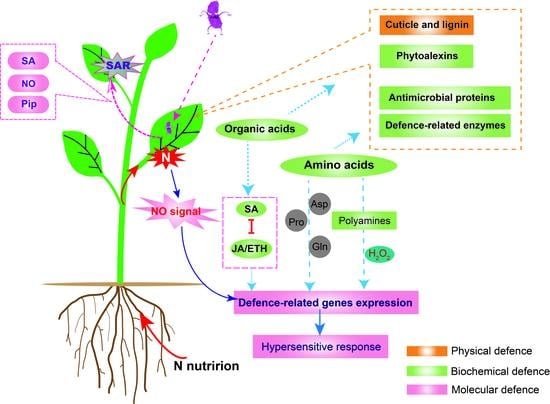
Unravelling the Roles of Nitrogen Nutrition in Plant Disease Defences
Abstract
1. Introduction
2. N Nutrition as It Impacts on the Pathogen
3. N Nutrition as It Impacts on Host Defence
3.1. Physical Defence Mechanisms
3.2. Plant Biochemical Defence Mechanism
3.2.1. Plant Metabolites and Biochemical Defence
3.2.2. Defence-Related Enzymes
3.3. Plant MOLECULAR Defence Mechanism
3.3.1. N Metabolism Links Hormones and Nitric Oxide Impacts on Defence
3.3.2. N Metabolism and SAR
4. Conclusions and Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Dietrich, R.; Ploss, K.; Heil, M. Constitutive and induced resistance to pathogens in Arabidopsis thaliana depends on nitrogen supply. Plant Cell Environ. 2004, 27, 896–906. [Google Scholar] [CrossRef]
- Serrano, M.; Coluccia, F.; Torres, M.; L’Haridon, F.; Metraux, J.P. The cuticle and plant defense to pathogens. Front. Plant Sci. 2014, 5, 274. [Google Scholar] [CrossRef] [PubMed]
- Grayer, R.J.; Kokubun, T. Plant–fungal interactions: The search for phytoalexins and other antifungal compounds from higher plants. Phytochemistry 2001, 56, 253–263. [Google Scholar] [CrossRef]
- Kumar, A.; Yogendra, K.N.; Karre, S.; Kushalappa, A.C.; Dion, Y.; Choo, T.M. WAX INDUCER1 (HvWIN1) transcription factor regulates free fatty acid biosynthetic genes to reinforce cuticle to resist Fusarium head blight in barley spikelets. J. Exp. Bot. 2016, 67, 4127–4139. [Google Scholar] [CrossRef] [PubMed]
- Sela, D.; Buxdorf, K.; Shi, J.X.; Feldmesser, E.; Schreiber, L.; Aharoni, A.; Levy, M. Overexpression of AtSHN1/WIN1 provokes unique defense responses. PLoS ONE 2013, 8, e70146. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, P.; Osbourn, A. Plant-microbe interactions: Chemical diversity in plant defense. Science 2009, 324, 746–748. [Google Scholar] [CrossRef]
- Bednarek, P. Chemical warfare or modulators of defence responses - the function of secondary metabolites in plant immunity. Curr. Opin. Plant Biol. 2012, 15, 407–414. [Google Scholar] [CrossRef]
- Conrath, U. Molecular aspects of defence priming. Trends Plant Sci. 2011, 16, 524–531. [Google Scholar] [CrossRef]
- Miller, R.N.; Costa Alves, G.S.; Van Sluys, M.A. Plant immunity: Unravelling the complexity of plant responses to biotic stresses. Ann. Bot. 2017, 119, 681–687. [Google Scholar] [CrossRef]
- Collier, S.M.; Moffett, P. NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 2009, 14, 521–529. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010, 13, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Baccelli, I.; Luna, E.; Flors, V. Defense priming: An adaptive part of induced resistance. Annu. Rev. Plant Biol. 2017, 68, 485–512. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.; Naylor, G.; Warner, S.A.; Sugars, J.M.; White, R.F.; Draper, J. Salicylic acid potentiates defence gene expression in tissue exhibiting acquired resistance to pathogen attack. Plant J. 1996, 9, 559–571. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Dong, X. Systemic Acquired Resistance: Turning Local Infection into Global Defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef]
- Scheible, W.-R.; Morcuende, R.; Czechowski, T.; Fritz, C.; Osuna, D.; Palacios-Rojas, N.; Schindelasch, D.; Thimm, O.; Udvardi, M.K.; Stitt, M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004, 136, 2483–2499. [Google Scholar] [CrossRef]
- Makino, A. Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiol. 2011, 155, 125–129. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G.; Hodges, M. Respiration and nitrogen assimilation: Targeting mitochondria-associated metabolism as a means to enhance nitrogen use efficiency. J. Exp. Bot. 2011, 62, 1467–1482. [Google Scholar] [CrossRef]
- Xuan, W.; Beeckman, T.; Xu, G. Plant nitrogen nutrition: Sensing and signaling. Curr. Opin. Plant Biol. 2017, 39, 57–65. [Google Scholar] [CrossRef]
- Mur, L.A.; Simpson, C.; Kumari, A.; Gupta, A.K.; Gupta, K.J. Moving nitrogen to the centre of plant defence against pathogens. Ann. Bot. 2017, 119, 703–709. [Google Scholar] [CrossRef]
- Bolton, M.D.; Thomma, B.P.H.J. The complexity of nitrogen metabolism and nitrogen-regulated gene expression in plant pathogenic fungi. Physiol. Mol. Plant Pathol. 2008, 72, 104–110. [Google Scholar] [CrossRef]
- Huang, H.; Nguyen Thi Thu, T.; He, X.; Gravot, A.; Bernillon, S.; Ballini, E.; Morel, J.B. Increase of Fungal Pathogenicity and Role of Plant Glutamine in Nitrogen-Induced Susceptibility (NIS) To Rice Blast. Front. Plant Sci. 2017, 8, 265. [Google Scholar] [CrossRef] [PubMed]
- Devadas, R.; Simpfendorfer, S.; Backhouse, D.; Lamb, D.W. Effect of stripe rust on the yield response of wheat to nitrogen. Crop J. 2014, 2, 201–206. [Google Scholar] [CrossRef]
- Ballini, E.; Nguyen, T.T.; Morel, J.B. Diversity and genetics of nitrogen-induced susceptibility to the blast fungus in rice and wheat. Rice 2013, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, F.; Tang, L.; Zheng, Y.; Li, Y.; Christie, P.; Li, L. Wheat powdery mildew and foliar N concentrations as influenced by N fertilization and belowground interactions with intercropped faba bean. Plant Soil 2007, 291, 1–13. [Google Scholar] [CrossRef]
- Brennan, R.F. Effect of ammonium chloride, ammonium sulphate, and sodium nitrate on take-all and grain yield of wheat grown on soils in South-western Australia. J. Plant Nutr. 1993, 16, 349–358. [Google Scholar] [CrossRef]
- Krupinsky, J.M.; Halvorson, A.D.; Tanaka, D.L.; Merrill, S.D. Nitrogen and Tillage Effects on Wheat Leaf Spot Diseases in the Northern Great Plains. Agron. J. 2007, 99, 562. [Google Scholar] [CrossRef]
- Brennan, R.F. Effect of superphosphate and nitrogen on yield and take-all of wheat. Fertil. Res. 1992, 31, 43–49. [Google Scholar] [CrossRef]
- Lecompte, F.; Ali Abro, M.; Nicot, P.C. Contrasted responses of Botrytis cinerea isolates developing on tomato plants grown under different nitrogen nutrition regimes. Plant Pathol. 2010, 59, 891–899. [Google Scholar] [CrossRef]
- Snoeijers, S.S.; Pérez-García, A.; Joosten, M.H.; De Wit, P.J. The effect of nitrogen on disease development and gene expression in bacterial and fungal plant pathogens. Eur. J. Plant Pathol. 2000, 106, 493–506. [Google Scholar] [CrossRef]
- Fagard, M.; Launay, A.; Clement, G.; Courtial, J.; Dellagi, A.; Farjad, M.; Krapp, A.; Soulie, M.C.; Masclaux-Daubresse, C. Nitrogen metabolism meets phytopathology. J. Exp. Bot. 2014, 65, 5643–5656. [Google Scholar] [CrossRef] [PubMed]
- Govrin, E.M.; Levine, A. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 2000, 10, 751–757. [Google Scholar] [CrossRef]
- Richard-Molard, C.; Wuillème, S.; Scheel, C.; Gresshoff, P.M.; Morot-Gaudry, J.F.; Limami, A.M. Nitrogen-induced changes in morphological development and bacterial susceptibility of Belgian endive (Cichorium intybus L.) are genotype-dependent. Planta 1999, 209, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Berry, P.M.; Kindred, D.R.; Olesen, J.E.; Jorgensen, L.N.; Paveley, N.D. Quantifying the effect of interactions between disease control, nitrogen supply and land use change on the greenhouse gas emissions associated with wheat production. Plant Pathol. 2010, 59, 753–763. [Google Scholar] [CrossRef]
- Brennan, R.F. The role of manganese and nitrogen nutrition in the susceptibility of wheat plants to take-all in Western Australia. Fertil. Res. 1992, 31, 35–41. [Google Scholar] [CrossRef]
- poae Landschoot, M. Nitrogen form and rate of nitrogen and chloride application for the control of summer patch in Kentucky bluegrass. Plant Dis. 1995, 79, 51. [Google Scholar]
- Elmer, W.H.; LaMondia, J.A. Influence of Ammonium Sulfate and Rotation Crops on Strawberry Black Root Rot. Plant Dis. 1999, 83, 119–123. [Google Scholar] [CrossRef]
- Morgan, K.T.; Timmer, L.W. Effect of inoculum density, nitrogen source and saprophytic fungi on Fusarium wilt of Mexican lime. Plant Soil 1984, 79, 203–210. [Google Scholar] [CrossRef]
- Wang, M.; Sun, Y.; Gu, Z.; Wang, R.; Sun, G.; Zhu, C.; Guo, S.; Shen, Q. Nitrate Protects Cucumber Plants Against Fusarium oxysporum by Regulating Citrate Exudation. Plant Cell Physiol. 2016, 57, 2001–2012. [Google Scholar] [CrossRef]
- Neumann, S.; Paveley, N.D.; Beed, F.D.; Sylvester-Bradley, R. Nitrogen per unit leaf area affects the upper asymptote of Puccinia striiformis f. sp. tritici epidemics in winter wheat. Plant Pathol. 2004, 53, 725–732. [Google Scholar]
- Dordas, C. Role of nutrients in controlling plant diseases in sustainable agriculture. A review. Agron. Sustain. Dev. 2008, 28, 33–46. [Google Scholar] [CrossRef]
- Solomon, P.S.; Oliver, R.P. Evidence that γ-aminobutyric acid is a major nitrogen source during Cladosporium fulvum infection of tomato. Planta 2002, 214, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Bönnighausen, J.; Gebhard, D.; Kröger, C.; Hadeler, B.; Tumforde, T.; Lieberei, R.; Bergemann, J.; Schäfer, W.; Bormann, J. Disruption of the GABA shunt affects mitochondrial respiration and virulence in the cereal pathogen Fusarium graminearum. Mol. Microbiol. 2015, 98, 1115–1132. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, M.; Sun, Y.; Gu, Z.; Wang, R.; Saydin, A.; Shen, Q.; Guo, S. Nitrate Increased Cucumber Tolerance to Fusarium Wilt by Regulating Fungal Toxin Production and Distribution. Toxins 2017, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Divon, H.H.; Ziv, C.; Davydov, O.; Yarden, O.; Fluhr, R. The global nitrogen regulator, FNR1, regulates fungal nutrition-genes and fitness during Fusarium oxysporum pathogenesis. Mol. Plant Pathol. 2006, 7, 485–497. [Google Scholar] [CrossRef]
- Tenuta, M.; Lazarovits, G. Ammonia and nitrous acid from nitrogenous amendments kill the microsclerotia of Verticillium dahliae. Phytopathology 2002, 92, 255–264. [Google Scholar] [CrossRef]
- Wang, L.; Bau, H.; Liao, H.; Chung, K. Factors affecting the production of elsinochrome phytotoxin by the citrus scab pathogen, Elsinoë fawcettii. Open Mycol. J. 2009, 3, 1–8. [Google Scholar] [CrossRef]
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002, 66, 447–459. [Google Scholar] [CrossRef]
- Datnoff, L.E.; Elmer, W.H.; Huber, D.M. Mineral Nutrition and Plant Disease; American Phytopathological Society: St. Paul, AK, USA, 2007. [Google Scholar]
- Pérez-García, A.; Snoeijers, S.S.; Joosten, M.H.; Goosen, T.; De Wit, P.J. Expression of the avirulence gene Avr9 of the fungal tomato pathogen Cladosporium fulvum is regulated by the global nitrogen response factor NRF1. Mol. Plant Microbe Interact. 2001, 14, 316–325. [Google Scholar] [CrossRef]
- Donofrio, N.M.; Oh, Y.; Lundy, R.; Pan, H.; Brown, D.E.; Jeong, J.S.; Coughlan, S.; Mitchell, T.K.; Dean, R.A. Global gene expression during nitrogen starvation in the rice blast fungus, Magnaporthe grisea. Fungal Genet. Biol. 2006, 43, 605–617. [Google Scholar] [CrossRef]
- Thomma, B.P.; HP, V.A.N.E.; Crous, P.W.; PJ, D.E.W. Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol. Plant Pathol. 2005, 6, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Prior, S.A.; Pritchard, S.G.; Runion, G.B.; Rogers, H.H.; Mitchell, R.J. Influence of atmospheric CO2 enrichment, soil N, and water stress on needle surface wax formation in Pinus palustris (Pinaceae). Am. J. Bot. 1997, 84, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Mrnka, L.; Tokárová, H.; Vosátka, M.; Matějka, P. Interaction of soil filamentous fungi affects needle composition and nutrition of Norway spruce seedlings. Trees 2009, 23, 887–897. [Google Scholar] [CrossRef]
- Plavcova, L.; Hacke, U.G.; Almeida-Rodriguez, A.M.; Li, E.; Douglas, C.J. Gene expression patterns underlying changes in xylem structure and function in response to increased nitrogen availability in hybrid poplar. Plant Cell Environ. 2013, 36, 186–199. [Google Scholar] [CrossRef]
- Camargo, E.L.O.; Nascimento, L.C.; Soler, M.; Salazar, M.M.; Lepikson-Neto, J.; Marques, W.L.; Alves, A.; Teixeira, P.J.P.L.; Mieczkowski, P.; Carazzolle, M.F.; et al. Contrasting nitrogen fertilization treatments impact xylem gene expression and secondary cell wall lignification in Eucalyptus. BMC Plant Biol. 2014, 14, 256. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, L.; Ding, Y.; Yao, X.; Wu, X.; Weng, F.; Li, G.; Liu, Z.; Tang, S.; Ding, C.; et al. Nitrogen fertilizer application affects lodging resistance by altering secondary cell wall synthesis in japonica rice (Oryza sativa). J. Plant Res. 2017, 130, 859–871. [Google Scholar] [CrossRef]
- Sun, Q.; Liu, X.; Yang, J.; Liu, W.; Du, Q.; Wang, H.; Fu, C.; Li, W. MicroRNA528 Affects Lodging Resistance of Maize by Regulating Lignin Biosynthesis under Nitrogen-Luxury Conditions. Mol. Plant 2018, 11, 806–814. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, L.; Wu, X.; Ding, Y.; Li, G.; Li, J.; Weng, F.; Liu, Z.; Tang, S.; Ding, C.; et al. Lodging Resistance of Japonica Rice (Oryza Sativa L.): Morphological and Anatomical Traits due to top-Dressing Nitrogen Application Rates. Rice 2016, 9, 31. [Google Scholar] [CrossRef]
- Kováčik, J.; Klejdus, B.; Bačkor, M.; Repčák, M. Phenylalanine ammonia-lyase activity and phenolic compounds accumulation in nitrogen-deficient Matricaria chamomilla leaf rosettes. Plant Sci. 2007, 172, 393–399. [Google Scholar] [CrossRef]
- Jauset, A.M.; Sarasua, M.J.; Avilla, J.; Albajes, R. Effect of nitrogen fertilization level applied to tomato on the greenhouse whitefly. Crop Prot. 2000, 19, 255–261. [Google Scholar] [CrossRef]
- Daane, K.; Johnson, R.; Michailides, T.; Crisosto, C.; Dlott, J.; Ramirez, H.; Morgan, D. Excess nitrogen raises nectarine susceptibility to disease and insects. Calif. Agric. 1995, 49, 13–18. [Google Scholar] [CrossRef]
- Tronchet, M.; Balague, C.; Kroj, T.; Jouanin, L.; Roby, D. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 2010, 11, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Nobuaki, M. The effect of ample nitrogen fertilizer on cell-wall materials and its significance to rice blast disease. Ann. Phytopathol. Soc. Jpn. 1975, 41, 56–61. [Google Scholar]
- Wilson, R.A.; Talbot, N.J. Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 2009, 7, 185. [Google Scholar] [CrossRef]
- Blanke, M.M.; Bacher, W.; Pring, R.J.; Baker, E.A. Ammonium nutrition enhances chlorophyll and glaucousness in kohlrabi. Ann. Bot. 1996, 78, 599–604. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, S.H.; Wang, P.F.; Li, W.; Lu, J. Effects of ammonium on the antioxidative response in Hydrilla verticillata (Lf) Royle plants. Ecotoxicol. Environ. Saf. 2010, 73, 189–195. [Google Scholar] [CrossRef]
- Massad, T.J.; Dyer, L.A.; Vega, C.G. Costs of defense and a test of the carbon-nutrient balance and growth-differentiation balance hypotheses for two co-occurring classes of plant defense. PLoS ONE 2012, 7, e47554. [Google Scholar] [CrossRef]
- Royer, M.; Larbat, R.; Le Bot, J.; Adamowicz, S.; Robin, C. Is the C:N ratio a reliable indicator of C allocation to primary and defence-related metabolisms in tomato? Phytochemistry 2013, 88, 25–33. [Google Scholar] [CrossRef]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing Plant Surfaces: Cuticular Wax Formation by Epidermal Cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef]
- Wang, M.; Gu, Z.; Wang, R.; Guo, J.; Ling, N.; Firbank, L.G.; Guo, S. Plant primary metabolism regulated by nitrogen contributes to plant-pathogen interactions. Plant Cell Physiol. 2018, 60, 329–342. [Google Scholar] [CrossRef]
- Zeier, J. New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ. 2013, 36, 2085–2103. [Google Scholar] [CrossRef] [PubMed]
- Ngadze, E.; Icishahayo, D.; Coutinho, T.A.; van der Waals, J.E. Role of Polyphenol Oxidase, Peroxidase, Phenylalanine Ammonia Lyase, Chlorogenic Acid, and Total Soluble Phenols in Resistance of Potatoes to Soft Rot. Plant Dis. 2012, 96, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Nelissen, I.; Eggermont, K.; Broekaert, W.F. Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 1999, 19, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Gnonlonfin, G.J.B.; Sanni, A.; Brimer, L. Review Scopoletin—A Coumarin Phytoalexin with Medicinal Properties. Crit. Rev. Plant Sci. 2012, 31, 47–56. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Abou-Mansour, E.; Buchala, A.; Mauch, F. Disease resistance of Arabidopsis to Phytophthora brassicae is established by the sequential action of indole glucosinolates and camalexin. Plant J. 2010, 62, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Kaplan, F.; Huffaker, A.; Dafoe, N.J.; Vaughan, M.M.; Ni, X.; Rocca, J.R.; Alborn, H.T.; Teal, P.E. Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc. Natl. Acad. Sci. USA 2011, 108, 5455–5460. [Google Scholar] [CrossRef]
- De Long, J.R.; Sundqvist, M.K.; Gundale, M.J.; Giesler, R.; Wardle, D.A.; Rasmann, S. Effects of elevation and nitrogen and phosphorus fertilization on plant defence compounds in subarctic tundra heath vegetation. Funct. Ecol. 2016, 30, 314–325. [Google Scholar] [CrossRef]
- Dormann, C.F. Consequences of manipulations in carbon and nitrogen supply for concentration of anti-herbivore defence compounds in Salix polaris. Écoscience 2016, 10, 312–318. [Google Scholar] [CrossRef]
- Khanna, A.Q.; Borowicz, V.A.; Jones, M.A. Effects of nitrogen fertilizer and defoliation on growth, foliar nitrogen and foliar coumestrol concentrations of soybean. Trans. Ill. State Acad. Sci. 1999, 92, 167–179. [Google Scholar]
- Tomova, L.; Braun, S.; Flückiger, W. The effect of nitrogen fertilization on fungistatic phenolic compounds in roots of beech (Fagus sylvatica) and Norway spruce (Picea abies). For. Pathol. 2005, 35, 262–276. [Google Scholar] [CrossRef]
- Bhaskar, C.V.; Rao, G.R.; Reddy, K.B. Effect of nitrogen and potassium nutrition on sheath rot incidence and phenol content in rice (Oryza sativa L.). Indian J. Plant Physiol. 2001, 6, 254–257. [Google Scholar]
- Keller, M.; Rogiers, S.Y.; Schultz, H.R. Nitrogen and ultraviolet radiation modify grapevines’ susceptibility to powdery mildew. Vitis-Geilweilerhof 2003, 42, 87–94. [Google Scholar]
- Thalineau, E.; Fournier, C.; Gravot, A.; Wendehenne, D.; Jeandroz, S.; Truong, H.N. Nitrogen modulation of Medicago truncatula resistance to Aphanomyces euteiches depends on plant genotype. Mol. Plant Pathol. 2018, 19, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Mittelstrass, K.; Treutter, D.; Plessl, M.; Heller, W.; Elstner, E.F.; Heiser, I. Modification of primary and secondary metabolism of potato plants by nitrogen application differentially affects resistance to Phytophthora infestans and Alternaria solani. Plant Biol. 2006, 8, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Leser, C.; Treutter, D. Effects of nitrogen supply on growth, contents of phenolic compounds and pathogen (scab) resistance of apple trees. Physiol. Plant. 2005, 123, 49–56. [Google Scholar] [CrossRef]
- Wu, C.H.; Dewir, Y.H.; Hahn, E.J.; Paek, K.Y. Optimization of culturing conditions for the production of biomass and phenolics from adventitious roots of Echinacea angustifolia. J. Plant Biol. 2006, 49, 193. [Google Scholar] [CrossRef]
- Cui, X.; Murthy, H.N.; Wu, C.; Paek, K. Adventitious root suspension cultures of Hypericum perforatum: Effect of nitrogen source on production of biomass and secondary metabolites. In Vitro Cell. Dev. Plant 2010, 46, 437–444. [Google Scholar] [CrossRef]
- Fallovo, C.; Schreiner, M.; Schwarz, D.; Colla, G.; Krumbein, A. Phytochemical changes induced by different nitrogen supply forms and radiation levels in two leafy Brassica species. J. Agric. Food Chem. 2011, 59, 4198–4207. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Zhong, J.J.; Yu, J.T. Effect of nitrogen source on cell growth and production of ginseng saponin and polysaccharide in suspension cultures of Panax notoginseng. Biotechnol. Prog. 1996, 12, 567–571. [Google Scholar] [CrossRef]
- Wang, J.W.; Tan, R.X. Artemisinin production in Artemisia annua hairy root cultures with improved growth by altering the nitrogen source in the medium. Biotechnol. Lett. 2002, 24, 1153–1156. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, W.; Wang, J.; Li, X.; Xiao, P. Improvement of growth and periplocin yield of Periploca sepium adventitious root cultures by altering nitrogen source supply. Chin. Herb. Med. 2011, 3, 226–231. [Google Scholar]
- Jin, X.; Hao, N.; Jiao, F.; Yang, Y.; Wang, D.; Xu, C.; Zhai, R. The effect of nitrogen supply on potato yield, tuber size and pathogen resistance in Solanum tuberosum exposed to Phytophthora infestans. Afr. J. Agric. Res. 2014, 9, 657–2663. [Google Scholar]
- Matros, A.; Amme, S.; Kettig, B.; Buck-Sorlin, G.H.; Sonnewald, U.W.E.; Mock, H.P. Growth at elevated CO2 concentrations leads to modified profiles of secondary metabolites in tobacco cv. SamsunNN and to increased resistance against infection with potato virus Y. Plant Cell Environ. 2006, 29, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Konishi, H.; Ishiguro, K.; Komatsu, S. A proteomics approach towards understanding blast fungus infection of rice grown under different levels of nitrogen fertilization. Proteomics 2001, 1, 1162–1171. [Google Scholar] [CrossRef]
- Thapa, S.; Prasanna, R.; Ramakrishnan, B.; Sheoran, N.; Kumar, A.; Velmourougane, K.; Kumar, A. Interactive effects of Magnaporthe inoculation and nitrogen doses on the plant enzyme machinery and phyllosphere microbiome of resistant and susceptible rice cultivars. Arch. Microbiol. 2018, 200, 1287–1305. [Google Scholar] [CrossRef]
- González-Hernández, A.I.; Fernández-Crespo, E.; Scalschi, L.; Hajirezaei, M.-R.; von Wirén, N.; García-Agustín, P.; Camañes, G. Ammonium mediated changes in carbon and nitrogen metabolisms induce resistance against Pseudomonas syringae in tomato plants. J. Plant Physiol. 2019, 239, 28–37. [Google Scholar] [CrossRef]
- Sarhan, A.R.T.; Barna, B.; Kiraly, Z. Effect of nitrogen nutrition on Fusarium wilt of tomato plants. Ann. Appl. Biol. 1982, 101, 245–250. [Google Scholar] [CrossRef]
- Anand, A.; Zhou, T.; Trick, H.N.; Gill, B.S.; Bockus, W.W.; Muthukrishnan, S. Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin-like protein, chitinase and glucanase against Fusarium graminearum. J. Exp. Bot. 2003, 54, 1101–1111. [Google Scholar] [CrossRef]
- Sarowar, S.; Kim, Y.J.; Kim, E.N.; Kim, K.D.; Hwang, B.K.; Islam, R.; Shin, J.S. Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stresses. Plant Cell Rep. 2005, 24, 216–224. [Google Scholar] [CrossRef]
- Lian, X.; Wang, S.; Zhang, J.; Feng, Q.; Zhang, L.; Fan, D.; Li, X.; Yuan, D.; Han, B.; Zhang, Q. Expression profiles of 10,422 genes at early stage of low nitrogen stress in rice assayed using a cDNA microarray. Plant Mol. Biol. 2006, 60, 617–631. [Google Scholar] [CrossRef]
- Vega, A.; Canessa, P.; Hoppe, G.; Retamal, I.; Moyano, T.C.; Canales, J.; Gutierrez, R.A.; Rubilar, J. Transcriptome analysis reveals regulatory networks underlying differential susceptibility to Botrytis cinerea in response to nitrogen availability in Solanum lycopersicum. Front. Plant Sci. 2015, 6, 911. [Google Scholar] [CrossRef] [PubMed]
- Kruse, C.; Jost, R.; Lipschis, M.; Kopp, B.; Hartmann, M.; Hell, R. Sulfur-enhanced defence: Effects of sulfur metabolism, nitrogen supply, and pathogen lifestyle. Plant Biol. 2007, 9, 608–619. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.L.; Valadares, N.F.; Moraes, D.I.; Rosa, J.C.; Araujo, H.S.; Beltramini, L.M. Physico-chemical and antifungal properties of protease inhibitors from Acacia plumosa. Phytochemistry 2009, 70, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Cipollini, D.F.; Bergelson, J. Plant density and nutrient availability constrain constitutive and wound-induced expression of trypsin inhibitors in Brassica napus. J. Chem. Ecol. 2001, 27, 593–610. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, H.B.; Bennett, J.O.; Kim, W.S.; Krishnan, A.H.; Mawhinney, T.P. Nitrogen lowers the sulfur amino acid content of soybean (Glycine max [L.] Merr.) by regulating the accumulation of Bowman—Birk protease inhibitor. J. Agric. Food Chem. 2005, 53, 6347–6354. [Google Scholar] [CrossRef] [PubMed]
- Stout, M.J.; Brovont, R.A.; Duffey, S.S. Effect of nitrogen availability on expression of constitutive and inducible chemical defenses in tomato, Lycopersicon esculentum. J. Chem. Ecol. 1998, 24, 945–963. [Google Scholar] [CrossRef]
- Solomon, P.S.; Tan, K.C.; Oliver, R.P. The nutrient supply of pathogenic fungi; a fertile field for study. Mol. Plant Pathol. 2003, 4, 203–210. [Google Scholar] [CrossRef]
- Påhlsson, A.M.B. Influence of nitrogen fertilization on minerals, carbohydrates, amino acids and phenolic compounds in beech (Fagus sylvatica L.) leaves. Tree Physiol. 1992, 10, 93–100. [Google Scholar] [CrossRef]
- Seifi, H.; De Vleesschauwer, D.; Aziz, A.; Hofte, M. Modulating plant primary amino acid metabolism as a necrotrophic virulence strategy: The immune-regulatory role of asparagine synthetase in Botrytis cinerea-tomato interaction. Plant Signal. Behav. 2014, 9, e27995. [Google Scholar] [CrossRef]
- Sonderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of glucosinolates--gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef]
- Yan, X.; Chen, S. Regulation of plant glucosinolate metabolism. Planta 2007, 226, 1343–1352. [Google Scholar] [CrossRef]
- Adio, A.M.; Casteel, C.L.; De Vos, M.; Kim, J.H.; Joshi, V.; Li, B.; Juery, C.; Daron, J.; Kliebenstein, D.J.; Jander, G. Biosynthesis and defensive function of Ndelta-acetylornithine, a jasmonate-induced Arabidopsis metabolite. Plant Cell 2011, 23, 3303–3318. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Y.; Wang, C.; Lu, W.; Jin, J.B.; Hua, X. Proline induces calcium-mediated oxidative burst and salicylic acid signaling. Amino Acids 2011, 40, 1473–1484. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.S.; An, S.H.; Hwang, B.K. Pepper asparagine synthetase 1 (CaAS1) is required for plant nitrogen assimilation and defense responses to microbial pathogens. Plant J. 2011, 67, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Brauc, S.; De Vooght, E.; Claeys, M.; Höfte, M.; Angenon, G. Influence of over-expression of cytosolic aspartate aminotransferase on amino acid metabolism and defence responses against Botrytis cinerea infection in Arabidopsis thaliana. J. Plant Physiol. 2011, 168, 1813–1819. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Ji, Y.; Bhuiyan, N.H.; Pilot, G.; Selvaraj, G.; Zou, J.; Wei, Y. Amino acid homeostasis modulates salicylic acid-associated redox status and defense responses in Arabidopsis. Plant Cell 2010, 22, 3845–3863. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Brotman, Y.; Segu, S.; Zeier, T.; Zeier, J.; Persijn, S.T.; Cristescu, S.M.; Harren, F.J.; Bauwe, H.; Fernie, A.R.; et al. The form of nitrogen nutrition affects resistance against Pseudomonas syringae pv. phaseolicola in tobacco. J. Exp. Bot. 2013, 64, 553–568. [Google Scholar] [CrossRef]
- Cowley, T.; Walters, D.R. Polyamine metabolism in barley reacting hypersensitively to the powdery mildew fungus Blumeria graminis f. sp. hordei. Plant Cell Environ. 2002, 25, 461–468. [Google Scholar] [CrossRef]
- Taler, D.; Galperin, M.; Benjamin, I.; Cohen, Y.; Kenigsbuch, D. Plant eR genes that encode photorespiratory enzymes confer resistance against disease. Plant Cell 2004, 16, 172–184. [Google Scholar] [CrossRef]
- Bloom, A.J. Photorespiration and nitrate assimilation: A major intersection between plant carbon and nitrogen. Photosynth. Res. 2015, 123, 117–128. [Google Scholar] [CrossRef]
- Dixon, R.A.; Achnine, L.; Kota, P.; Liu, C.J.; Reddy, M.S.; Wang, L. The phenylpropanoid pathway and plant defence—A genomics perspective. Mol. Plant Pathol. 2002, 3, 371–390. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Bi, Y.; Zhu, T.; Rothstein, S.J. Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol. Biol. 2007, 65, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hou, M.; Mur, L.A.; Yang, Y.; Zhang, T.; Xu, X.; Huang, S.; Tong, H. Nitrogen drives plant growth to the detriment of leaf sugar and steviol glycosides metabolisms in Stevia (Stevia rebaudiana Bertoni). Plant Physiol. Biochem. 2019, 141, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Baldwin, I.T. Nitrogen supply influences herbivore-induced direct and indirect defenses and transcriptional responses in Nicotiana attenuata. Plant Physiol. 2004, 135, 496–506. [Google Scholar] [CrossRef] [PubMed]
- De Gara, L.; de Pinto, M.C.; Tommasi, F. The antioxidant systems vis-à-vis reactive oxygen species during plant–pathogen interaction. Plant Physiol. Biochem. 2003, 41, 863–870. [Google Scholar] [CrossRef]
- Yao, X.; Liu, Q. Changes in photosynthesis and antioxidant defenses of Picea asperata seedlings to enhanced ultraviolet-B and to nitrogen supply. Physiol. Plant. 2006, 129, 364–374. [Google Scholar] [CrossRef]
- Campos, F.G.; Vieira, M.A.R.; Amaro, A.C.E.; delaCruz-Chacon, I.; Marques, M.O.M.; Ferreira, G.; Boaro, C.S.F. Nitrogen in the defense system of Annona emarginata (Schltdl.) H. Rainer. PLoS ONE 2019, 14, e0217930. [Google Scholar] [CrossRef]
- Lin, Y.; Chao, Y.; Huang, W.; Kao, C. Effect of nitrogen deficiency on antioxidant status and Cd toxicity in rice seedlings. Plant Growth Regul. 2011, 64, 263–273. [Google Scholar] [CrossRef]
- Kovacik, J.; Klejdus, B.; Backor, M. Nitric oxide signals ROS scavenger-mediated enhancement of PAL activity in nitrogen-deficient Matricaria chamomilla roots: Side effects of scavengers. Free Radic. Biol. Med. 2009, 46, 1686–1893. [Google Scholar] [CrossRef]
- Kong, L.; Wang, F.; Si, J.; Feng, B.; Zhang, B.; Li, S.; Wang, Z. Increasing in ROS levels and callose deposition in peduncle vascular bundles of wheat (Triticum aestivum L.) grown under nitrogen deficiency. J. Plant Interact. 2013, 8, 109–116. [Google Scholar] [CrossRef]
- Ali, S.; Farooq, M.A.; Jahangir, M.M.; Abbas, F.; Bharwana, S.A.; Zhang, G.P. Effect of chromium and nitrogen form on photosynthesis and anti-oxidative system in barley. Biol. Plant. 2013, 57, 758–763. [Google Scholar] [CrossRef]
- Rios-Gonzalez, K.; Erdei, L.; Lips, S.H. The activity of antioxidant enzymes in maize and sunflower seedlings as affected by salinity and different nitrogen sources. Plant Sci. 2002, 162, 923–930. [Google Scholar] [CrossRef]
- Begara-Morales, J.C.; Sanchez-Calvo, B.; Chaki, M.; Valderrama, R.; Mata-Perez, C.; Padilla, M.N.; Corpas, F.J.; Barroso, J.B. Antioxidant Systems are Regulated by Nitric Oxide-Mediated Post-translational Modifications (NO-PTMs). Front. Plant Sci. 2016, 7, 152. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Valdivia, M.D.; Aparicio-Tejo, P.M.; Lamsfus, C.; Cruz, C.; Martins-Loucao, M.A.; Moran, J.F. Nitrogen nutrition and antioxidant metabolism in ammonium-tolerant and -sensitive plants. Physiol. Plant 2008, 132, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Devadas, S.K.; Enyedi, A.; Raina, R. The Arabidopsis hrl1 mutation reveals novel overlapping roles for salicylic acid, jasmonic acid and ethylene signalling in cell death and defence against pathogens. Plant J. 2002, 30, 467–480. [Google Scholar] [CrossRef]
- Yaeno, T.; Iba, K. BAH1/NLA, a RING-type ubiquitin E3 ligase, regulates the accumulation of salicylic acid and immune responses to Pseudomonas syringae DC3000. Plant Physiol. 2008, 148, 1032–1041. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, S.; Sato, T.; Asada, Y.; Yasuda, S.; Yoshida, M.; Chiba, Y.; Yamaguchi, J. The Arabidopsis ubiquitin ligases ATL31 and ATL6 control the defense response as well as the carbon/nitrogen response. Plant Mol. Biol. 2012, 79, 217–227. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308. [Google Scholar] [CrossRef]
- Scalschi, L.; Sanmartín, M.; Camañes, G.; Troncho, P.; Sánchez-Serrano, J.J.; García-Agustín, P.; Vicedo, B. Silencing of OPR3 in tomato reveals the role of OPDA in callose deposition during the activation of defense responses against Botrytis cinerea. Plant J. 2015, 81, 304–315. [Google Scholar] [CrossRef]
- Farjad, M.; Rigault, M.; Pateyron, S.; Martin-Magniette, M.L.; Krapp, A.; Meyer, C.; Fagard, M. Nitrogen Limitation Alters the Response of Specific Genes to Biotic Stress. Int. J. Mol. Sci. 2018, 19, 3364. [Google Scholar] [CrossRef]
- Berens, M.L.; Berry, H.M.; Mine, A.; Argueso, C.T.; Tsuda, K. Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 2017, 55, 401–425. [Google Scholar] [CrossRef] [PubMed]
- Oka, M.; Shimoda, Y.; Sato, N.; Inoue, J.; Yamazaki, T.; Shimomura, N.; Fujiyama, H. Abscisic acid substantially inhibits senescence of cucumber plants (Cucumis sativus) grown under low nitrogen conditions. J. Plant Physiol. 2012, 169, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Garnica, M.; Houdusse, F.; Zamarreno, A.M.; Garcia-Mina, J.M. The signal effect of nitrate supply enhances active forms of cytokinins and indole acetic content and reduces abscisic acid in wheat plants grown with ammonium. J. Plant Physiol. 2010, 167, 1264–1272. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.; Prats, E.; Pierre, S.; Hall, M.A.; Hebelstrup, K.H. Integrating nitric oxide into salicylic acid and jasmonic acid/ ethylene plant defense pathways. Front. Plant Sci. 2013, 4, 215. [Google Scholar] [CrossRef]
- Mur, L.A.; Santosa, I.E.; Laarhoven, L.J.; Holton, N.J.; Harren, F.J.; Smith, A.R. Laser photoacoustic detection allows in planta detection of nitric oxide in tobacco following challenge with avirulent and virulent Pseudomonas syringae Pathovars. Plant Physiol. 2005, 138, 1247–1258. [Google Scholar] [CrossRef]
- Chen, J.; Vandelle, E.; Bellin, D.; Delledonne, M. Detection and function of nitric oxide during the hypersensitive response in Arabidopsis thaliana: Where there’s a will there’s a way. Nitric Oxide 2014, 43, 81–88. [Google Scholar] [CrossRef]
- Wang, Y.; Loake, G.J.; Chu, C. Cross-talk of nitric oxide and reactive oxygen species in plant programed cell death. Front. Plant Sci. 2013, 4, 314. [Google Scholar] [CrossRef]
- Modolo, L.V. Nitric Oxide Synthase-Mediated Phytoalexin Accumulation in Soybean Cotyledons in Response to the Diaporthe phaseolorum f. sp. meridionalis Elicitor. Plant Physiol. 2002, 130, 1288–1297. [Google Scholar] [CrossRef]
- Modolo, L.V.; Augusto, O.; Almeida, I.M.G.; Magalhaes, J.R.; Salgado, I. Nitrite as the major source of nitric oxide production by Arabidopsis thalianain response to Pseudomonas syringae. FEBS Lett. 2005, 579, 3814–3820. [Google Scholar] [CrossRef]
- Modolo, L.V.; Augusto, O.; Almeida, I.M.G.; Pinto-Maglio, C.A.F.; Oliveira, H.C.; Seligman, K.; Salgado, I. Decreased arginine and nitrite levels in nitrate reductase-deficient Arabidopsis thaliana plants impair nitric oxide synthesis and the hypersensitive response to Pseudomonas syringae. Plant Sci. 2006, 171, 34–40. [Google Scholar] [CrossRef]
- Vitor, S.C.; Duarte, G.T.; Saviani, E.E.; Vincentz, M.G.; Oliveira, H.C.; Salgado, I. Nitrate reductase is required for the transcriptional modulation and bactericidal activity of nitric oxide during the defense response of Arabidopsis thaliana against Pseudomonas syringae. Planta 2013, 238, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Camañes, G.; Pastor, V.; Cerezo, M.; García-Andrade, J.; Vicedo, B.; García-Agustín, P.; Flors, V.A. Deletion in NRT2.1 Attenuates Pseudomonas syringae-Induced Hormonal Perturbation, Resulting in Primed Plant Defenses. Plant Physiol. 2012, 158, 1054–1066. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lu, Y.; Liu, P.; Wen, W.; Zhang, J.; Ge, X.; Xia, Y. The ammonium/nitrate ratio is an input signal in the temperature-modulated, SNC1-mediated and EDS1-dependent autoimmunity of nudt6–2 nudt7. Plant J. 2013, 73, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.; Kumari, A.; Brotman, Y.; Zeier, J.; Mandon, J.; Cristescu, S.M.; Harren, F.; Kaiser, W.M.; Fernie, A.R.; Gupta, K.J. Nitrite and nitric oxide are important in the adjustment of primary metabolism during the hypersensitive response in tobacco. J. Exp. Bot. 2019, 70, 4571–4582. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.; Sivakumaran, A.; Mandon, J.; Cristescu, S.M.; Harren, F.J.; Hebelstrup, K.H. Haemoglobin modulates salicylate and jasmonate/ethylene-mediated resistance mechanisms against pathogens. J. Exp. Bot. 2012, 63, 4375–4387. [Google Scholar] [CrossRef]
- Chun, H.J.; Park, H.C.; Koo, S.C.; Lee, J.H.; Park, C.Y.; Choi, M.S.; Kang, C.H.; Baek, D.; Cheong, Y.H.; Yun, D.J. Constitutive expression of mammalian nitric oxide synthase in tobacco plants triggers disease resistance to pathogens. Mol. Cells 2012, 34, 463–471. [Google Scholar] [CrossRef]
- Lindermayr, C.; Sell, S.; Müller, B.; Leister, D.; Durner, J. Redox Regulation of the NPR1-TGA1 System of Arabidopsis thaliana by Nitric Oxide. Plant Cell 2010, 22, 2894–2907. [Google Scholar] [CrossRef]
- Zimerman-Lax, N.; Shenker, M.; Tamir-Ariel, D.; Perl-Treves, R.; Burdman, S. Effects of nitrogen nutrition on disease development caused by Acidovorax citrulli on melon foliage. Eur. J. Plant Pathol. 2015, 145, 125–137. [Google Scholar] [CrossRef]
- Zhang, F.; Wan, X.; Zheng, Y.; Sun, L.; Chen, Q.; Zhu, X.; Guo, Y.; Liu, M. Effects of nitrogen on the activity of antioxidant enzymes and gene expression in leaves of Populus plants subjected to cadmium stress. J. Plant Interact. 2014, 9, 599–609. [Google Scholar] [CrossRef]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, W.; Liu, L.; Chen, T.; Zhou, F.; Lin, Y. Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol. 2013, 13, 132. [Google Scholar] [CrossRef]
- Seifi, H.S.; Van Bockhaven, J.; Angenon, G.; Hofte, M. Glutamate metabolism in plant disease and defense: Friend or foe? Mol. Plant Microbe Interact. 2013, 26, 475–485. [Google Scholar] [CrossRef]
- Pastor, V.; Gamir, J.; Camañes, G.; Cerezo, M.; Sánchez-Bel, P.; Flors, V. Disruption of the ammonium transporter AMT1.1 alters basal defenses generating resistance against Pseudomonas syringae and Plectosphaerella cucumerina. Front. Plant Sci. 2014, 5, 231. [Google Scholar] [CrossRef] [PubMed]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Mallick, N.; Mitra, A. Salicylic acid-induced resistance to Fusarium oxysporum f. sp. lycopersici in tomato. Plant Physiol. Biochem. 2009, 47, 642–649. [Google Scholar] [CrossRef] [PubMed]
- PiterkovÁ, J.; PetŘIvalskÝ, M.; LuhovÁ, L.; MieslerovÁ, B.; SedlÁŘOvÁ, M.; Lebeda, A. Local and systemic production of nitric oxide in tomato responses to powdery mildew infection. Mol. Plant Pathol. 2009, 10, 501–513. [Google Scholar] [CrossRef]
- Wang, C.; El-Shetehy, M.; Shine, M.B.; Yu, K.; Navarre, D.; Wendehenne, D.; Kachroo, A.; Kachroo, P. Free radicals mediate systemic acquired resistance. Cell Rep. 2014, 7, 348–355. [Google Scholar] [CrossRef]
- Yang, H.; Ludewig, U. Lysine catabolism, amino acid transport, and systemic acquired resistance: What is the link? Plant Signal. Behav. 2014, 9, e28933. [Google Scholar] [CrossRef]
- Bernsdorff, F.; Doring, A.C.; Gruner, K.; Schuck, S.; Brautigam, A.; Zeier, J. Pipecolic Acid Orchestrates Plant Systemic Acquired Resistance and Defense Priming via Salicylic Acid-Dependent and -Independent Pathways. Plant Cell 2016, 28, 102–129. [Google Scholar] [CrossRef]
- Navarova, H.; Bernsdorff, F.; Doring, A.C.; Zeier, J. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 2012, 24, 5123–5141. [Google Scholar] [CrossRef]

| Disease Incidence | Effect of Nitrogen in the Form of | ||
|---|---|---|---|
| Unspecified N | NH4+ | NO3− | |
| Cases | 73 | 19 | 22 |
| Increase in disease | 40 | 9 | 13 |
| Decrease in disease | 25 | 9 | 8 |
| No effect or variable | 8 | 1 | 1 |
| Host | Disease | Pathogen | Defence-Related Enzymes/Compounds | Reference |
|---|---|---|---|---|
| Apple tree | Scab disease | Venturia inaequalis | Procyanidins, Flavonols | [86] |
| Grapevines | Powdery mildew | Uncinula necator | Flavonol glycosides, Cinnamic acid | [83] |
| Medicago truncatula | Root rot | Aphanomyces euteiches | Soluble phenolics, Phenylalanine ammonia lyase | [84] |
| Potato | Early blight | Alternaria solani | Chlorogenic acid, Flavonols, Neochlorogenic acid | [85] |
| Late blight | Phytophthora infestans | Phenylalanine ammonia lyase, Polyphenol oxidase, chitinase, Flavonols | [93] | |
| Leaf necrosis | potato virus Y | Phenylalanine ammonia lyase | [94] | |
| Rice | Rice blast | Magnaporthe grisea | Sulfur-rich thaumatin-like protein | [95] |
| Magnaporthe oryzae | Phenylalanine ammonia lyase, Superoxide dismutase, Glucanases, Chitosanase, Phenylalanine ammonia lyase, | [24,96] | ||
| Sheath rot | Sarocladium oryzae | Phenols | [82] | |
| Tomato | Bacterial speck | Pseudomonas syringae | Superoxide dismutase | [97] |
| Fusarium wilt | Fusarium oxysporum | Phenols, peroxidase | [98] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Wang, M.; Mur, L.A.J.; Shen, Q.; Guo, S. Unravelling the Roles of Nitrogen Nutrition in Plant Disease Defences. Int. J. Mol. Sci. 2020, 21, 572. https://doi.org/10.3390/ijms21020572
Sun Y, Wang M, Mur LAJ, Shen Q, Guo S. Unravelling the Roles of Nitrogen Nutrition in Plant Disease Defences. International Journal of Molecular Sciences. 2020; 21(2):572. https://doi.org/10.3390/ijms21020572
Chicago/Turabian StyleSun, Yuming, Min Wang, Luis Alejandro Jose Mur, Qirong Shen, and Shiwei Guo. 2020. "Unravelling the Roles of Nitrogen Nutrition in Plant Disease Defences" International Journal of Molecular Sciences 21, no. 2: 572. https://doi.org/10.3390/ijms21020572
APA StyleSun, Y., Wang, M., Mur, L. A. J., Shen, Q., & Guo, S. (2020). Unravelling the Roles of Nitrogen Nutrition in Plant Disease Defences. International Journal of Molecular Sciences, 21(2), 572. https://doi.org/10.3390/ijms21020572