Divergent Evolution of Eukaryotic CC- and A-Adding Enzymes
Abstract
1. Introduction
2. Results
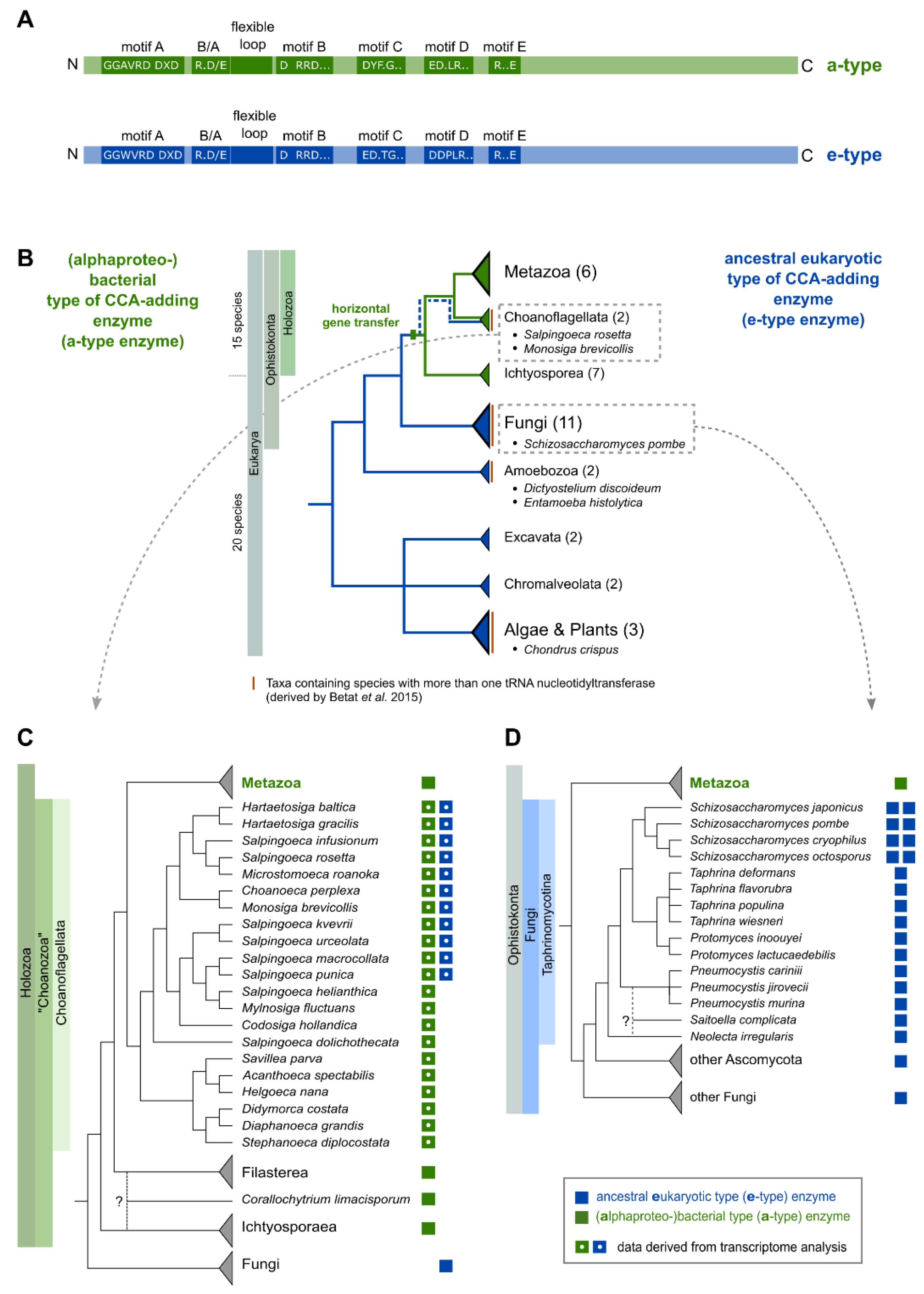
2.1. The Occurrence of More Than One tRNA Nucleotidyltransferase Is Also Common in Eukaryotes
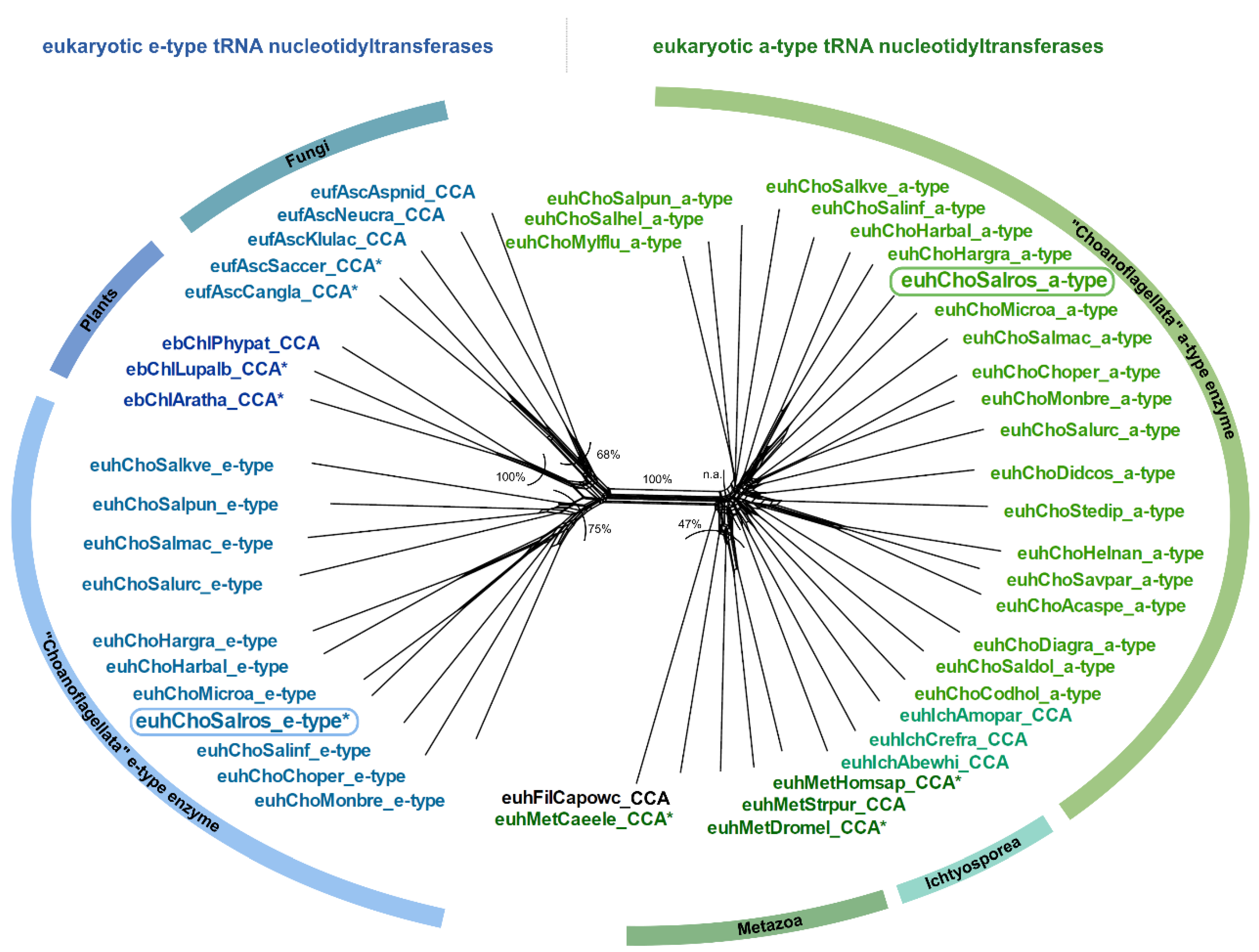
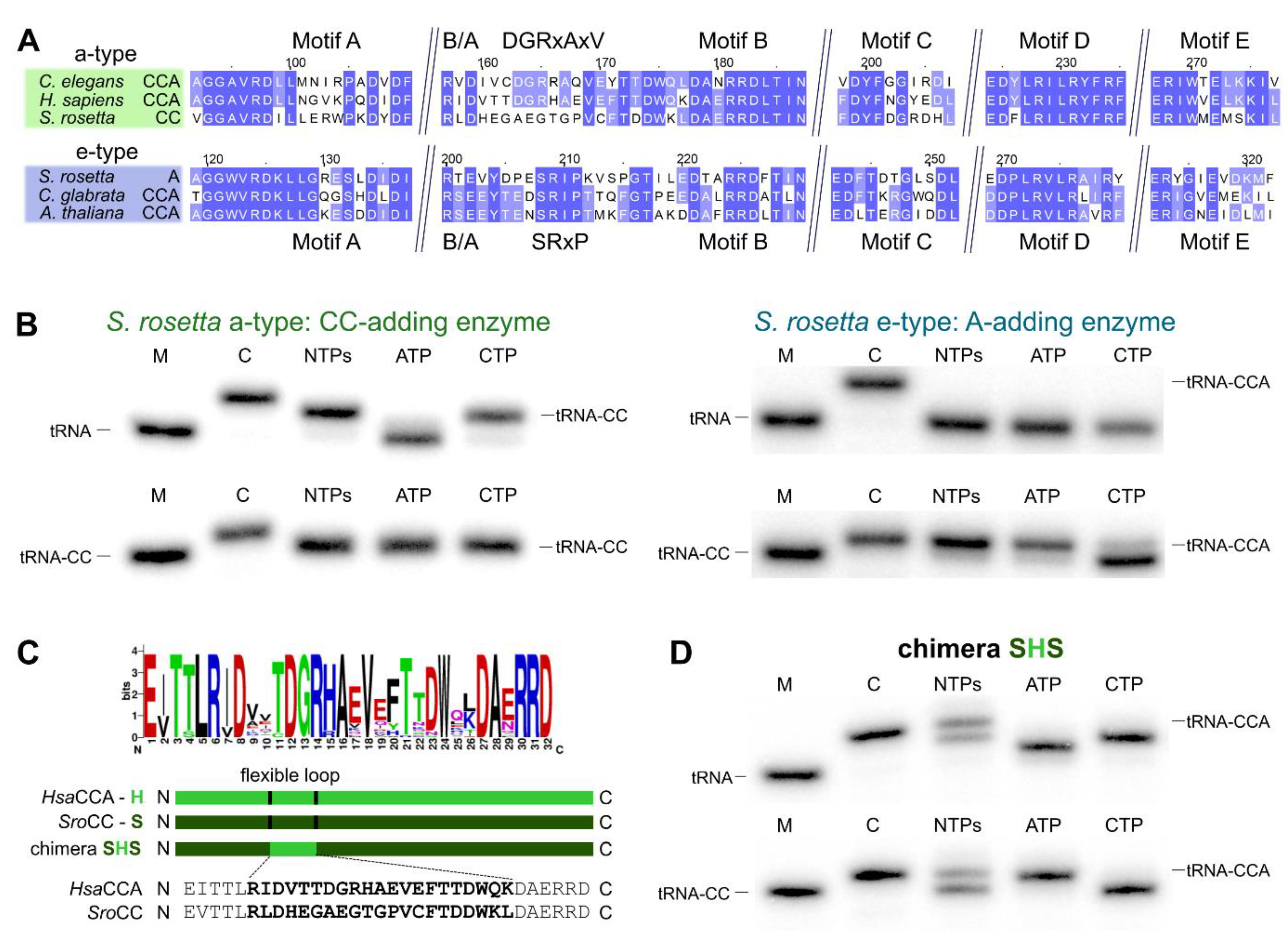
2.2. CCA-Addition in S. rosetta Requires Collaboration of CC- and A-Adding Enzymes of Different Evolutionary Origin
2.3. Introduction of a Conserved Loop Region Restores CCA-Adding Activity of S. rosetta CC-Adding Enzyme
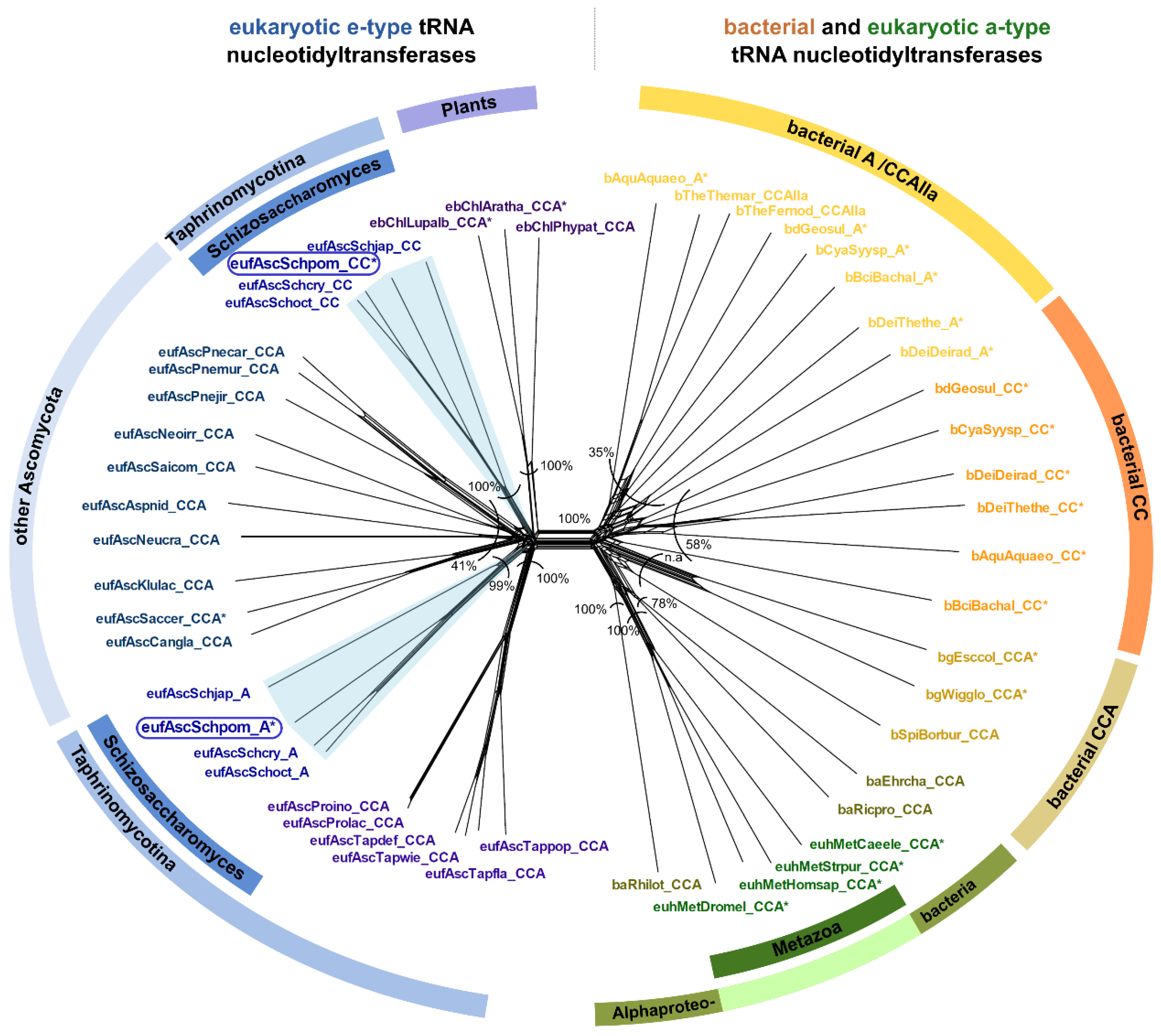
2.4. Schizosaccharomyces pombe Has Two Ancestral Eukaryotic Nucleotidyltransferases with Separate Activities
3. Discussion
3.1. Restricted Activities for CC- and A-Addition Are Widely Distributed in Bacteria as Well as in Eukaryotes
3.2. Salpingoeca rosetta Carries CC and A-Adding Enzymes of Different Evolutionary Origins
3.3. CC-and A-Adding Enzymes in S. pombe Are the Result of a Recent Gene Duplication Event in the Common Ancestor of Schizosaccharomyces
3.4. The Flexible Loop—Equally Important for the Evolution of CC-Adding Enzymes in Bacteria as Well as in Eukaryotes
4. Material and Methods
4.1. mRNA Extraction (Salpingoeca rosetta) and cDNA Source (Schizosaccharomyces pombe)
4.2. Cloning, Overexpression, and Purification of Recombinant tRNA Nucleotidyltransferases
4.3. Preparation of tRNA Substrates
4.4. In Vitro Nucleotide Incorporation Assay
4.5. Identification of Candidate tRNA Nucleotidyltransferases from Choanoflagellate Transcriptomes
4.6. Identification of Candidate tRNA Nucleotidyltransferases from Taphrinomycotina
4.7. Alignments and Phylogenetic Networks
4.8. Species Trees Topology
4.9. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Deutscher, M.P. 7 tRNA Nucleotidyltransferase. In The Enzymes: Volume XV: Nucleic Acids, Part B, 3rd ed.; Boyer, P.D., Ed.; Academic Press: New York, NY, USA, 1982; pp. 183–215. ISBN 9780121227159. [Google Scholar]
- Green, R.; Noller, H.F. Ribosomes and translation. Annu. Rev. Biochem. 1997, 66, 679–716. [Google Scholar] [CrossRef] [PubMed]
- Sprinzl, M.; Cramer, F. The -C-C-A End of tRNA and Its Role in Protein Biosynthesis. In Progress in Nucleic Acid Research and Molecular Biology; Cohn, W.E., Ed.; Academic Press: New York, NY, USA; London, UK, 1979; pp. 1–69. ISBN 9780125400220. [Google Scholar]
- Marck, C.; Grosjean, H. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 2002, 8, 1189–1232. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009, 37, D93–D97. [Google Scholar] [CrossRef] [PubMed]
- Jühling, F.; Mörl, M.; Hartmann, R.K.; Sprinzl, M.; Stadler, P.F.; Pütz, J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009, 37, D159–D162. [Google Scholar] [CrossRef]
- Abe, T.; Ikemura, T.; Sugahara, J.; Kanai, A.; Ohara, Y.; Uehara, H.; Kinouchi, M.; Kanaya, S.; Yamada, Y.; Muto, A.; et al. tRNADB-CE 2011: tRNA gene database curated manually by experts. Nucleic Acids Res. 2011, 39, D210–D213. [Google Scholar] [CrossRef]
- Betat, H.; Mede, T.; Tretbar, S.; Steiner, L.; Stadler, P.F.; Mörl, M.; Prohaska, S.J. The ancestor of modern Holozoa acquired the CCA-adding enzyme from Alphaproteobacteria by horizontal gene transfer. Nucleic Acids Res. 2015, 43, 6739–6746. [Google Scholar] [CrossRef]
- Deutscher, M.P. Ribonucleases, tRNA nucleotidyltransferase, and the 3’ processing of tRNA. Prog. Nucleic Acid Res. Mol. Biol. 1990, 39, 209–240. [Google Scholar]
- Li, F.; Xiong, Y.; Wang, J.; Cho, H.D.; Tomita, K.; Weiner, A.M.; Steitz, T.A. Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell 2002, 111, 815–824. [Google Scholar] [CrossRef]
- Betat, H.; Rammelt, C.; Mörl, M. tRNA nucleotidyltransferases: Ancient catalysts with an unusual mechanism of polymerization. Cell. Mol. Life Sci. 2010, 67, 1447–1463. [Google Scholar] [CrossRef]
- Deutscher, M.P.; Lin, J.J.-C.; Evans, J.A. Transfer RNA metabolism in Escherichia coli cells deficient in tRNA nucleotidyltransferase. J. Mol. Biol. 1977, 117, 1081–1094. [Google Scholar] [CrossRef]
- Zhu, L.; Deutscher, M.P. tRNA nucleotidyltransferase is not essential for Escherichia coli viability. EMBO J. 1987, 6, 2473–2477. [Google Scholar] [CrossRef] [PubMed]
- Dupasquier, M.; Kim, S.; Halkidis, K.; Gamper, H.; Hou, Y.-M. tRNA integrity is a prerequisite for rapid CCA addition: Implication for quality control. J. Mol. Biol. 2008, 379, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Whipple, J.M.; Phizicky, E.M.; Sharp, P.A. tRNAs marked with CCACCA are targeted for degradation. Science 2011, 334, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Wellner, K.; Czech, A.; Ignatova, Z.; Betat, H.; Mörl, M. Examining tRNA 3’-ends in Escherichia coli: teamwork between CCA-adding enzyme, RNase T, and RNase R. RNA 2018, 24, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Sander, C. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 1995, 20, 345–347. [Google Scholar] [CrossRef]
- Aravind, L. DNA polymerase beta-like nucleotidyltransferase superfamily: Identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999, 27, 1609–1618. [Google Scholar] [CrossRef]
- Yue, D.; Maizels, N.; Weiner, A.M. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: Characterization of the CCA-adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae. RNA 1996, 2, 895–908. [Google Scholar]
- Yue, D.; Weiner, A.M.; Maizels, N. The CCA-adding Enzyme Has a Single Active Site. J. Biol. Chem. 1998, 273, 29693–29700. [Google Scholar] [CrossRef]
- Martin, G.; Keller, W. RNA-specific ribonucleotidyl transferases. RNA 2007, 13, 1834–1849. [Google Scholar] [CrossRef]
- Neuenfeldt, A.; Just, A.; Betat, H.; Mörl, M. Evolution of tRNA nucleotidyltransferases: A small deletion generated CC-adding enzymes. Proc. Natl. Acad. Sci. USA 2008, 105, 7953–7958. [Google Scholar] [CrossRef]
- Just, A.; Butter, F.; Trenkmann, M.; Heitkam, T.; Mörl, M.; Betat, H. A comparative analysis of two conserved motifs in bacterial poly(A) polymerase and CCA-adding enzyme. Nucleic Acids Res. 2008, 36, 5212–5220. [Google Scholar] [CrossRef] [PubMed]
- Toh, Y.; Takeshita, D.; Numata, T.; Fukai, S.; Nureki, O.; Tomita, K. Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme. EMBO J. 2009, 28, 3353–3365. [Google Scholar] [CrossRef] [PubMed]
- Hoffmeier, A.; Betat, H.; Bluschke, A.; Günther, R.; Junghanns, S.; Hofmann, H.-J.; Mörl, M. Unusual evolution of a catalytic core element in CCA-adding enzymes. Nucleic Acids Res. 2010, 38, 4436–4447. [Google Scholar] [CrossRef]
- Franz, P.; Betat, H.; Mörl, M. Genotyping bacterial and fungal pathogens using sequence variation in the gene for the CCA-adding enzyme. BMC Microbiol. 2016, 16, 47. [Google Scholar] [CrossRef]
- Tomita, K.; Fukai, S.; Ishitani, R.; Ueda, T.; Takeuchi, N.; Vassylyev, D.G.; Nureki, O. Structural basis for template-independent RNA polymerization. Nature 2004, 430, 700–704. [Google Scholar] [CrossRef]
- Tomita, K.; Weiner, A.M. Collaboration between CC- and A-adding enzymes to build and repair the 3’-terminal CCA of tRNA in Aquifex aeolicus. Science 2001, 294, 1334–1336. [Google Scholar] [CrossRef]
- Tomita, K.; Weiner, A.M. Closely related CC- and A-adding enzymes collaborate to construct and repair the 3’-terminal CCA of tRNA in Synechocystis sp. and Deinococcus radiodurans. J. Biol. Chem. 2002, 277, 48192–48198. [Google Scholar] [CrossRef]
- Bralley, P.; Chang, S.A.; Jones, G.H. A phylogeny of bacterial RNA nucleotidyltransferases: Bacillus halodurans contains two tRNA nucleotidyltransferases. J. Bacteriol. 2005, 187, 5927–5936. [Google Scholar] [CrossRef]
- Bralley, P.; Cozad, M.; Jones, G.H. Geobacter sulfurreducens contains separate C- and A-adding tRNA nucleotidyltransferases and a poly(A) polymerase. J. Bacteriol. 2009, 191, 109–114. [Google Scholar] [CrossRef]
- Tretbar, S.; Neuenfeldt, A.; Betat, H.; Mörl, M. An inhibitory C-terminal region dictates the specificity of A-adding enzymes. Proc. Natl. Acad. Sci. USA 2011, 108, 21040–21045. [Google Scholar] [CrossRef]
- Jones, G.H. Phylogeny and Evolution of RNA 3’-Nucleotidyltransferases in Bacteria. J. Mol. Evol. 2019, 87, 254–270. [Google Scholar] [CrossRef]
- Yamashita, S.; Martinez, A.; Tomita, K. Measurement of Acceptor-TΨC Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme. Structure 2015, 23, 830–842. [Google Scholar] [CrossRef]
- Leibovitch, M.; Bublak, D.; Hanic-Joyce, P.J.; Tillmann, B.; Flinner, N.; Amsel, D.; Scharf, K.-D.; Mirus, O.; Joyce, P.B.M.; Schleiff, E. The folding capacity of the mature domain of the dual-targeted plant tRNA nucleotidyltransferase influences organelle selection. Biochem. J. 2013, 453, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Betat, H.; Mörl, M. The CCA-adding enzyme: A central scrutinizer in tRNA quality control. Bioessays 2015, 37, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Reid, N.E.; Ngou, J.S.; Joyce, P.B.M. Schizosaccharomyces pombe contains separate CC- and A-adding tRNA nucleotidyltransferases. Biochem. Biophys. Res. Commun. 2019, 508, 785–790. [Google Scholar] [CrossRef]
- Preston, M.A.; Porter, D.F.; Chen, F.; Buter, N.; Lapointe, C.P.; Keles, S.; Kimble, J.; Wickens, M. Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase. Nat. Methods 2019, 16, 437–445. [Google Scholar] [CrossRef]
- Richter, D.J.; Fozouni, P.; Eisen, M.B.; King, N. Gene family innovation, conservation and loss on the animal stem lineage. Elife 2018, 7. [Google Scholar] [CrossRef]
- Claros, M.G.; Vincens, P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996, 241, 779–786. [Google Scholar] [CrossRef]
- Hou, Y.M. Unusual synthesis by the Escherichia coli CCA-adding enzyme. RNA 2000, 6, 1031–1043. [Google Scholar] [CrossRef][Green Version]
- Betat, H.; Rammelt, C.; Martin, G.; Mörl, M. Exchange of regions between bacterial poly(A) polymerase and the CCA-adding enzyme generates altered specificities. Mol. Cell 2004, 15, 389–398. [Google Scholar] [CrossRef]
- Tomari, Y.; Suzuki, T.; Ueda, T. tRNA recognition by CCA-adding enzyme. Nucleic Acids Res. Suppl. 2002, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Reichert, A.S.; Thurlow, D.L.; Mörl, M. A eubacterial origin for the human tRNA nucleotidyltransferase? Biol. Chem. 2001, 382, 1431–1438. [Google Scholar] [CrossRef] [PubMed]
- Augustin, M.A.; Reichert, A.S.; Betat, H.; Huber, R.; Mörl, M.; Steegborn, C. Crystal Structure of the Human CCA-adding Enzyme: Insights into Template-independent Polymerization. J. Mol. Biol. 2003, 328, 985–994. [Google Scholar] [CrossRef]
- Hanic-Joyce, P.J.; Joyce, P.B.M. Characterization of a gene encoding tRNA nucleotidyltransferase from Candida glabrata. Yeast 2002, 19, 1399–1411. [Google Scholar] [CrossRef]
- von Braun, S.S.; Sabetti, A.; Hanic-Joyce, P.J.; Gu, J.; Schleiff, E.; Joyce, P.B.M. Dual targeting of the tRNA nucleotidyltransferase in plants: Not just the signal. J. Exp. Bot. 2007, 58, 4083–4093. [Google Scholar] [CrossRef]
- Shanmugam, K.; Hanic-Joyce, P.J.; Joyce, P.B.M. Purification and characterization of a tRNA nucleotidyltransferase from Lupinus albus and functional complementation of a yeast mutation by the corresponding cDNA. Plant Mol. Biol. 1996, 30, 281–295. [Google Scholar] [CrossRef]
- Chen, J.Y.; Kirchner, G.; Aebi, M.; Martin, N.C. Purification and properties of yeast ATP (CTP):tRNA nucleotidyltransferase from wild type and overproducing cells. J. Biol. Chem. 1990, 265, 16221–16224. [Google Scholar]
- Kimura, M. The neutral theory of molecular evolution: A review of recent evidence. Jpn. J. Genet. 1991, 66, 367–386. [Google Scholar] [CrossRef]
- King, J.L.; Jukes, T.H. Non-Darwinian evolution. Science 1969, 164, 788–798. [Google Scholar] [CrossRef]
- Pöhler, M.-T.; Roach, T.M.; Betat, H.; Jackman, J.E.; Mörl, M. A Temporal Order in 5’- and 3’- Processing of Eukaryotic tRNAHis. Int. J. Mol. Sci. 2019, 20, 1384. [Google Scholar] [CrossRef]
- Wende, S.; Bonin, S.; Götze, O.; Betat, H.; Mörl, M. The identity of the discriminator base has an impact on CCA addition. Nucleic Acids Res. 2015, 43, 5617–5629. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.; Obermaier-Kusser, B.; Jacobs, M.; Milles, C.; Mörl, M.; von Pein, H.D.; Grau, A.J.; Bauer, M.F. A new mitochondrial point mutation in the transfer RNA(Lys) gene associated with progressive external ophthalmoplegia with impaired respiratory regulation. J. Neurol. Sci. 2012, 316, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Friedman, R. Parallel evolution by gene duplication in the genomes of two unicellular fungi. Genome Res. 2003, 13, 794–799. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hoffman, C.S.; Wood, V.; Fantes, P.A. An Ancient Yeast for Young Geneticists: A Primer on the Schizosaccharomyces pombe Model System. Genetics 2015, 201, 403–423. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Aime, M.C.; Grigoriev, I.V.; Martin, F.; Stajich, J.E.; Blackwell, M. The Fungal Tree of Life: from Molecular Systematics to Genome-Scale Phylogenies. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Ernst, F.G.M.; Rickert, C.; Bluschke, A.; Betat, H.; Steinhoff, H.-J.; Mörl, M. Domain movements during CCA-addition: A new function for motif C in the catalytic core of the human tRNA nucleotidyltransferases. RNA Biol. 2015, 12, 435–446. [Google Scholar] [CrossRef]
- Wijn R, d.e.; Hennig, O.; Ernst, F.G.M.; Lorber, B.; Betat, H.; Mörl, M.; Sauter, C. Combining crystallogenesis methods to produce diffraction-quality crystals of a psychrophilic tRNA-maturation enzyme. Acta Crystallogr. F Struct. Biol. Commun. 2018, 74, 747–753. [Google Scholar] [CrossRef]
- Schürer, H.; Lang, K.; Schuster, J.; Mörl, M. A universal method to produce in vitro transcripts with homogeneous 3’ ends. Nucleic Acids Res. 2002, 30, 56. [Google Scholar] [CrossRef]
- Mörl, M.; Hartmann, R.K. Production of RNAs with Homogeneous 5’- and 3’-Ends. In Handbook of RNA Biochemistry, 2nd, Completely Revised and Enlarged ed.; Hartmann, R.K., Bindereif, A., Schön, A., Westhof, E., Eds.; Wiley-VCH: Weinheim, Germany, 2014; pp. 29–44. ISBN 9783527647064. [Google Scholar]
- Goring, M.E.; Leibovitch, M.; Gea-Mallorqui, E.; Karls, S.; Richard, F.; Hanic-Joyce, P.J.; Joyce, P.B.M. The ability of an arginine to tryptophan substitution in Saccharomyces cerevisiae tRNA nucleotidyltransferase to alleviate a temperature-sensitive phenotype suggests a role for motif C in active site organization. Biochim. Biophys. Acta 2013, 1834, 2097–2106. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Philippe, H.; Derelle, R.; Lopez, P.; Pick, K.; Borchiellini, C.; Boury-Esnault, N.; Vacelet, J.; Renard, E.; Houliston, E.; Quéinnec, E.; et al. Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 2009, 19, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Burki, F.; Kaplan, M.; Tikhonenkov, D.V.; Zlatogursky, V.; Minh, B.Q.; Radaykina, L.V.; Smirnov, A.; Mylnikov, A.P.; Keeling, P.J. Untangling the early diversification of eukaryotes: A phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc. Biol. Sci. 2016, 283. [Google Scholar] [CrossRef] [PubMed]
- Carr, M.; Richter, D.J.; Fozouni, P.; Smith, T.J.; Jeuck, A.; Leadbeater, B.S.C.; Nitsche, F. A six-gene phylogeny provides new insights into choanoflagellate evolution. Mol. Phylogenet. Evol. 2017, 107, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Shalchian-Tabrizi, K.; Minge, M.A.; Espelund, M.; Orr, R.; Ruden, T.; Jakobsen, K.S.; Cavalier-Smith, T. Multigene phylogeny of choanozoa and the origin of animals. PLoS ONE 2008, 3, e2098. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erber, L.; Franz, P.; Betat, H.; Prohaska, S.; Mörl, M. Divergent Evolution of Eukaryotic CC- and A-Adding Enzymes. Int. J. Mol. Sci. 2020, 21, 462. https://doi.org/10.3390/ijms21020462
Erber L, Franz P, Betat H, Prohaska S, Mörl M. Divergent Evolution of Eukaryotic CC- and A-Adding Enzymes. International Journal of Molecular Sciences. 2020; 21(2):462. https://doi.org/10.3390/ijms21020462
Chicago/Turabian StyleErber, Lieselotte, Paul Franz, Heike Betat, Sonja Prohaska, and Mario Mörl. 2020. "Divergent Evolution of Eukaryotic CC- and A-Adding Enzymes" International Journal of Molecular Sciences 21, no. 2: 462. https://doi.org/10.3390/ijms21020462
APA StyleErber, L., Franz, P., Betat, H., Prohaska, S., & Mörl, M. (2020). Divergent Evolution of Eukaryotic CC- and A-Adding Enzymes. International Journal of Molecular Sciences, 21(2), 462. https://doi.org/10.3390/ijms21020462




