Hemmule: A Novel Structure with the Properties of the Stem Cell Niche
Abstract
1. Introduction
2. Results
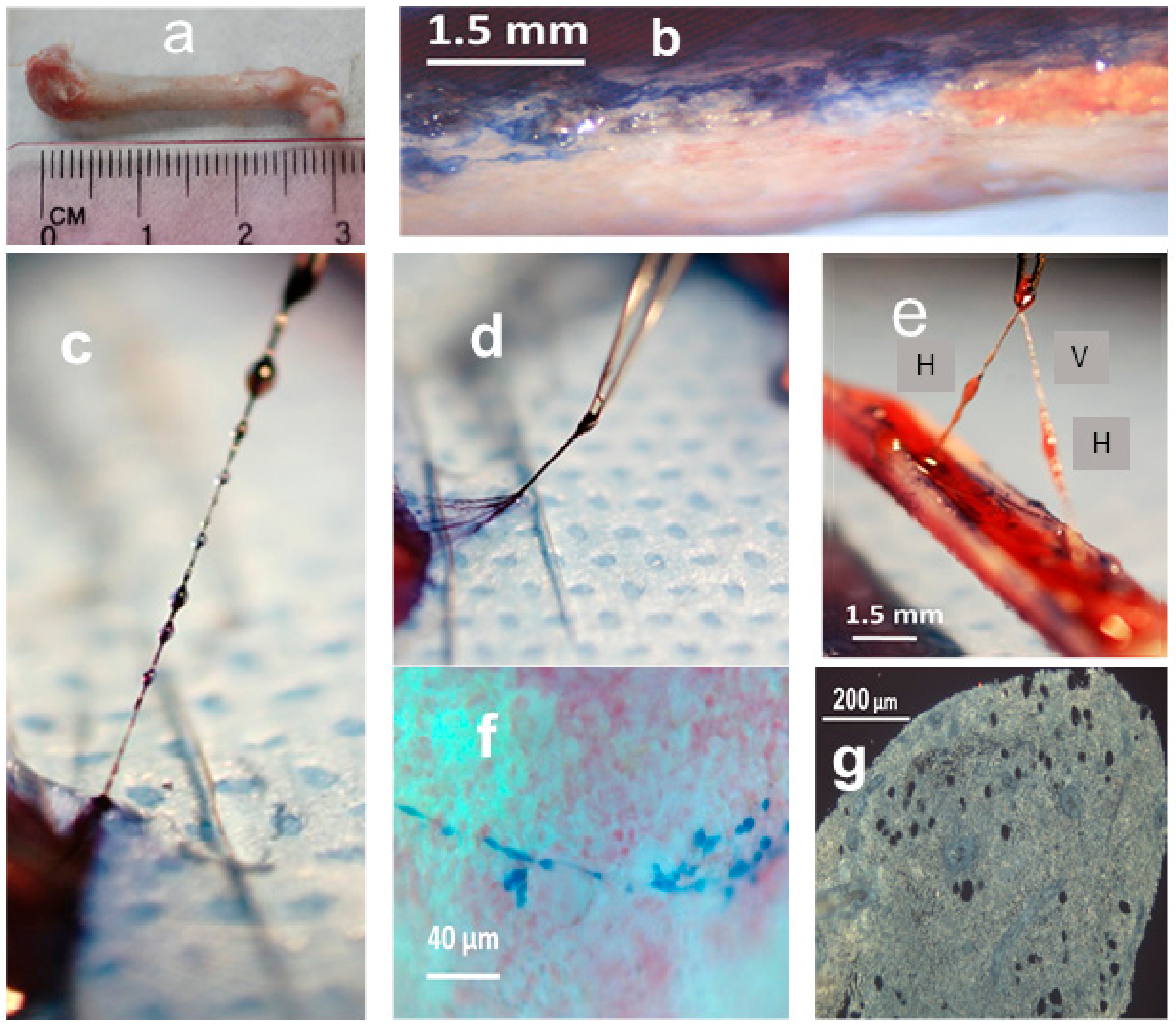
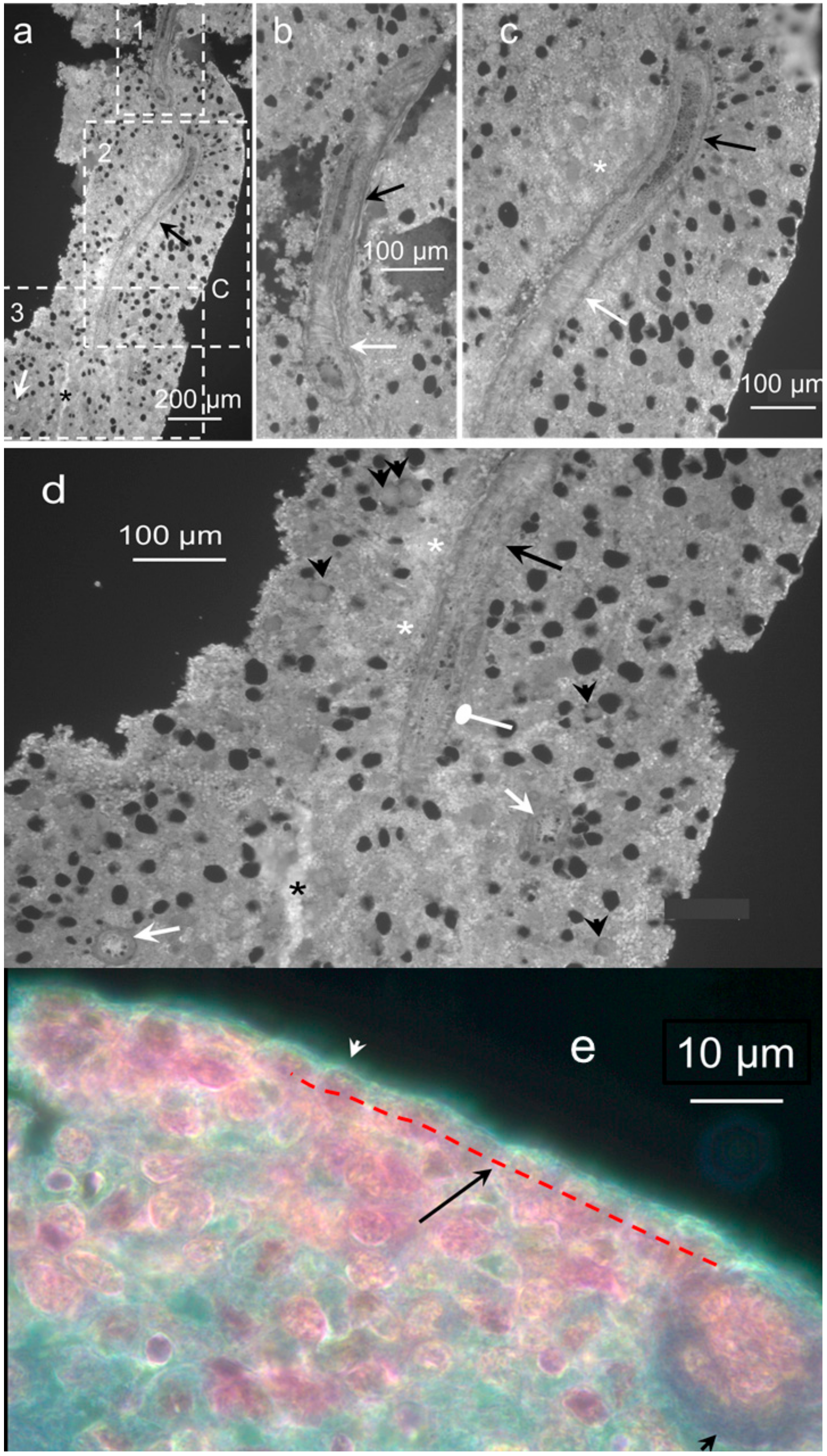
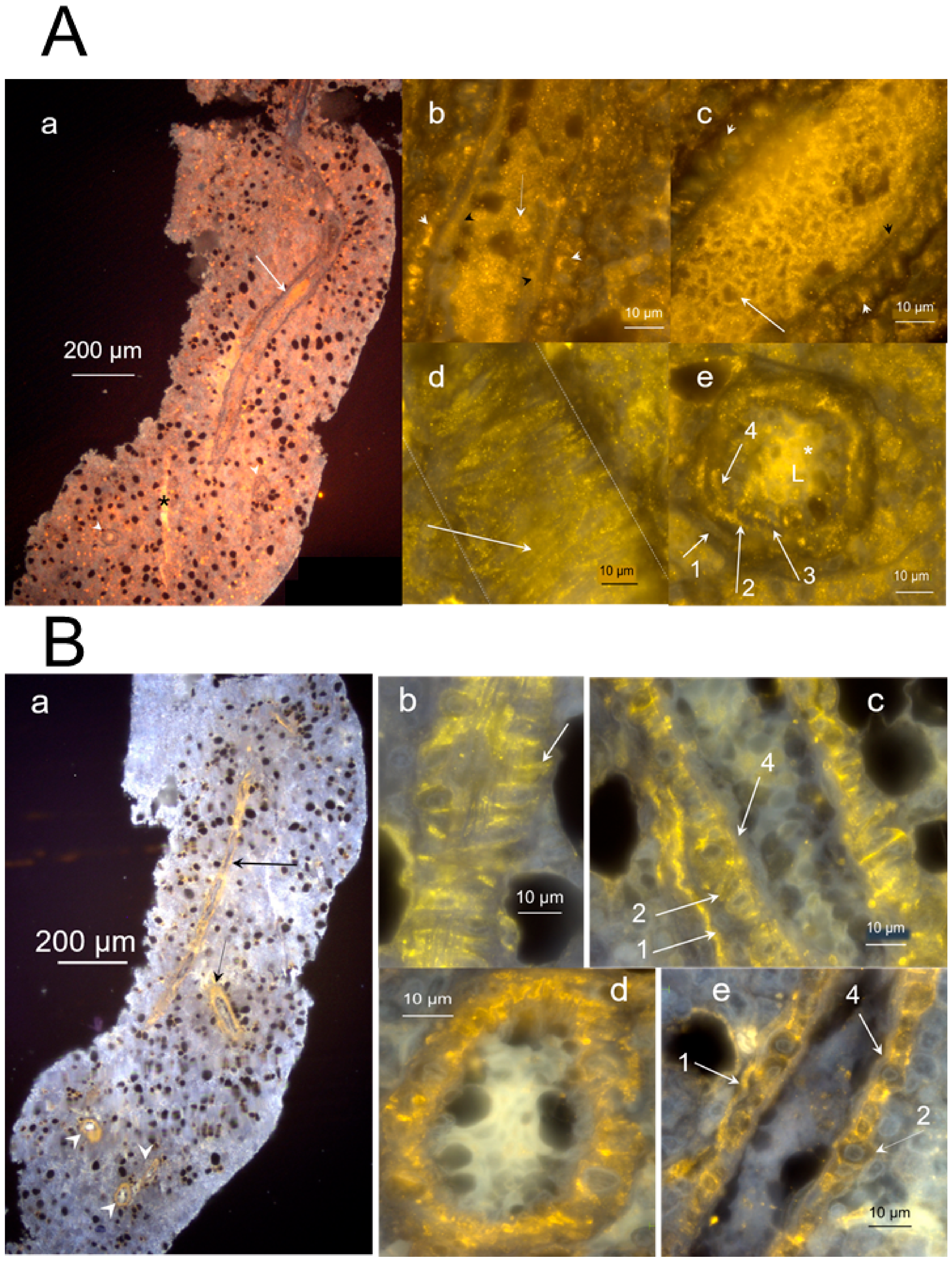
2.1. Morphology and Fine Structure of Hemmule
2.2. Nature of Hemmule Vessels
2.2.1. Comparison of Hemmule Vessels with Blood and Lymphatic Vessels
2.2.2. RNA Expression Patterns in the Hemmule, Bone Marrow, Lymph Node, and Blood Vessel
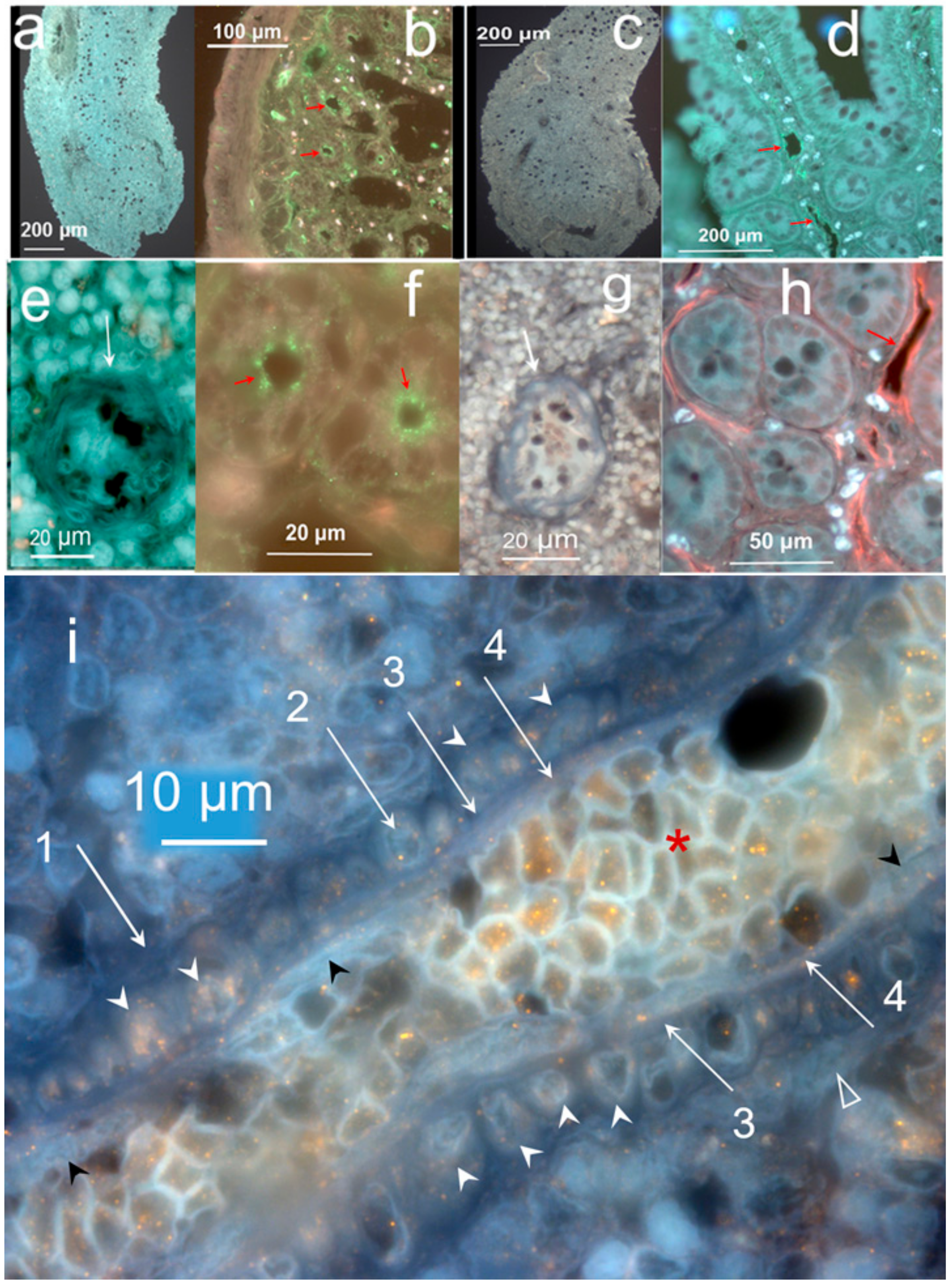
2.3. Cells Found in the Hemmule
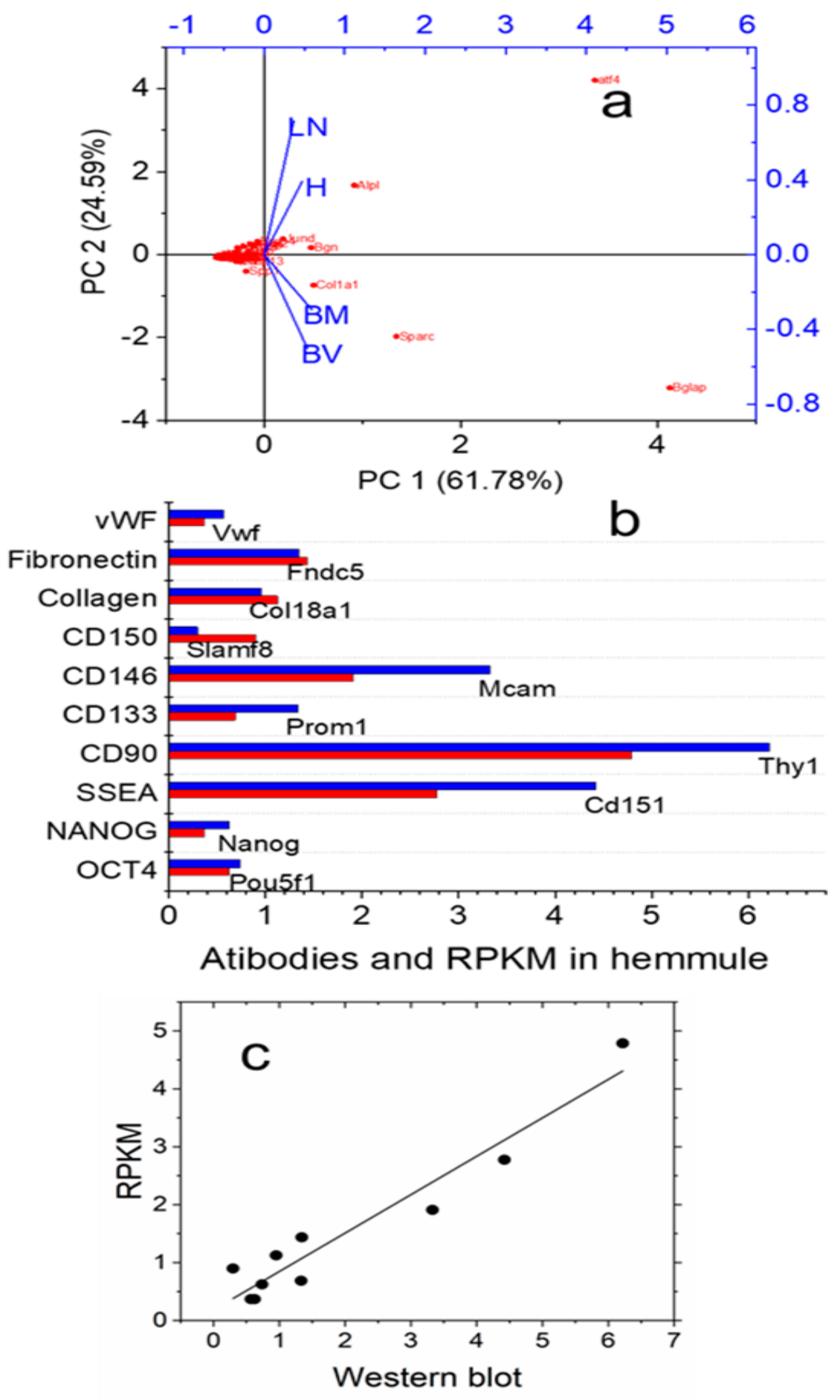
2.3.1. Stem Cells and Gene Presentations in Hemmule by Immunoblotting and RNA-Seq
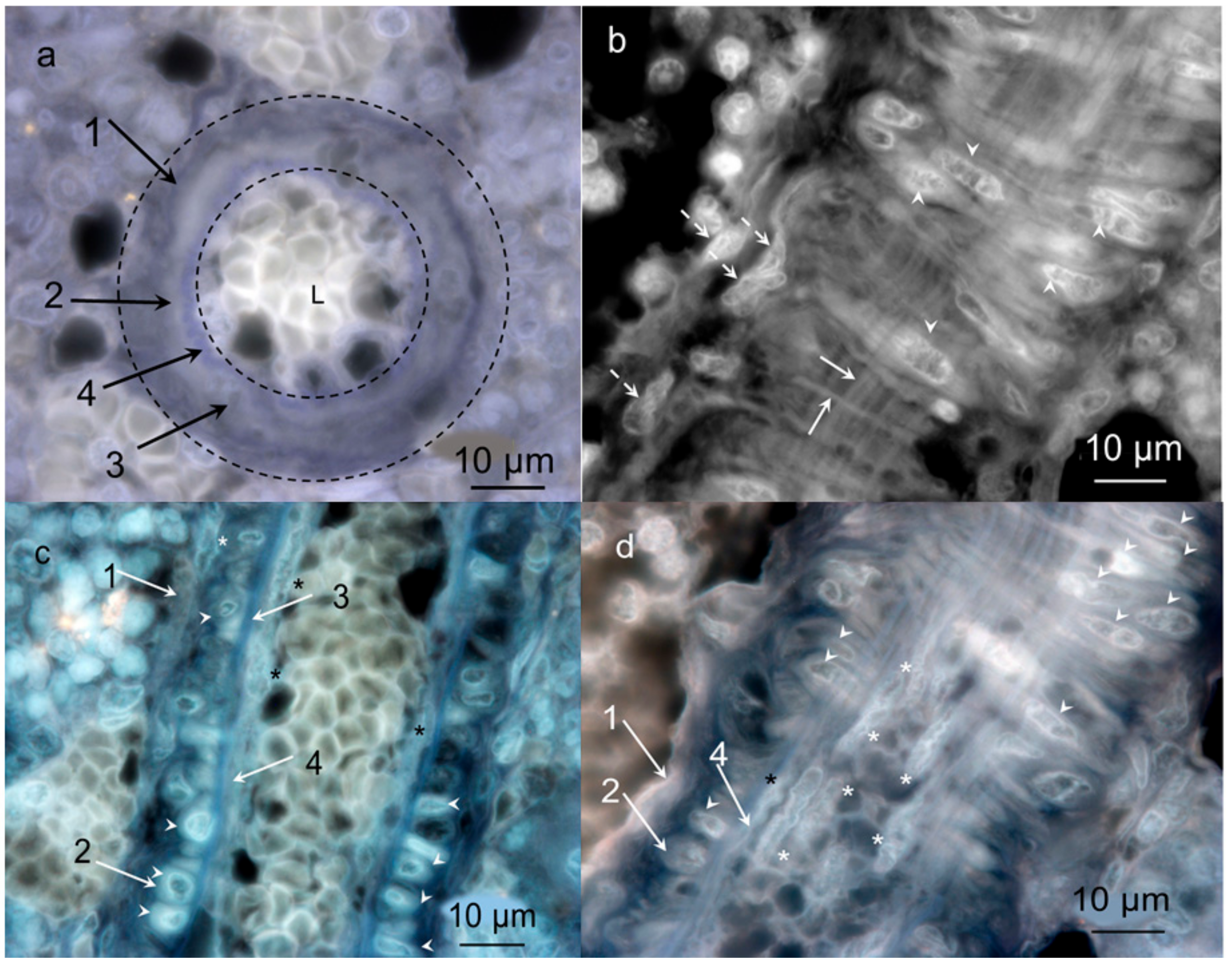
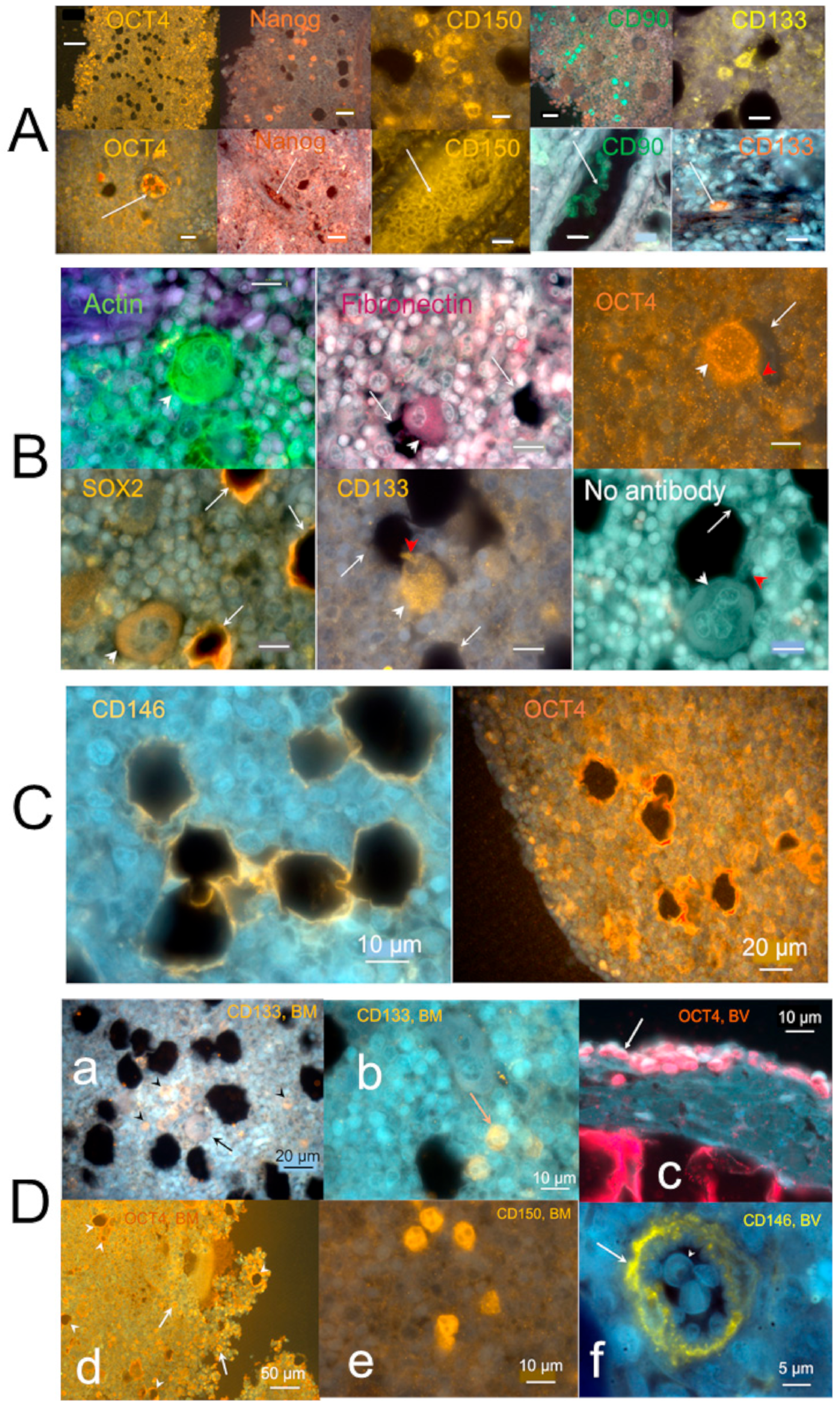
2.3.2. Layered Structure and Cells in Hemmule Vessels Revealed by Immunohistochemistry
2.3.3. Individual Cells Stained by Stem Cell Antibodies in Hemmule
2.3.4. Large Size Cells and Their Footprints
2.3.5. Protein Allocation in Hemmule and Vessels Revealed by Immunohistochemistry and RNA-Seq Analysis
2.3.6. Genes Related to Functional Regulation of Stem Cells in the Hemmule Found by RNA-Seq Analysis
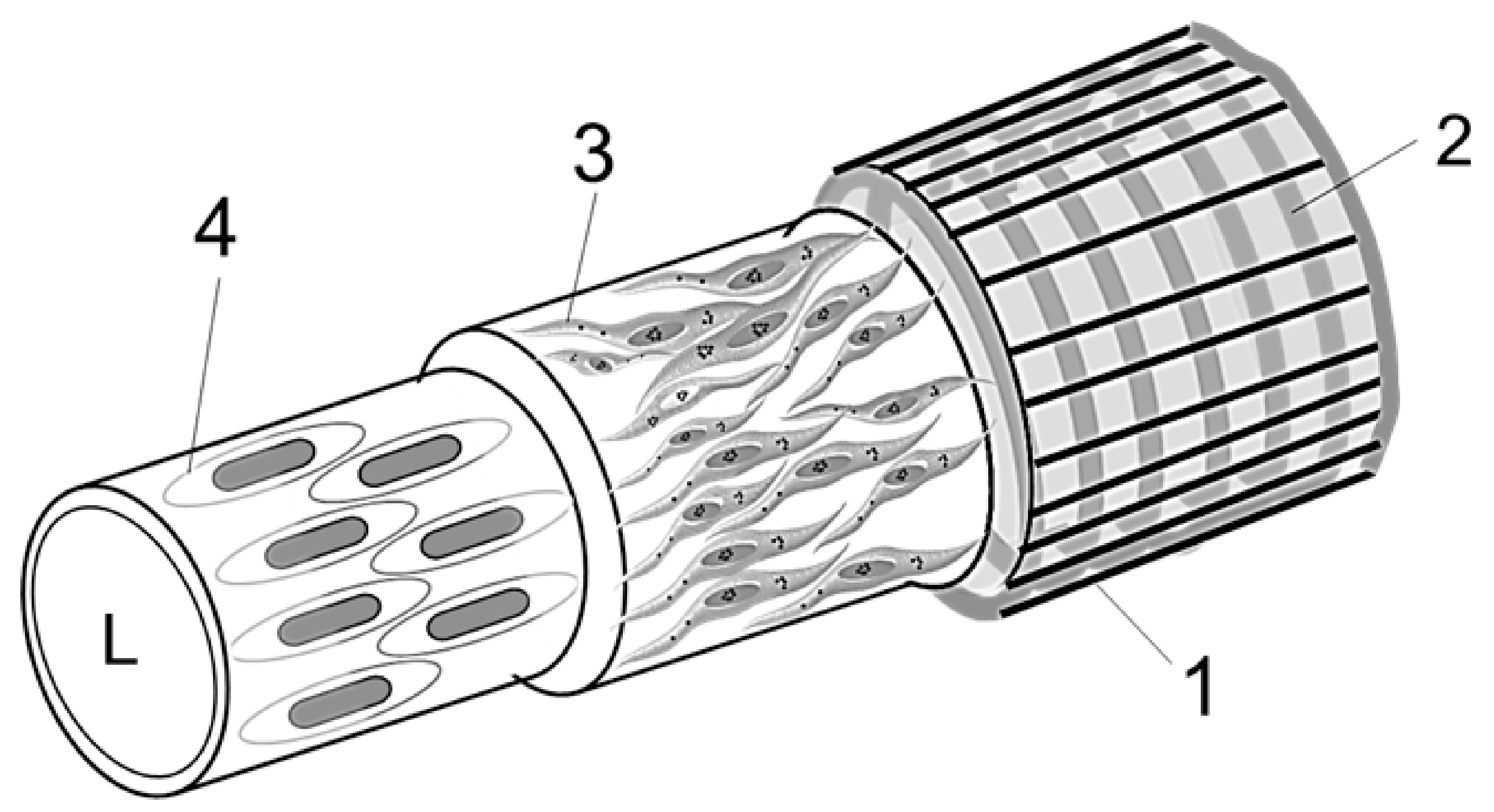
2.3.7. Diagram of a Vessel Inside a Hemmule
3. Discussion
Hemmule as a Stem Cell Niche
4. Materials and Methods
4.1. Animals
4.2. Microdissection and Extraction of Hemmules
4.3. Immunohistochemistry
4.4. Western Blots
4.5. RNA Analysis
4.6. High-Resolution Fluorescent Microscopy
4.7. Statistical Analysis
4.8. Principal Component Analysis Plot
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of variance |
| BM | Bone marrow |
| BV | Blood vessel |
| H | Hemmule |
| LN | Lymph node |
| RPKM | Reads assigned per kilobase per million mapped reads |
References
- Davis, H.; Guo, X.; Lambert, S.; Stancescu, M.; Hickman, J.J. Small Molecule Induction of Human Umbilical Stem Cells into Myelin Basic Protein Positive Oligodendrocytes in a Defined Three-Dimensional Environment. ACS Chem. Neurosci. 2011, 3, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Scadden, D.T. The stem-cell niche as an entity of action. Nature 2006, 441, 1075–1079. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xie, T. Stem cell niche: Structure and function. Annu. Rev. Cell Dev. Biol. 2005, 21, 605–631. [Google Scholar] [CrossRef] [PubMed]
- Adams, G.B.; Scadden, D.T. The hematopoietic stem cell in its place. Nat. Immunol. 2006, 7, 333–337. [Google Scholar] [CrossRef]
- Hooper, A.T.; Butler, J.M.; Nolan, D.J.; Kranz, A.; Iida, K.; Kobayashi, M.; Kopp, H.G.; Shido, K.; Petit, I.; Yanger, K.; et al. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell Stem Cell 2009, 4, 263–274. [Google Scholar] [CrossRef]
- Kunisaki, Y.; Bruns, I.; Scheiermann, C.; Ahmed, J.; Pinho, S.; Zhang, D.; Mizoguchi, T.; Wei, Q.; Lucas, D.; Ito, K.; et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature 2013, 502, 637–643. [Google Scholar] [CrossRef]
- Nombela-Arrieta, C.; Pivarnik, G.; Winkel, B.; Canty, K.J.; Harley, B.; Mahoney, J.E.; Park, S.Y.; Lu, J.; Protopopov, A.; Silberstein, L.E. Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nat. Cell Biol. 2013, 15, 533–543. [Google Scholar] [CrossRef]
- Blazsek, I. The haematon: Discovery of the basic, physiological, structural-proliferative unit involving the stem cell niche in vertebrate haematopoietic tissues. Acta Physiol. Hung. 2010, 97, 93. [Google Scholar]
- Blazsek, I.; Misset, J.L.; Comisso, M.; Mathe, G. Hematon—A multicellular functional unit in primary hematopoiesis. Biomed. Pharmacother. 1988, 42, 661–668. [Google Scholar]
- Blazsek, I.; Chagraoui, J.; Péault, B. Ontogenic emergence of the hematon, a morphogenetic stromal unit that supports multipotential hematopoietic progenitors in mouse bone marrow. Blood 2000, 96, 3763–3771. [Google Scholar] [CrossRef]
- Blazsek, I.; Clay, D.; Leclerc, P.; Blazsek, J.; Candelier, J.-J.; Dobo-Nagy, C.; Khazaal, I.; Péault, B.; Uzan, G.; Le Bousse-Kerdilès, M.-C. Purification and processing of blood-forming tissue units, the haematons, in searching for mammalian stem cell niches. Protoc. Exch. 2013. [Google Scholar] [CrossRef]
- Charbord, P.; Tavian, M.; Humeau, L.; Peault, B. Early ontogeny of the human marrow from long bones: An immunohistochemical study of hematopoiesis and its microenvironment. Blood 1996, 87, 4109–4119. [Google Scholar] [CrossRef] [PubMed]
- Péault, B.; Touraine, J.-L.; Charbord, P. Haematopoietic stem cell emergence and development in the human embryo and fetus; perspectives for blood cell therapies in utero. Semin. Neonatol. 1999, 4, 55–66. [Google Scholar] [CrossRef]
- Tavian, M.; Peault, B. Embryonic development of the human hematopoietic system. Int. J. Dev. Biol. 2005, 49, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Benedito, R.; Bixel, M.G.; Zeuschner, D.; Stehling, M.; Savendahl, L.; Haigh, J.J.; Snippert, H.; Clevers, H.; Breier, G.; et al. Identification of a clonally expanding haematopoietic compartment in bone marrow. EMBO J. 2013, 32, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Ugarte, F.; Forsberg, E.C. Haematopoietic stem cell niches: New insights inspire new questions. EMBO J. 2013, 32, 2535–2547. [Google Scholar] [CrossRef] [PubMed]
- Ahmadbeigi, N.; Soleimani, M.; Vasei, M.; Gheisari, Y.; Mortazavi, Y.; Azadmanesh, K.; Omidkhoda, A.; Janzamin, E.; Nardi, N.B. Isolation, characterization, and transplantation of bone marrow-derived cell components with hematopoietic stem cell niche properties. Stem Cells Dev. 2013, 22, 3052–3061. [Google Scholar] [CrossRef]
- Roche, B.; David, V.; Vanden-Bossche, A.; Peyrin, F.; Malaval, L.; Vico, L.; Lafage-Proust, M.H. Structure and quantification of microvascularisation within mouse long bones: What and how should we measure? Bone 2012, 50, 390–399. [Google Scholar] [CrossRef]
- Cairns, J. Mutation selection and the natural history of cancer. Nature 1975, 255, 197–200. [Google Scholar] [CrossRef]
- Fliedner, T.M.; Graessle, D.; Paulsen, C.; Reimers, K. Structure and function of bone marrow hemopoiesis: Mechanisms of response to ionizing radiation exposure. Cancer Biother. Radiopharm. 2002, 17, 405–426. [Google Scholar] [CrossRef]
- Morrison, S.J.; Scadden, D.T. The bone marrow niche for haematopoietic stem cells. Nature 2014, 505, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Schofield, R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 1978, 4, 7–25. [Google Scholar] [PubMed]
- Banerji, S.; Ni, J.; Wang, S.-X.; Clasper, S.; Su, J.; Tammi, R.; Jones, M.; Jackson, D.G. LYVE-1, a New Homologue of the CD44 Glycoprotein, Is a Lymph-specific Receptor for Hyaluronan. J. Cell Biol. 1999, 144, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Liu, H.; He, Y.; Liu, Y.; Yang, C.; Zhou, M.; Wang, W.; Cui, L.; Hu, J.; Gao, F. The Interaction between LYVE-1 with Hyaluronan on the Cell Surface May Play a Role in the Diversity of Adhesion to Cancer Cells. PLoS ONE 2013, 8, e63463. [Google Scholar] [CrossRef]
- Zhang, H.; Tse, J.; Hu, X.; Witte, M.; Bernas, M.; Kang, J.; Tilahun, F.; Hong, Y.-K.; Qiu, M.; Chen, L. Novel discovery of LYVE-1 expression in the hyaloid vascular system. Investig. Ophthalmol. Vis. Sci. 2010, 51, 6157–6161. [Google Scholar] [CrossRef]
- Calvi, L.M.; Link, D.C. Cellular complexity of the bone marrow hematopoietic stem cell niche. Calcif. Tissue Int. 2014, 94, 112–124. [Google Scholar] [CrossRef]
- Trost, A.; Bruckner, D.; Kaser-Eichberger, A.; Motloch, K.; Bogner, B.; Runge, C.; Strohmaier, C.; Couillard-Despres, S.; Reitsamer, H.A.; Schroedl, F. Lymphatic and vascular markers in an optic nerve crush model in rat. Exp. Eye Res. 2017, 159, 30–39. [Google Scholar] [CrossRef]
- Lv, F.-J.; Tuan, R.S.; Cheung, K.M.C.; Leung, V.Y.L. Concise Review: The Surface Markers and Identity of Human Mesenchymal Stem Cells. Stem Cells 2014, 32, 1408–1419. [Google Scholar] [CrossRef]
- Oguro, H.; Ding, L.; Morrison, S.J. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell 2013, 13, 102–116. [Google Scholar] [CrossRef]
- Riekstina, U.; Cakstina, I.; Parfejevs, V.; Hoogduijn, M.; Jankovskis, G.; Muiznieks, I.; Muceniece, R.; Ancans, J. Embryonic stem cell marker expression pattern in human mesenchymal stem cells derived from bone marrow, adipose tissue, heart and dermis. Stem Cell Rev. 2009, 5, 378–386. [Google Scholar] [CrossRef]
- Maier, T.; Güell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef] [PubMed]
- Payne, S.H. The utility of protein and mRNA correlation. Trends Biochem. Sci. 2015, 40, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, T.; Kobayashi, Y.; Nakajima, H.; Yamagishi, Y. Generation of robust vascular networks from cardiovascular blast populations derived from human induced pluripotent stem cells in vivo and ex vivo organ culture system. Biochem. Biophys. Res. Commun. 2013, 441, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Powell, D.W.; Pinchuk, I.V.; Saada, J.I.; Chen, X.; Mifflin, R.C. Mesenchymal cells of the intestinal lamina propria. Annu. Rev. Physiol. 2011, 73, 213–237. [Google Scholar] [CrossRef]
- Dominguez, R.; Holmes, K.C. Actin structure and function. Annu. Rev. Biophys. 2011, 40, 169–186. [Google Scholar] [CrossRef]
- Owens, G.K. Regulation of differentiation of vascular smooth muscle cells. Physiol. Rev. 1995, 75, 487–517. [Google Scholar] [CrossRef]
- Jones, P.A.; Scott-Burden, T.; Gevers, W. Glycoprotein, elastin, and collagen secretion by rat smooth muscle cells. Proc. Natl. Acad. Sci. USA 1979, 76, 353–357. [Google Scholar] [CrossRef]
- Kiel, M.J.; Yilmaz, Ö.H.; Iwashita, T.; Yilmaz, O.H.; Terhorst, C.; Morrison, S.J. SLAM Family Receptors Distinguish Hematopoietic Stem and Progenitor Cells and Reveal Endothelial Niches for Stem Cells. Cell 2005, 121, 1109–1121. [Google Scholar] [CrossRef]
- Matic, I.; Antunovic, M.; Brkic, S.; Josipovic, P.; Mihalic, K.C.; Karlak, I.; Ivkovic, A.; Marijanovic, I. Expression of OCT-4 and SOX-2 in Bone Marrow-Derived Human Mesenchymal Stem Cells during Osteogenic Differentiation. Open Access Maced. J. Med Sci. 2016, 4, 9–16. [Google Scholar] [CrossRef]
- Seo, E.; Basu-Roy, U.; Zavadil, J.; Basilico, C.; Mansukhani, A. Distinct functions of Sox2 control self-renewal and differentiation in the osteoblast lineage. Mol. Cell. Biol. 2011, 31, 4593–4608. [Google Scholar] [CrossRef]
- Basu-Roy, U.; Ambrosetti, D.; Favaro, R.; Nicolis, S.K.; Mansukhani, A.; Basilico, C. The transcription factor Sox2 is required for osteoblast self-renewal. Cell Death Differ. 2010, 17, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Freeman, B.T.; Jung, J.P.; Ogle, B.M. Single-Cell RNA-Seq of Bone Marrow-Derived Mesenchymal Stem Cells Reveals Unique Profiles of Lineage Priming. PLoS ONE 2015, 10, e0136199. [Google Scholar] [CrossRef] [PubMed]
- Blair, H.C.; Larrouture, Q.C.; Li, Y.; Lin, H.; Beer-Stoltz, D.; Liu, L.; Tuan, R.S.; Robinson, L.J.; Schlesinger, P.H.; Nelson, D.J. Osteoblast differentiation and bone matrix formation in vivo and in vitro. Tissue Eng. 2017, 23, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Harkness, L.; Zaher, W.; Ditzel, N.; Isa, A.; Kassem, M. CD146/MCAM defines functionality of human bone marrow stromal stem cell populations. Stem Cell Res. Ther. 2016, 7, 4. [Google Scholar] [CrossRef]
- Tripodo, C.; Di Bernardo, A.; Ternullo, M.P.; Guarnotta, C.; Porcasi, R.; Ingrao, S.; Gianelli, U.; Boveri, E.; Iannitto, E.; Franco, G.; et al. CD146(+) bone marrow osteoprogenitors increase in the advanced stages of primary myelofibrosis. Haematologica 2009, 94, 127–130. [Google Scholar] [CrossRef][Green Version]
- Calvi, L.M.; Adams, G.B.; Weibrecht, K.W.; Weber, J.M.; Olson, D.P.; Knight, M.C.; Martin, R.P.; Schipani, E.; Divieti, P.; Bringhurst, F.R.; et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 2003, 425, 841–846. [Google Scholar] [CrossRef]
- Gulati, G. Blood Cells: Morphology and Clinical Relevance, 2nd ed.; American Society of Clinical Oncology: Chicago, IL, USA, 2014. [Google Scholar]
- Boque, C.; Pujolmoix, N.; Linde, M.A.; Murcia, C.; Guanyabens, C.; Soler, J. Use of monoclonal anti-actin as a megakaryocyte marker in paraffin wax embedded bone-marrow biopsy specimens. J. Clin. Pathol. 1989, 42, 982–984. [Google Scholar] [CrossRef]
- Wang, J.; Chen, S.; Zhang, C.; Stegeman, S.; Pfaff-Amesse, T.; Zhang, Y.; Zhang, W.; Amesse, L.; Chen, Y. Human Endometrial Stromal Stem Cells Differentiate into Megakaryocytes with the Ability to Produce Functional Platelets. PLoS ONE 2012, 7, e44300. [Google Scholar] [CrossRef]
- Zarif, M.N.; Soleimani, M.; Abolghasemi, H.; Amirizade, N.; Arefian, E.; Rahimian, A. Megakaryocytic differentiation of CD133+ hematopoietic stem cells by down-regulation of microRNA-10a. Hematology 2013, 18, 93–100. [Google Scholar] [CrossRef]
- Malara, A.; Currao, M.; Gruppi, C.; Celesti, G.; Viarengo, G.; Buracchi, C.; Laghi, L.; Kaplan, D.L.; Balduini, A. Megakaryocytes Contribute to the Bone Marrow-Matrix Environment by Expressing Fibronectin, Type IV Collagen, and Laminin. Stem Cells 2014, 32, 926–937. [Google Scholar] [CrossRef]
- Bessis, M. Cytology of the Blood and Blood-Forming Organs; Grune & Stratton: New York, NY, USA, 1956; p. 629. [Google Scholar]
- Hartenstein, V. Blood cells and blood cell development in the animal kingdom. Annu. Rev. Cell Dev. Biol. 2006, 22, 677–712. [Google Scholar] [CrossRef] [PubMed]
- Maximow, A. A Text-Book of Histology; Bloom, W., Ed.; W.B. Sounders Company: London, UK, 1931; p. 833. [Google Scholar]
- Mendez-Ferrer, S.; Michurina, T.V.; Ferraro, F.; Mazloom, A.R.; MacArthur, B.D.; Lira, S.A.; Scadden, D.T.; Ma’ayan, A.; Enikolopov, G.N.; Frenette, P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 2010, 466, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Mierzejewska, K.; Heo, J.; Kang, J.W.; Kang, H.; Ratajczak, J.; Ratajczak, M.Z.; Kucia, M.; Shin, D.M. Genome-wide analysis of murine bone marrow-derived very small embryonic-like stem cells reveals that mitogenic growth factor signaling pathways play a crucial role in the quiescence and ageing of these cells. Int. J. Mol. Med. 2013, 32, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, M.Z.; Zuba-Surma, E.; Wojakowski, W.; Suszynska, M.; Mierzejewska, K.; Liu, R.; Ratajczak, J.; Shin, D.M.; Kucia, M. Very small embryonic-like stem cells (VSELs) represent a real challenge in stem cell biology: Recent pros and cons in the midst of a lively debate. Leukemia 2013, 28, 473. [Google Scholar] [CrossRef]
- Boltin, D.; Kamenetsky, Z.; Perets, T.T.; Snir, Y.; Sapoznikov, B.; Schmilovitz-Weiss, H.; Ablin, J.N.; Dickman, R.; Niv, Y. Circulating Bone Marrow-Derived CD45-/CD34+/CD133+/VEGF+ Endothelial Progenitor Cells in Adults with Crohn’s Disease. Dig. Dis. Sci. 2017, 62, 633–638. [Google Scholar] [CrossRef]
- Quirici, N.; Soligo, D.; Caneva, L.; Servida, F.; Bossolasco, P.; Deliliers, G.L. Differentiation and expansion of endothelial cells from human bone marrow CD133(+) cells. Br. J. Haematol. 2001, 115, 186–194. [Google Scholar] [CrossRef]
- Yang, J.; Ii, M.; Kamei, N.; Alev, C.; Kwon, S.-M.; Kawamoto, A.; Akimaru, H.; Masuda, H.; Sawa, Y.; Asahara, T. CD34+ Cells Represent Highly Functional Endothelial Progenitor Cells in Murine Bone Marrow. PLoS ONE 2011, 6, e20219. [Google Scholar] [CrossRef]
- Singh, P.; Schwarzbauer, J.E. Fibronectin and stem cell differentiation—Lessons from chondrogenesis. J. Cell Sci. 2012, 125, 3703–3712. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Galmiche, M.C.; Koteliansky, V.E.; Briere, J.; Herve, P.; Charbord, P. Stromal cells from human long-term marrow cultures are mesenchymal cells that differentiate following a vascular smooth muscle differentiation pathway. Blood 1993, 82, 66–76. [Google Scholar] [CrossRef]
- Tanokashira, D.; Morita, T.; Hayashi, K.; Mayanagi, T.; Fukumoto, K.; Kubota, Y.; Yamashita, T.; Sobue, K. Glucocorticoid suppresses dendritic spine development mediated by down-regulation of caldesmon expression. J. Neurosci. 2012, 32, 14583–14591. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; He, Y.; Yang, J.; Fu, W.; Xu, X.; Si, Y. MYBPH inhibits vascular smooth muscle cell migration and attenuates neointimal hyperplasia in a rat carotid balloon-injury model. Exp. Cell Res. 2017, 359, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.F.; Bhatti, S.; Pyne-Geithman, G.J.; Farjah, M.; Manaves, V.; Walker, L.; Franks, R.; Strauch, A.R.; Paul, R.J. Expression and function of COOH-terminal myosin heavy chain isoforms in mouse smooth muscle. Am. J. Physiol. Cell Physiol. 2007, 293, C238–C245. [Google Scholar] [CrossRef] [PubMed]
- Azuma, K.; Ichimura, K.; Mita, T.; Nakayama, S.; Jin, W.L.; Hirose, T.; Fujitani, Y.; Sumiyoshi, K.; Shimada, K.; Daida, H.; et al. Presence of alpha-smooth muscle actin-positive endothelial cells in the luminal surface of adult aorta. Biochem. Biophys. Res. Commun. 2009, 380, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Fuscoe, J.C.; Zhao, C.; Guo, C.; Jia, M.; Qing, T.; Bannon, D.I.; Lancashire, L.; Bao, W.; Du, T.; et al. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat. Commun. 2014, 5, 3230. [Google Scholar] [CrossRef]
- Khaldoyanidi, S.; Moll, J.; Karakhanova, S.; Herrlich, P.; Ponta, H. Hyaluronate-Enhanced Hematopoiesis: Two Different Receptors Trigger the Release of Interleukin-1β and Interleukin-6 From Bone Marrow Macrophages. Blood 1999, 94, 940–949. [Google Scholar] [CrossRef]
- Poulos, M.G.; Guo, P.; Kofler, N.M.; Pinho, S.; Gutkin, M.C.; Tikhonova, A.; Aifantis, I.; Frenette, P.S.; Kitajewski, J.; Rafii, S.; et al. Endothelial Jagged-1 is necessary for homeostatic and regenerative hematopoiesis. Cell Rep. 2013, 4, 1022–1034. [Google Scholar] [CrossRef]
- Arai, F.; Hirao, A.; Ohmura, M.; Sato, H.; Matsuoka, S.; Takubo, K.; Ito, K.; Koh, G.Y.; Suda, T. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 2004, 118, 149–161. [Google Scholar] [CrossRef]
- Murphy, M.J.; Wilson, A.; Trumpp, A. More than just proliferation: Myc function in stem cells. Trends Cell Biol. 2005, 15, 128–137. [Google Scholar] [CrossRef]
- Suda, T.; Arai, F.; Hirao, A. Hematopoietic stem cells and their niche. Trends Immunol. 2005, 26, 426–433. [Google Scholar] [CrossRef]
- Wilson, A.; Murphy, M.J.; Oskarsson, T.; Kaloulis, K.; Bettess, M.D.; Oser, G.M.; Pasche, A.C.; Knabenhans, C.; Macdonald, H.R.; Trumpp, A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004, 18, 2747–2763. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Trumpp, A. Bone-marrow haematopoietic-stem-cell niches. Nat. Rev. Immunol. 2006, 6, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Kohara, H.; Noda, M.; Nagasawa, T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 2006, 25, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Saunders, T.L.; Enikolopov, G.; Morrison, S.J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 2012, 481, 457–462. [Google Scholar] [CrossRef]
- Yamazaki, S.; Ema, H.; Karlsson, G.; Yamaguchi, T.; Miyoshi, H.; Shioda, S.; Taketo, M.M.; Karlsson, S.; Iwama, A.; Nakauchi, H. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell 2011, 147, 1146–1158. [Google Scholar] [CrossRef]
- Bruns, I.; Lucas, D.; Pinho, S.; Ahmed, J.; Lambert, M.P.; Kunisaki, Y.; Scheiermann, C.; Schiff, L.; Poncz, M.; Bergman, A.; et al. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat. Med. 2014, 20, 1315. [Google Scholar] [CrossRef]
- Gasser, T.C.; Ogden, R.W.; Holzapfel, G.A. Hyperelastic modelling of arterial layers with distributed collagen fibre orientations. J. R. Soc. Interface 2006, 3, 15–35. [Google Scholar] [CrossRef]
- Erbel, R.; Eggebrecht, H. Aortic dimensions and the risk of dissection. Heart 2006, 92, 137–142. [Google Scholar] [CrossRef]
- Carlson, E.C.; Burrows, M.E.; Johnson, P.C. Electron microscopic studies of cat mesenteric arterioles: A structure-function analysis. Microvasc. Res. 1982, 24, 123–141. [Google Scholar] [CrossRef]
- Sacchetti, B.; Funari, A.; Michienzi, S.; Di Cesare, S.; Piersanti, S.; Saggio, I.; Tagliafico, E.; Ferrari, S.; Robey, P.G.; Riminucci, M.; et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell 2007, 131, 324–336. [Google Scholar] [CrossRef]
- Pietras, E.M.; Warr, M.R.; Passegué, E. Cell cycle regulation in hematopoietic stem cells. J. Cell Biol. 2011, 195, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Perry, J.M.; Marshall, H.; Venkatraman, A.; Qian, P.; He, X.C.; Ahamed, J.; Li, L. Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat. Med. 2014, 20, 1321–1326. [Google Scholar] [CrossRef] [PubMed]
- Comazzetto, S.; Murphy, M.M.; Berto, S.; Jeffery, E.; Zhao, Z.; Morrison, S.J. Restricted Hematopoietic Progenitors and Erythropoiesis Require SCF from Leptin Receptor+ Niche Cells in the Bone Marrow. Cell Stem Cell 2019, 24, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro Gomes, A.; Hara, T.; Lim, V.Y.; Herndler-Brandstetter, D.; Nevius, E.; Sugiyama, T.; Tani-Ichi, S.; Schlenner, S.; Richie, E.; Rodewald, H.R.; et al. Hematopoietic Stem Cell Niches Produce Lineage-Instructive Signals to Control Multipotent Progenitor Differentiation. Immunity 2016, 45, 1219–1231. [Google Scholar] [CrossRef]
- Balzano, M.; De Grandis, M.; Vu Manh, T.P.; Chasson, L.; Bardin, F.; Farina, A.; Serge, A.; Bidaut, G.; Charbord, P.; Herault, L.; et al. Nidogen-1 Contributes to the Interaction Network Involved in Pro-B Cell Retention in the Peri-sinusoidal Hematopoietic Stem Cell Niche. Cell Rep. 2019, 26, 3257–3271. [Google Scholar] [CrossRef]
- Kurth, I.; Franke, K.; Pompe, T.; Bornhäuser, M.; Werner, C. Extracellular Matrix Functionalized Microcavities to Control Hematopoietic Stem and Progenitor Cell Fate. Macromol. Biosci. 2011, 11, 739–747. [Google Scholar] [CrossRef]
- Vainrub, A.; Pustovyy, O.; Vodyanoy, V. Resolution of 90 nm (lambda/5) in an optical transmission microscope with an annular condenser. Opt. Lett. 2006, 31, 2855–2857. [Google Scholar] [CrossRef]
- Vodyanoy, V.J.; Pustovyy, O.M. Simultaneous Observation of Darkfield Images and Fluorescence Using Filter and Diaphragm. U.S. Patent 7,688,505, 30 March 2010. [Google Scholar]
| Antibody | Layer 1 1 | Layer 2 2 | Layer 3 3 | Layer 4 4 | Inside 5 | Large Cells 6 | Footprints 7 | Scattered 8 |
|---|---|---|---|---|---|---|---|---|
| α-Actin | − | + | − | − | − | − | − | − |
| Actin | − | + | + | + | + | + | − | + |
| CD146 | − | + | − | +− | + | −+ | −+ | − |
| CD90 | − | −+ | − | − | + | − | − | + |
| CD133 | − | − | − | − | + | + | − | + |
| CD150 | − | + | − | +− | + | − | − | + |
| Collagen-1 | + | − | − | +− | + | − | − | − |
| Fibronectin | + | + | − | − | − | + | − | − |
| LYVE-1 | − | − | − | − | + | − | − | − |
| Nanog | + | + | − | − | + | − | − | + |
| OCT4 | − | − | − | − | + | + | + | + |
| RECA-1 | − | − | − | − | − | − | − | − |
| REXO1 | − | − | − | − | − | − | − | + |
| SOX2 | −+ | + | − | +− | − | +− | + | + |
| SSEA-1 | − | − | − | − | − | − | − | + |
| vWF | − | + | − | − | + | − | − | + |
| Gene | H | BM | Protein | Ref. |
|---|---|---|---|---|
| RPKM | RPKM | |||
| Cald1 | 52.00 | 54.23 | Caldesmon 1 | [64] |
| Mybph | 0.902 | 0.629 | Myosin binding protein | [65] |
| Myh11 | 0.846 | 0.538 | Myosin, heavy chain 11, smooth muscle | [66] |
| Acta2 | 15.61 | 10.842 | Actin, alpha 2, smooth muscle, aorta | [67] |
| Actg2 | 0.409 | 0.083 | Actin, gamma 2, smooth muscle, enteric | [68] |
| Cnn3 | 44.46 | 57.44 | Calponin 3 | [68] |
| Kptn | 2.650 | 0.26 | Kaptin (actin binding protein) | [68] |
| Gsn | 15.650 | 29.215 | Gelsolin | [68] |
| Eln | 7.223 | 1.427 | Elastin | [68] |
| Col4a1 | 7.223 | 4.451 | Collagen type IV alpha 1 chain | [68] |
| Col4a2 | 3.880 | 2.974 | Collagen type IV alpha 2 chain | [68] |
| Gene | BV | BM | LN | H | Protein |
|---|---|---|---|---|---|
| Tek | 1.90 | 3.50 | 0.46 | 2.42 | TEK receptor tyrosine kinase, TIE2 |
| Cdh2 | 1.14 | 0.10 | 0.14 | 1.06 | N-cadherin |
| Kdr | 3.11 | 2.90 | 1.04 | 2.96 | Vegfr-2, vascular endothelial growth factor |
| Notch1 | 0.91 | 0.74 | 0.90 | 0.84 | Notch signaling protein |
| Myc | 3.69 | 7.14 | 21.6 | 3.93 | Myelocytomatosis oncogene protein |
| Krcc1 | 22.8 | 67.6 | 21.6 | 28.4 | P21—cyclin-dependent-kinase |
| Ctnnb1 | 50.8 | 79.8 | 9.90 | 56.4 | β-catenin |
| Cxcl12 | 361 | 516 | 11.7 | 463 | Chemokine (C-X-C motif) receptor 4 |
| Scf | 16.1 | 13.9 | 0.35 | 18.2 | Ligand of the tyrosine-kinase receptor |
| Tgf-β1 | 29.3 | 62.1 | 32.5 | 36.7 | Transforming growth factor beta-1 |
| Gfap | 0.72 | 1.1 | 0.13 | 0.75 | Glial fibrillary acidic protein |
| Cxcl4 | 265 | 796 | 6.1 | 204 | Platelet factor 4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vodyanoy, V.; Pustovyy, O.; Globa, L.; Kulesza, R.J., Jr.; Sorokulova, I. Hemmule: A Novel Structure with the Properties of the Stem Cell Niche. Int. J. Mol. Sci. 2020, 21, 539. https://doi.org/10.3390/ijms21020539
Vodyanoy V, Pustovyy O, Globa L, Kulesza RJ Jr., Sorokulova I. Hemmule: A Novel Structure with the Properties of the Stem Cell Niche. International Journal of Molecular Sciences. 2020; 21(2):539. https://doi.org/10.3390/ijms21020539
Chicago/Turabian StyleVodyanoy, Vitaly, Oleg Pustovyy, Ludmila Globa, Randy J. Kulesza, Jr., and Iryna Sorokulova. 2020. "Hemmule: A Novel Structure with the Properties of the Stem Cell Niche" International Journal of Molecular Sciences 21, no. 2: 539. https://doi.org/10.3390/ijms21020539
APA StyleVodyanoy, V., Pustovyy, O., Globa, L., Kulesza, R. J., Jr., & Sorokulova, I. (2020). Hemmule: A Novel Structure with the Properties of the Stem Cell Niche. International Journal of Molecular Sciences, 21(2), 539. https://doi.org/10.3390/ijms21020539





