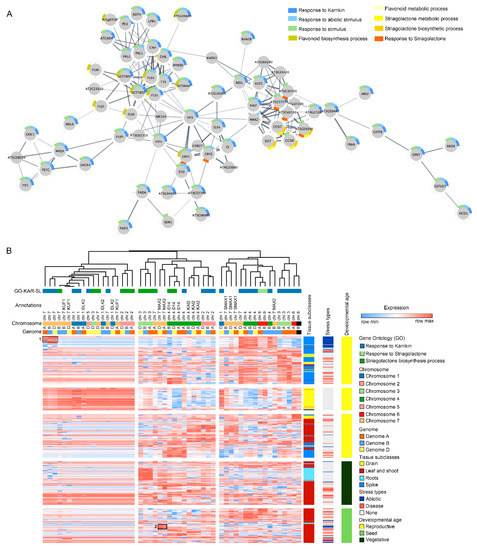
Figure 2.
Genes associated with the known KAR and strigolactone (SL) cross talk can be identified in wheat and related crop species. (A) The dynamics of selected GO categories for KAR and SL proteins were captured in a protein–protein interaction map using proteins related to KAR and SL-associated GO terms in A. thaliana. The network built using the sequences tagged with the KAR and SL gene- associated GO terms provides evidence for the direct relationship between the two hormone signalling pathways. Node colouring in the graph depicts the significantly enriched GO terms for KAR and SL. (B) Gene expression patterns of homologous genes associated with KAR and SL-metabolism or signalling GO terms in wheat were analysed using transcriptome data acquired at different plant development stages and abiotic and biotic stress conditions. Dendrograms show the clustering of GO terms associated with KAR and SL, gene annotations, chromosomal locations and the corresponding common bread wheat (hexaploid) genomes A, B and D. The heat map depicts the relative transcript level of KAR- and SL-related gene expression across wheat tissues, stress subtypes and developmental stages. High expression levels in abiotic stress conditions were primarily enriched in chromosome group 7 (marked as 1); MAX2-dependent abiotic stress tolerance and KAR signalling pathways (marked as 2).
To further investigate the wheat gene orthologues identified from interaction networks, the expression data for the gene list (known karrikin response, strigolactone response, strigolactone biosynthesis and the annotated DLK2, MAX2 orthologs and SL function-associated genes) were downloaded from publicly available wheat gene expression databases [108,109]. To understand how KAR and SL play a role in stress response in plants, expression data for samples collected from abiotic or biotic stress experiments were examined. Notably, genes with high expression levels in abiotic stress conditions were primarily enriched in chromosome group 7 (marked as 1; Figure 2B). Likewise, another set of enrichment was observed for genes in the chromosome 4 (Figure 2B).
In Arabidopsis, KAI2 mutants have shown osmotic stress tolerance by activating DLK2 and Karrikin Upregulated F-BOX1 (KUF1) genes while inhibiting the hypocotyl elongation [13]. Although, a positive correlation has been observed for KAR and SL-related genes with the abiotic stress tolerance, the results herein provide evidence that most of the gene expression was observed in the wheat spike tissue. It is possible that due to the under-representation of stressed root-specific transcriptome data, the identification of any positive correlation with the root and KAR and SL genes was not observed herein. Future transcriptome studies may be performed on wheat roots with or without KAR or SL treatment to understand their roles in abiotic stress tolerance in agronomically important plant species.
Interestingly, we identified another cluster of genes associated with MAX2-dependent abiotic stress tolerance and KAR signalling pathways (marked as 2; Figure 2B). Studies have shown that KAR and SL play a strong role in plant responses to drought and salt stress through MAX2 pathways in Arabidopsis [43,45]. Here, the interaction network and the clustering analysis have shown that KAR-dependent stress response pathways are active in Arabidopsis. Further exploring the members of the KAR and SL signalling pathways and investigating their orthologues in wheat will enable us to gain a better insight into the molecular mechanisms underlying their roles in crop adaptation to abiotic stress responses. The mapping results presented here should be taken cautiously as these are the tentative leads for KAR and SL pathways in cereal crops. Future follow-up studies should be designed to validate the target genes from cereal crops and elucidate their functional variations in a genome content and structure on crop performance. Extensive genomic profiling with transcriptomics, proteomics and metabolomics across KAR and SL mutants for cereal species would greatly expand the knowledgebase in this area. The results obtained through ‘omics data mining can then provide us with an avenue for developing improved stress-tolerant crop traits.
8. Concluding Remarks and Future Perspectives
Comprehensive molecular and structural analyses along with plant phenotype experiments have been performed on SL and KAR signalling pathways in recent decades. However, challenges remain regarding how to translate this new knowledge from model species and apply it to commercial crops. Advancements in acquiring omics data and integrative system biology analyses have enabled researchers to look beyond model plants and explore the activation or regulatory signalling mechanisms in specific ecological contexts. Comparative omics analyses are likely to be the most promising strategies for gaining insights into critical KAR pathway residues, types of regulation and their application feasibility for crops (Figure 1).
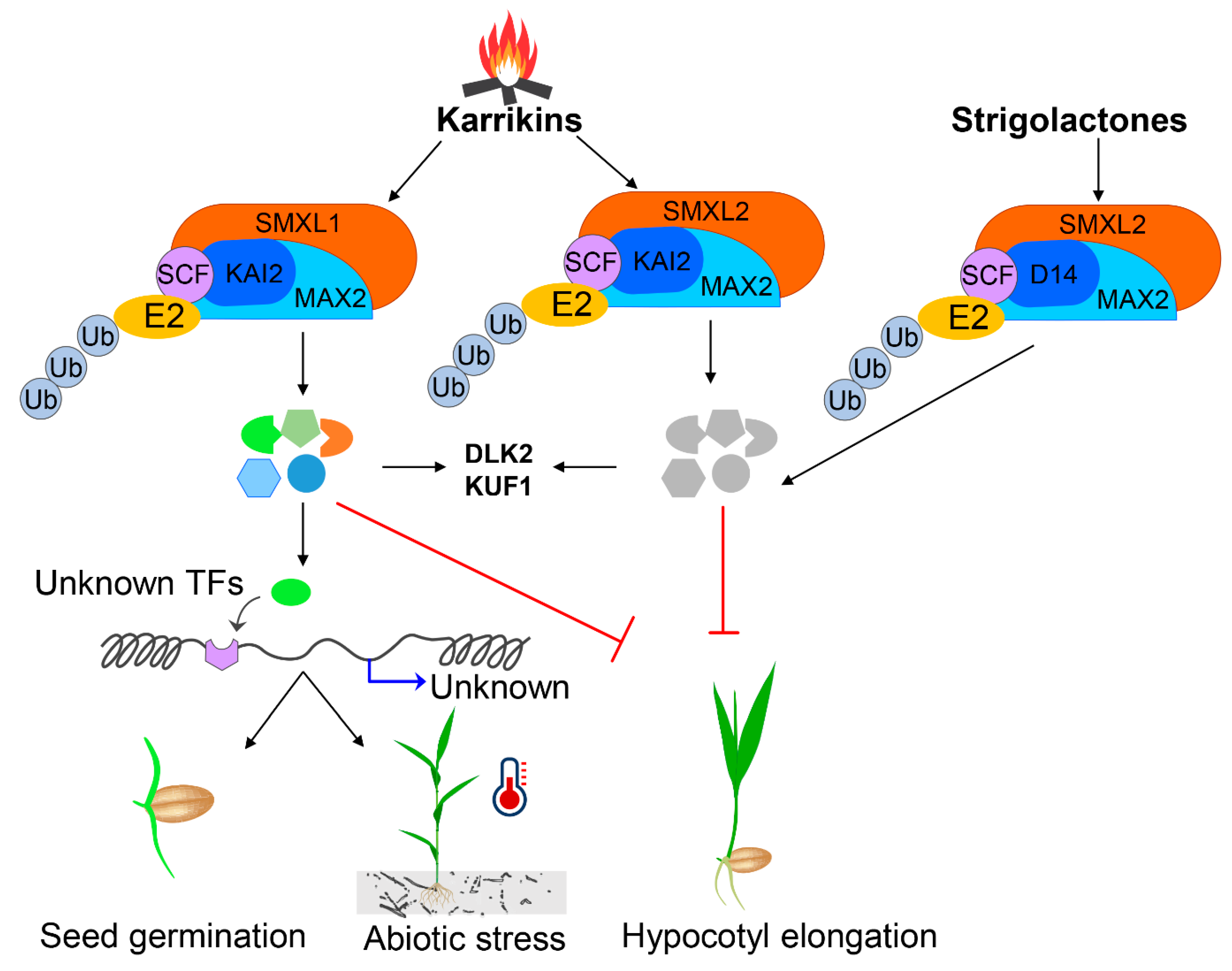
KARs and plant phytohormones such as ABA, GA and auxin can control seed germination and seedling development. However, there are knowledge gaps as to how KARs can interact and precisely synchronise the key signalling pathways during germination, in particular for auxin. The questions remain: why and how different plants species perceive KAR responses (Figure 3); how do different SMXL proteins regulate various downstream signalling responses; besides KAI2 activation, how are KARs metabolised into the active ligand; what is the precise mechanism of action; and, additionally, what is the evolutionary context of the SMXL family that should be considered in order to decode the ligand-receptor activation machineries.
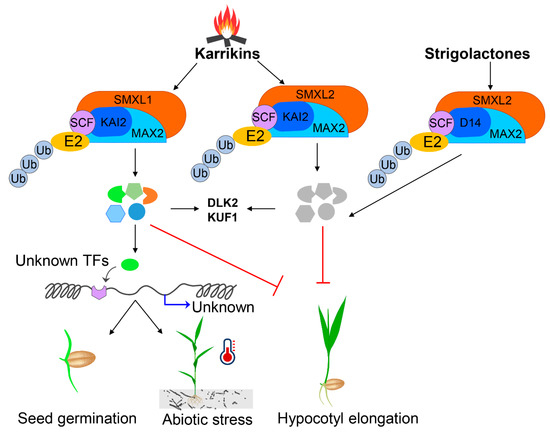
Figure 3.
Proposed model of strigolactone and karrikin-assisted germination and abiotic stress tolerance pathways for agricultural crops. The figure has been re-drawn and modified from (Wang et al., 2020; Yao and Waters, 2020; Morffy et al., 2016).
The advancement of next generation sequencing technologies, mass spectrometry-based proteomics and data mining strategies has enabled the identification and functional characterisation of the genetic factors controlling superior agronomic traits in crops. The integration and application of post-genomic profiling platforms have helped to unravel the complex phenotypes and genetic diversity in plant species [18,110,111]. As we have seen, the response to KARs can be diverse across crop species. Large-scale ‘pan-omics’ approaches can be used as an option to decode the unknown regulatory mechanisms for KAR-dependent pathways. The ‘proteogenomic’ approach, which exploits the information gathered from genomic and proteomic studies, widely used in clinical research [102], can be introduced to explore the KAR and SL-dependent pathways in plants. The pan-genomic and comparative genomics analysis approaches may also be used to identify and screen for the diversity of KAR receptor genes present across different plant species. Comparative -omics and data mining strategies can also be applied to mutants and/or different plant species. Upon the identification of target genes, pathways and the molecular processes for KAR signalling pathways, marker-assisted selection, transgenic, and genome editing approaches may be applied to develop the seeds and plants with enhanced agronomic traits. Multidisciplinary translational research is required to apply these scientific discoveries into useful benefits for crop development and seedling vigour.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/20/7512/s1. Table S1: GO-terms associated with karrikin, response to strigolactone, strigolactone metabolic process and strigolactone biosynthetic process.
Author Contributions
Conceptualization, M.L.C. and S.K.; formal analysis, A.J. and U.B.; investigation, A.J.; resources, M.L.C. and S.K.; writing—original draft preparation, U.B.; writing—review and editing, A.J., J.A.B., S.K. and M.L.C.; visualization, A.J. and U.B.; supervision, M.L.C. and S.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ABA | Abscisic acid |
| Chr | Chromosome |
| CRL | Cullin-RING ubiquitin ligase |
| DLK2 | DWARF 14-like2 |
| GA | Gibberellic acid |
| GO | Gene ontology |
| GWAS | Genome-wide association studies |
| IAA | Indole-3-acetic acid |
| JA | Jasmonic acid |
| KAI2 | Karrikin insensitive 2 |
| KAR | Karrikin |
| KL | KAI2-ligand |
| LRR | Leucine-rich repeat containing domain |
| MAX2 | More Auxiliary Branches |
| NO2 | Nitrogen oxide |
| SCF | Skp-Cullin-F-box |
| SL | Strigolactone |
| SMXL | Suppressor of MAX-like |
| TMB | Trimethylbutanolide |
| TF | Transcription factor |
| QTL | Quantitative trait locus |
References
- Adkins, S.; Peters, N. Smoke derived from burnt vegetation stimulates germination of arable weeds. Seed Sci. Res. 2001, 11, 213–222. [Google Scholar] [CrossRef]
- Aslam, M.M.; Rehman, S.; Khatoon, A.; Jamil, M.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Li, X.; Sunohara, Y.; Matsumoto, H. Molecular responses of maize shoot to a plant derived smoke solution. Int. J. Mol. Sci. 2019, 20, 1319. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; ur Rehman, S.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Yamaguchi, T.; Sunohara, Y.; Matsumoto, H.; Komatsu, S. Proteomic analysis of the effect of plant-derived smoke on soybean during recovery from flooding stress. J. Proteom. 2018, 181, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.C.; Flematti, G.R.; Ghisalberti, E.L.; Dixon, K.W.; Smith, S.M. Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annu. Rev. Plant Biol. 2012, 63, 107–130. [Google Scholar] [CrossRef] [PubMed]
- Alboresi, A.; Gestin, C.; Leydecker, M.T.; Bedu, M.; Meyer, C.; Truong, H.N. Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ. 2005, 28, 500–512. [Google Scholar] [CrossRef] [PubMed]
- Flematti, G.R.; Merritt, D.J.; Piggott, M.J.; Trengove, R.D.; Smith, S.M.; Dixon, K.W.; Ghisalberti, E.L. Burning vegetation produces cyanohydrins that liberate cyanide and stimulate seed germination. Nat. Commun. 2011, 2, 1–6. [Google Scholar] [CrossRef]
- Flematti, G.R.; Ghisalberti, E.L.; Dixon, K.W.; Trengove, R.D. A compound from smoke that promotes seed germination. Science 2004, 305, 977. [Google Scholar] [CrossRef] [PubMed]
- Van Staden, J.; Jager, A.; Light, M.; Burger, B. Isolation of the major germination cue from plant-derived smoke. S. Afr. J. Bot. 2004, 70, 654–659. [Google Scholar] [CrossRef]
- Meng, Y.; Shuai, H.; Luo, X.; Chen, F.; Zhou, W.; Yang, W.; Shu, K. Karrikins: Regulators involved in phytohormone signaling networks during seed germination and seedling development. Front. Plant Sci. 2017, 7, 2021. [Google Scholar] [CrossRef]
- Morffy, N.; Faure, L.; Nelson, D.C. Smoke and hormone mirrors: Action and evolution of karrikin and strigolactone signaling. Trend Genet. 2016, 32, 176–188. [Google Scholar] [CrossRef]
- Nelson, D.C.; Scaffidi, A.; Dun, E.A.; Waters, M.T.; Flematti, G.R.; Dixon, K.W.; Beveridge, C.A.; Ghisalberti, E.L.; Smith, S.M. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 8897–8902. [Google Scholar] [CrossRef] [PubMed]
- Villaécija-Aguilar, J.A.; Hamon-Josse, M.; Carbonnel, S.; Kretschmar, A.; Schmid, C.; Dawid, C.; Bennett, T.; Gutjahr, C. SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PLoS Genet. 2019, 15, e1008327. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Waters, M.T.; Smith, S.M. Karrikin-KAI2 signalling provides Arabidopsis seeds with tolerance to abiotic stress and inhibits germination under conditions unfavourable to seedling establishment. New Phytol. 2018, 219, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Elsadek, M.A.; Yousef, E.A. Smoke-water enhances germination and seedling growth of four horticultural crops. Plants 2019, 8, 104. [Google Scholar] [CrossRef]
- Conn, C.E.; Nelson, D.C. Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front. Plant Sci. 2016, 6, 1219. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef]
- Hilhorst, H.W. Definitions and hypotheses of seed dormancy. Annu. Plant Rev. Online 2007, 27, 50–71. [Google Scholar] [CrossRef]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59. [Google Scholar] [CrossRef]
- Allen, P.S.; Benech-Arnold, R.L.; Batlla, D.; Bradford, K.J. Modeling of seed dormancy. In Seed Development, Dormancy and Germination; Bradford, K.J., Nonogaki, H., Eds.; Blackwell Publishing Ltd: Oxford, UK, 2007; pp. 72–112. [Google Scholar] [CrossRef]
- Baskin, J.M.; Baskin, C.C. A classification system for seed dormancy. Seed Sci. Res. 2004, 14, 1–16. [Google Scholar] [CrossRef]
- Nonogaki, H.; Bassel, G.W.; Bewley, J.D. Germination—still a mystery. Plant Sci. 2010, 179, 574–581. [Google Scholar] [CrossRef]
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed germination and vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S. Seed dormancy and germination. Curr. Biol. 2017, 27, R874–R878. [Google Scholar] [CrossRef] [PubMed]
- Finch-Savage, W.E.; Bassel, G.W. Seed vigour and crop establishment: Extending performance beyond adaptation. J. Exp. Bot. 2016, 67, 567–591. [Google Scholar] [CrossRef] [PubMed]
- Catusse, J.; Meinhard, J.; Job, C.; Strub, J.M.; Fischer, U.; Pestsova, E.; Westhoff, P.; Van Dorsselaer, A.; Job, D. Proteomics reveals potential biomarkers of seed vigor in sugarbeet. Proteomics 2011, 11, 1569–1580. [Google Scholar] [CrossRef]
- Bewley, J.D.; Bradford, K.J.; Hilhorst, H.W.; Nonogaki, H. Environmental regulation of dormancy and germination. In Seeds; Springer: New York, NY, USA, 2013; pp. 299–339. [Google Scholar]
- Dixon, K.; Merritt, D.; Flematti, G.; Ghisalberti, E. Karrikinolide–a phytoreactive compound derived from smoke with applications in horticulture, ecological restoration and agriculture. Acta Hortic. 2009, 813, 155–170. [Google Scholar] [CrossRef]
- Van Staden, J.; Drewes, F.; Brown, N. Some chromatographic characteristics of germination stimulants in plant-derived smoke extracts. Plant Growth Regul. 1995, 17, 241–249. [Google Scholar] [CrossRef]
- Brown, N.; Van Staden, J. Smoke as a germination cue: A review. Plant Growth Regul. 1997, 22, 115–124. [Google Scholar] [CrossRef]
- Staden, J.V.; Brown, N.A.; Jäger, A.K.; Johnson, T.A. Smoke as a germination cue. Plant Spec. Biol. 2008, 15, 167–178. [Google Scholar] [CrossRef]
- Rundel, P.W.; Arroyo, M.T.; Cowling, R.M.; Keeley, J.E.; Lamont, B.B.; Pausas, J.G.; Vargas, P. Fire and plant diversification in Mediterranean-climate regions. Front. Plant Sci. 2018, 9, 851. [Google Scholar] [CrossRef]
- Dixon, K.; Roche, S. The role of combustion products (smoke) in stimulating ex-situ and in-situ germination of Western Australian plants. Proc. Int. Plant Propagators Soc. 1995, 45, 53–56. [Google Scholar]
- De Lange, J.; Boucher, C. Autecological studies on Audouinia capitata (Bruniaceae). I. Plant-derived smoke as a seed germination cue. S. Afr. J. Bot. 1990, 56, 700–703. [Google Scholar] [CrossRef]
- Keeley, J.E.; Fotheringham, C. Smoke-induced seed germination in California chaparral. Ecology 1998, 79, 2320–2336. [Google Scholar] [CrossRef]
- Çatav, Ş.S.; Küçükakyüz, K.; Akbaş, K.; Tavşanoğlu, Ç. Smoke-enhanced seed germination in Mediterranean Lamiaceae. Seed Sci. Res. 2014, 24, 257–264. [Google Scholar] [CrossRef]
- Kulkarni, M.; Sparg, S.; Light, M.; Van Staden, J. Stimulation of rice (Oryza sativa L.) seedling vigour by smoke-water and butenolide. J. Arron. Crop. Sci. 2006, 192, 395–398. [Google Scholar] [CrossRef]
- Gupta, S.; Plačková, L.; Kulkarni, M.G.; Doležal, K.; Van Staden, J. Role of smoke stimulatory and inhibitory biomolecules in phytochrome-regulated seed germination of Lactuca sativa. Plant Physiol. 2019, 181, 458–470. [Google Scholar] [CrossRef] [PubMed]
- Mojzes, A.; Csontos, P.; Kalapos, T. Is the positive response of seed germination to plant-derived smoke associated with plant traits? Acta Oecol. 2015, 65, 24–31. [Google Scholar] [CrossRef]
- Senaratna, T.; Dixon, K.; Bunn, E.; Touchell, D. Smoke-saturated water promotes somatic embryogenesis in geranium. Plant Growth Regul. 1999, 28, 95–99. [Google Scholar] [CrossRef]
- Brits, G.; Calitz, F.; Brown, N.; Manning, J. Desiccation as the active principle in heat-stimulated seed germination of Leucospermum R. Br.(Proteaceae) in fynbos. New Phytol. 1993, 125, 397–403. [Google Scholar] [CrossRef]
- Soós, V.; Sebestyén, E.; Juhász, A.; Pintér, J.; Light, M.E.; Van Staden, J.; Balázs, E. Stress-related genes define essential steps in the response of maize seedlings to smoke-water. Funct. Integr. Genom. 2009, 9, 231–242. [Google Scholar] [CrossRef]
- Yao, J.; Waters, M.T. Perception of karrikins by plants: A continuing enigma. J. Exp. Bot. 2020, 71, 1774–1781. [Google Scholar] [CrossRef]
- Li, W.; Nguyen, K.H.; Chu, H.D.; Ha, C.V.; Watanabe, Y.; Osakabe, Y.; Leyva-González, M.A.; Sato, M.; Toyooka, K.; Voges, L. The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana. PLoS Genet. 2017, 13, e1007076. [Google Scholar] [CrossRef] [PubMed]
- Baxter, B.; Granger, J.; Van Staden, J. Plant-derived smoke and seed germination: Is all smoke good smoke? That is the burning question. S. Afr. J. Bot. 1995, 61, 275–277. [Google Scholar] [CrossRef]
- Li, W.; Tran, L.-S.P. Are karrikins involved in plant abiotic stress responses? Trends Plant Sci. 2015, 20, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Otori, M.; Murashita, Y.; Ur Rehman, S.; Komatsu, S. Proteomic study to understand promotive effects of plant-derived smoke on soybean (Glycine max L.) root growth under flooding stress. Plant Mol. Biol. Rep. 2020, 1–10. [Google Scholar] [CrossRef]
- Gupta, S.; Hrdlička, J.; Ngoroyemoto, N.; Nemahunguni, N.K.; Gucký, T.; Novák, O.; Kulkarni, M.G.; Doležal, K.; Van Staden, J. Preparation and standardisation of smoke-water for seed germination and plant growth stimulation. J. Plant Growth Regul. 2019. [Google Scholar] [CrossRef]
- Jäger, A.; Light, M.; Van Staden, J. Effects of source of plant material and temperature on the production of smoke extracts that promote germination of light-sensitive lettuce seeds. Environ. Exp. Bot. 1996, 36, 421–429. [Google Scholar] [CrossRef]
- Burger, B.; Pošta, M.; Light, M.; Kulkarni, M.; Viviers, M.; Van Staden, J. More butenolides from plant-derived smoke with germination inhibitory activity against karrikinolide. S. Afr. J. Bot. 2018, 115, 256–263. [Google Scholar] [CrossRef]
- Flematti, G.R.; Ghisalberti, E.L.; Dixon, K.W.; Trengove, R.D. Identification of alkyl substituted 2 H-furo [2¨C-c] pyran-2-ones as germination stimulants present in smoke. J. Agr. Food Chem. 2009, 57, 9475–9480. [Google Scholar] [CrossRef] [PubMed]
- Flematti, G.R.; Ghisalberti, E.L.; Dixon, K.W.; Trengove, R.D. Germination stimulant in smoke: Isolation and identification. In Bioactive Natural Products: Detection, Isolation, and Structural Determination; Colegate, S.M., Molyneux, R.J., Eds.; CRC Press: Boca Raton, FL, USA, 2008; pp. 531–554. [Google Scholar]
- Flematti, G.R.; Waters, M.T.; Scaffidi, A.; Merritt, D.J.; Ghisalberti, E.L.; Dixon, K.W.; Smith, S.M. Karrikin and cyanohydrin smoke signals provide clues to new endogenous plant signaling compounds. Mol. Plant 2013, 6, 29–37. [Google Scholar] [CrossRef]
- Keeley, J.E.; Fotheringham, C. Trace gas emissions and smoke-induced seed germination. Science 1997, 276, 1248–1250. [Google Scholar] [CrossRef]
- Keeley, J.; Pausas, J. Evolution of ‘smoke’ induced seed germination in pyroendemic plants. S. Afr. J. Bot. 2018, 115, 251–255. [Google Scholar] [CrossRef]
- Soós, V.; Sebestyén, E.; Posta, M.; Kohout, L.; Light, M.E.; Van Staden, J.; Balázs, E. Molecular aspects of the antagonistic interaction of smoke-derived butenolides on the germination process of G rand R apids lettuce (Lactuca sativa) achenes. New Phytol. 2012, 196, 1060–1073. [Google Scholar] [CrossRef] [PubMed]
- Papenfus, H.B.; Kulkarni, M.G.; Pošta, M.; Finnie, J.F.; Van Staden, J. Smoke-isolated trimethylbutenolide inhibits seed germination of different weed species by reducing amylase activity. Weed Sci. 2015, 63, 312–320. [Google Scholar] [CrossRef]
- Baldwin, I.T.; Staszak-Kozinski, L.; Davidson, R. Up in smoke: I. Smoke-derived germination cues for postfire annual, Nicotiana attenuata torr. Ex. Watson. J. Chem. Ecol. 1994, 20, 2345–2371. [Google Scholar] [CrossRef]
- Nelson, D.C.; Flematti, G.R.; Riseborough, J.-A.; Ghisalberti, E.L.; Dixon, K.W.; Smith, S.M. Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 7095–7100. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Chen, F.; Shuai, H.; Luo, X.; Ding, J.; Tang, S.; Xu, S.; Liu, J.; Liu, W.; Du, J. Karrikins delay soybean seed germination by mediating abscisic acid and gibberellin biogenesis under shaded conditions. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Scaffidi, A.; Sun, Y.K.; Flematti, G.R.; Smith, S.M. The karrikin response system of Arabidopsis. Plant J. 2014, 79, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef] [PubMed]
- Seto, Y.; Sado, A.; Asami, K.; Hanada, A.; Umehara, M.; Akiyama, K.; Yamaguchi, S. Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. USA 2014, 111, 1640–1645. [Google Scholar] [CrossRef]
- Végh, A.; Incze, N.; Fábián, A.; Huo, H.; Bradford, K.J.; Balázs, E.; Soós, V. Comprehensive analysis of DWARF14-LIKE2 (DLK2) reveals its functional divergence from strigolactone-related paralogs. Front. Plant Sci. 2017, 8, 1641. [Google Scholar] [CrossRef]
- Bennett, T.; Leyser, O. Strigolactone signalling: Standing on the shoulders of DWARFs. Curr. Opin. Plant Biol. 2014, 22, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Nelson, D.C.; Scaffidi, A.; Flematti, G.R.; Sun, Y.K.; Dixon, K.W.; Smith, S.M. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 2012, 139, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Koltai, H. Strigolactones are regulators of root development. New Phytol. 2011, 190, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Soto, M.J.; Fernández-Aparicio, M.; Castellanos-Morales, V.; García-Garrido, J.M.; Ocampo, J.A.; Delgado, M.J.; Vierheilig, H. First indications for the involvement of strigolactones on nodule formation in alfalfa (Medicago sativa). Soil Biol. Biochem. 2010, 42, 383–385. [Google Scholar] [CrossRef]
- Brewer, P.B.; Koltai, H.; Beveridge, C.A. Diverse roles of strigolactones in plant development. Mol. Plant 2013, 6, 18–28. [Google Scholar] [CrossRef]
- Murase, K.; Hirano, Y.; Sun, T.-p.; Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, T.; Wang, M.; Liu, Y.; Yuan, S.; Gao, Y.; Yin, L.; Sun, W.; Peng, L.; Zhang, W. DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching. Plant Cell Physiol. 2014, 55, 1096–1109. [Google Scholar] [CrossRef]
- Zhou, F.; Lin, Q.; Zhu, L.; Ren, Y.; Zhou, K.; Shabek, N.; Wu, F.; Mao, H.; Dong, W.; Gan, L. D14–SCF D3-dependent degradation of D53 regulates strigolactone signalling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, X.; Xiong, G.; Liu, H.; Chen, F.; Wang, L.; Meng, X.; Liu, G.; Yu, H.; Yuan, Y. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 2013, 504, 401–405. [Google Scholar] [CrossRef]
- Arite, T.; Umehara, M.; Ishikawa, S.; Hanada, A.; Maekawa, M.; Yamaguchi, S.; Kyozuka, J. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009, 50, 1416–1424. [Google Scholar] [CrossRef] [PubMed]
- Soundappan, I.; Bennett, T.; Morffy, N.; Liang, Y.; Stanga, J.P.; Abbas, A.; Leyser, O.; Nelson, D.C. SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 2015, 27, 3143–3159. [Google Scholar] [CrossRef] [PubMed]
- Janssen, B.J.; Snowden, K.C. Strigolactone and karrikin signal perception: Receptors, enzymes, or both? Front. Plant Sci. 2012, 3, 296. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, S.; Zhu, W.; Jia, K.; Yang, H.; Wang, X. Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev. Cell 2013, 27, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Stanga, J.P.; Morffy, N.; Nelson, D.C. Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta 2016, 243, 1397–1406. [Google Scholar] [CrossRef]
- Rusnac, D.V.; Zheng, N. Overview of protein degradation in plant hormone signaling. In Plant structural biology: Hormonal regulations; Hejátko, J., Hakoshima, T., Eds.; Springer: Cham, Switzerland, 2018; pp. 11–30. [Google Scholar]
- Nelson, D.C.; Riseborough, J.-A.; Flematti, G.R.; Stevens, J.; Ghisalberti, E.L.; Dixon, K.W.; Smith, S.M. Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 2009, 149, 863–873. [Google Scholar] [CrossRef]
- Gilkerson, J.; Kelley, D.R.; Tam, R.; Estelle, M.; Callis, J. Lysine residues are not required for proteasome-mediated proteolysis of the auxin/indole acidic acid protein IAA1. Plant Physiol. 2015, 168, 708–720. [Google Scholar] [CrossRef]
- Bernardo-García, S.; de Lucas, M.; Martínez, C.; Espinosa-Ruiz, A.; Davière, J.-M.; Prat, S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014, 28, 1681–1694. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF COI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Hakoshima, T. Overview of proteins in plant hormone signaling. In Plant Structural Biology: Hormonal Regulations; Springer: Cham, Switzerland, 2018; pp. 3–10. [Google Scholar] [CrossRef]
- Stanga, J.P.; Smith, S.M.; Briggs, W.R.; Nelson, D.C. Suppressor of more axillary growth 2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 2013, 163, 318–330. [Google Scholar] [CrossRef]
- Hayward, A.; Stirnberg, P.; Beveridge, C.; Leyser, O. Interactions between auxin and strigolactone in shoot branching control. Plant Physiol. 2009, 151, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Nodzyński, T.; Vanneste, S.; Zwiewka, M.; Pernisová, M.; Hejátko, J.; Friml, J. Enquiry into the topology of plasma membrane-localized PIN auxin transport components. Mol. Plant 2016, 9, 1504–1519. [Google Scholar] [CrossRef]
- Shen, H.; Zhu, L.; Bu, Q.-Y.; Huq, E. MAX2 affects multiple hormones to promote photomorphogenesis. Mol. Plant 2012, 5, 750–762. [Google Scholar] [CrossRef]
- Bose, U.; Broadbent, J.A.; Byrne, K.; Blundell, M.J.; Howitt, C.A.; Colgrave, M.L. Proteome analysis of hordein-null barley lines reveals storage protein synthesis and compensation mechanisms. J. Agric. Food Chem. 2020, 68, 5763–5775. [Google Scholar] [CrossRef]
- Komatsu, S.; Hiraga, S.; Yanagawa, Y. Proteomics techniques for the development of flood tolerant crops. J. Proteome Res. 2012, 11, 68–78. [Google Scholar] [CrossRef]
- Wu, X.; Gong, F.; Cao, D.; Hu, X.; Wang, W. Advances in crop proteomics: PTMs of proteins under abiotic stress. Proteomics 2016, 16, 847–865. [Google Scholar] [CrossRef]
- Ahmad, P.; Abdel Latef, A.A.; Rasool, S.; Akram, N.A.; Ashraf, M.; Gucel, S. Role of proteomics in crop stress tolerance. Front. Plant Sci. 2016, 7, 1336. [Google Scholar] [CrossRef]
- Rehman, A.; ur Rehman, S.; Khatoon, A.; Qasim, M.; Itoh, T.; Iwasaki, Y.; Wang, X.; Sunohara, Y.; Matsumoto, H.; Komatsu, S. Proteomic analysis of the promotive effect of plant-derived smoke on plant growth of chickpea. J. Proteom. 2018, 176, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Kobayashi, T.; Zhu, W.; Imai, H.; Zhao, R.; Ohno, T.; ur Rehman, S.; Uemura, M.; Tian, J.; Komatsu, S. Plant-derived smoke enhances plant growth through ornithine-synthesis pathway and ubiquitin-proteasome pathway in soybean. J. Proteom. 2020, 103781. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, Q.; Yu, H.; Ma, H.; Li, X.; Yang, J.; Chu, J.; Xie, Q.; Wang, Y.; Smith, S.M. Strigolactone and karrikin signaling pathways elicit ubiquitination and proteolysis of SMXL2 to regulate hypocotyl elongation in Arabidopsis. Plant Cell 2020, 32, 2251–2270. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zhao, X.; Mace, E.; Henry, R.; Jordan, D. Exploring and exploiting pan-genomics for crop improvement. Mol. Plant 2019, 12, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Bayer, P.E.; Golicz, A.A.; Scheben, A.; Batley, J.; Edwards, D. Plant pan-genomes are the new reference. Nat. Plants 2020, 6, 914–920. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiong, G.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.; Zeng, L.; Xu, E.; Xu, J. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015, 47, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Sun, H.; Xu, D.; Chen, Q.; Liang, Y.; Wang, X.; Xu, G.; Tian, J.; Wang, C.; Li, D. ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. USA 2018, 115, E334–E341. [Google Scholar] [CrossRef]
- Wang, W.; Pan, Q.; Tian, B.; He, F.; Chen, Y.; Bai, G.; Akhunova, A.; Trick, H.N.; Akhunov, E. Gene editing of the wheat homologs of TONNEAU 1-recruiting motif encoding gene affects grain shape and weight in wheat. Plant J. 2019, 100, 251–264. [Google Scholar] [CrossRef]
- Zhang, B.; Whiteaker, J.R.; Hoofnagle, A.N.; Baird, G.S.; Rodland, K.D.; Paulovich, A.G. Clinical potential of mass spectrometry-based proteogenomics. Nat. Rev. Clin. Oncol. 2019, 16, 256–268. [Google Scholar] [CrossRef]
- Colinas, M.; Goossens, A. Combinatorial transcriptional control of plant specialized metabolism. Trends Plant Sci. 2018, 23, 324–336. [Google Scholar] [CrossRef]
- Bemer, M.; van Dijk, A.D.; Immink, R.G.; Angenent, G.C. Cross-family transcription factor interactions: An additional layer of gene regulation. Trends Plant Sci. 2017, 22, 66–80. [Google Scholar] [CrossRef]
- Taniguchi, S.; HOSOKAWA-SHINONAGA, Y.; Tamaoki, D.; Yamada, S.; Akimitsu, K.; Gomi, K. Jasmonate induction of the monoterpene linalool confers resistance to rice bacterial blight and its biosynthesis is regulated by JAZ protein in rice. Plant Cell Environ. 2014, 37, 451–461. [Google Scholar] [CrossRef]
- Nisha, S.; Revathi, K.; Chandrasekaran, R.; Kirubakaran, S.A.; Sathish-Narayanan, S.; Stout, M.J.; Senthil-Nathan, S. Effect of plant compounds on induced activities of defense-related enzymes and pathogenesis related protein in bacterial blight disease susceptible rice Plant Physiol. Mol. Plant Pathol. 2012, 80, 1–9. [Google Scholar] [CrossRef]
- Yamagami, A.; Nakazawa, M.; Matsui, M.; Tujimoto, M.; Sakuta, M.; Asami, T.; Nakano, T. Chemical genetics reveal the novel transmembrane protein BIL4, which mediates plant cell elongation in brassinosteroid signaling. Biosci. Biotechnol. Biochem. 2009, 73, 415–421. [Google Scholar] [CrossRef] [PubMed]
- IWGSC. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-González, R.; Borrill, P.; Lang, D.; Harrington, S.; Brinton, J.; Venturini, L. The transcriptional landscape of hexaploid wheat across tissues and cultivars. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef]
- Tanner, G.J.; Blundell, M.J.; Colgrave, M.L.; Howitt, C.A. Creation of the first ultra-low gluten barley (Hordeum vulgare L.) for coeliac and gluten-intolerant populations. Plant Biotechnol. J. 2016, 14, 1139–1150. [Google Scholar] [CrossRef]
- Tanner, G.J.; Colgrave, M.L.; Blundell, M.J.; Howitt, C.A.; Bacic, A. Hordein accumulation in developing barley grains. Front. Plant Sci. 2019, 10, 649. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).