A New Cold-Adapted and Salt-Tolerant Glutathione Reductase from Antarctic Psychrophilic Bacterium Psychrobacter sp. and Its Resistance to Oxidation
Abstract
1. Introduction
2. Results and Discussion
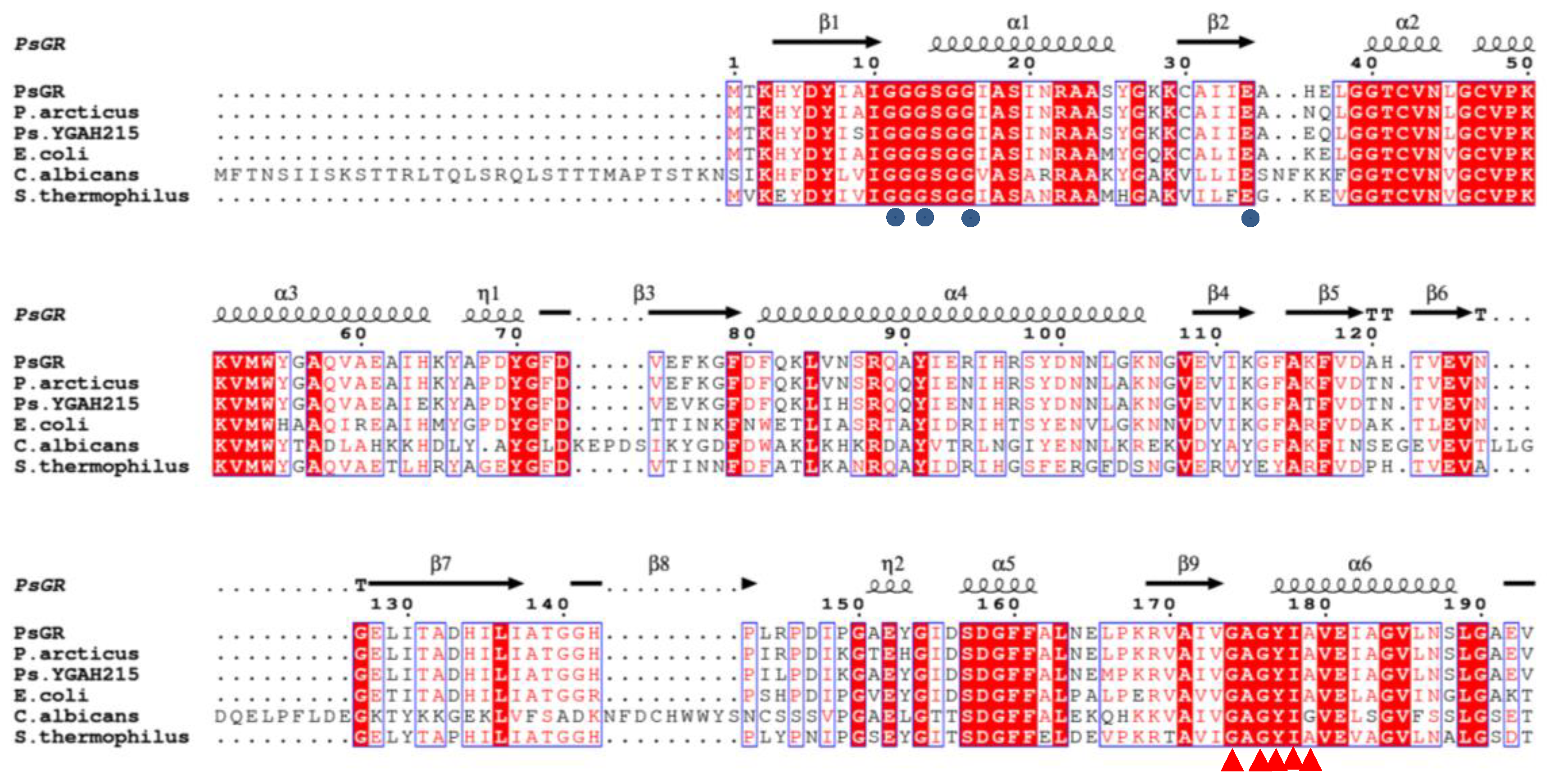
2.1. Identification of the psgr Gene
2.2. Homology Modeling Analysis and Binding Energy Interactions of PsGR
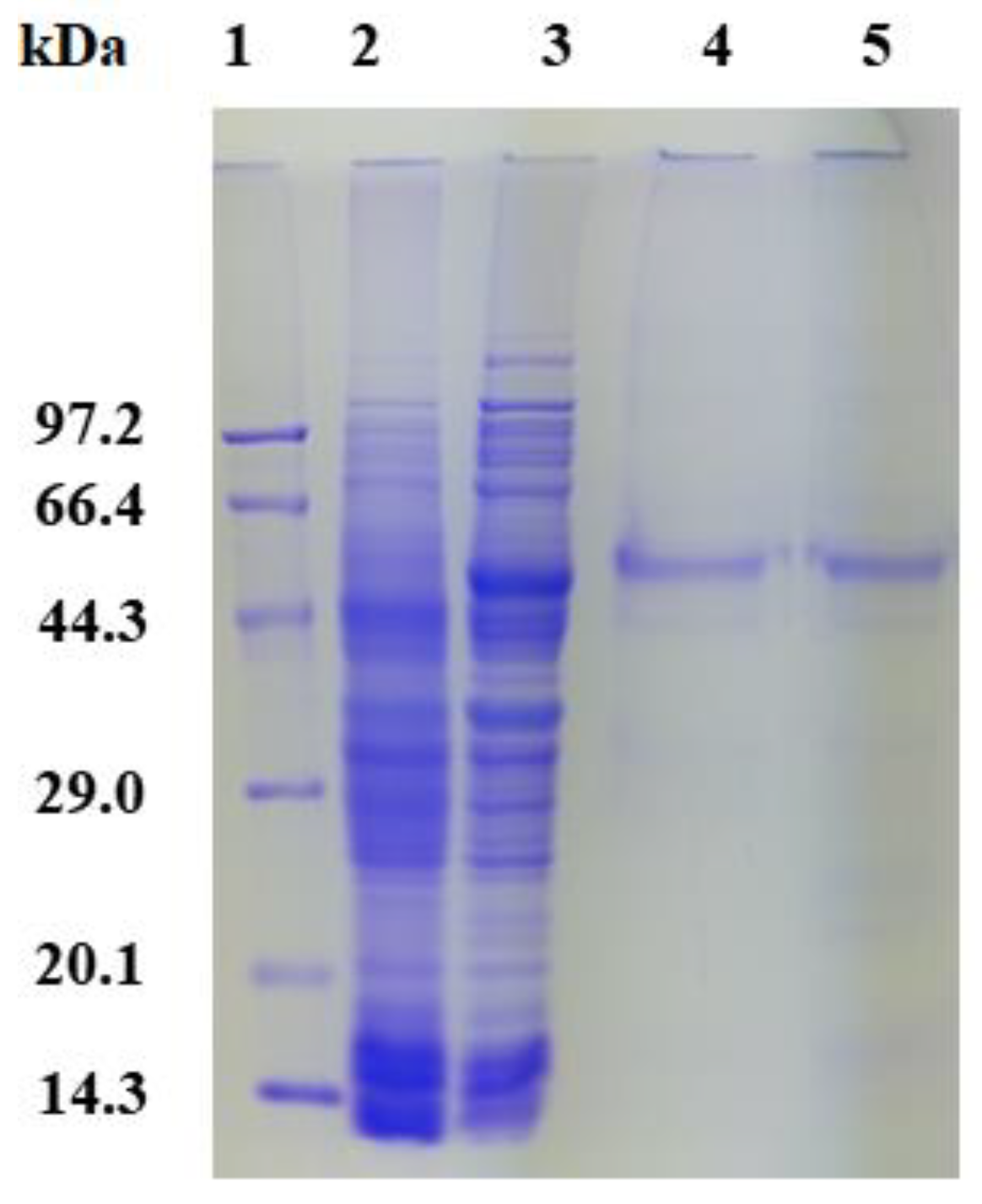
2.3. Expression, Purification, and Enzyme Assays of PsGR
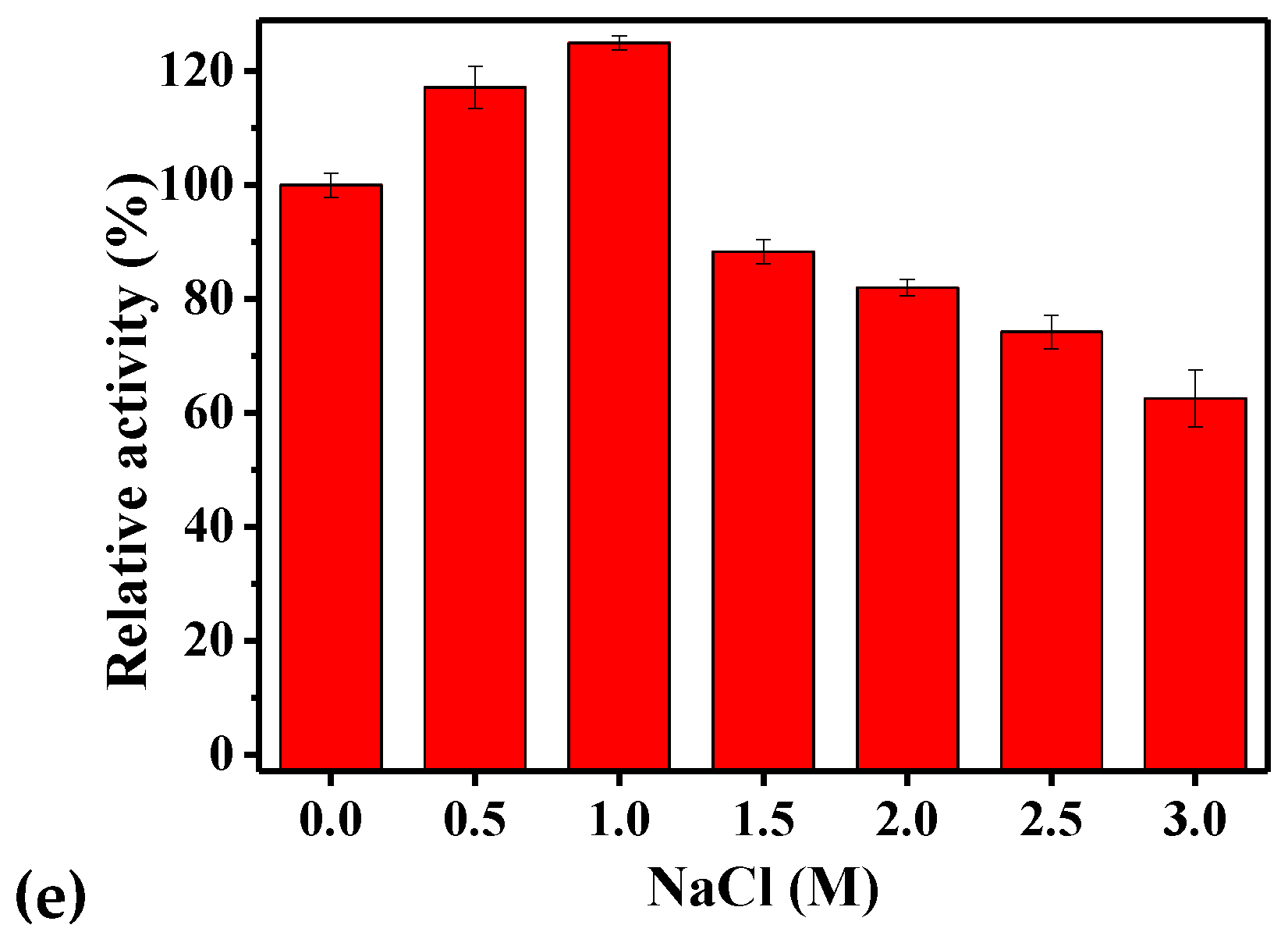
2.4. Biochemical Characteristics of rPsGR
2.5. Kinetics and Thermodynamics Parameters
2.6. Disk Diffusion Assay
3. Materials and Methods
3.1. Bacterial Strains and Plasmids
3.2. Gene Cloning and Bioinformatics Analysis of PsGR
3.3. Homology Modeling and Binding Energy Interactions of PsGR
3.4. Expression and Purification of PsGR
3.5. GR Activity Assays
3.6. Biochemical Characteristics of rPsGR
3.7. Kinetics and Thermodynamics Parameters of rPsGR
3.8. Disk Diffusion Assay
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| psgr | A glutathione reductase gene from Psychrobacter sp. ANT206 |
| E. coli | Escherichia coli |
| GR | Glutathione reductase |
| PsGR | Protein expressed by psgr |
| rPsGR | recombinant PsGR |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| GSH | Reduced glutathione |
| GSSG | Oxidized glutathione |
| ORF | Open reading frame |
| H2O2 | Oxidizing agent |
| P. arcticus | Psychrobacter arcticus 273-4 |
| C. psychrerythraea | Colwellia psychrerythraea |
| C. albicans | Candida albicans |
| S. thermophilus | Streptococcus thermophilus |
| Ps. YGAH215 | Psychrobacter sp. YGAH215 |
| E. faecalis | Enterococcus faecalis |
| X. campestris | Xanthomonas campestris |
| S. sp. | Sphingomonas sp. |
| R. rubrum | Rhodospirillum rubrum |
| S. pneumoniae | Streptococcus pneumoniae |
| P. aeruginosa | Pseudomonas aeruginosa |
| C. vinosum | Chromatium vinosum |
| pHMB | para-hydroximercuribenzoate |
| IPTG | Isopropyl-β-d-thiogalactopyranoside |
| BL21/pET-28a(+) | E. coli BL21 with pET-28a(+) |
| BL21/pET-28a(+)-rPsGR | E. coli BL21 with pET-28a(+) containing GR |
| LB medium | Luria Bertani medium |
| OD600 | Optical density at 600 nm |
References
- Piette, F.; D’Amico, S.; Mazzucchelli, G.; Danchin, A.; Leprince, P.; Feller, G. Life in the cold: A proteomic study of cold-repressed proteins in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Appl. Environ. Microbiol. 2011, 77, 3881–3883. [Google Scholar] [CrossRef]
- Morgan-Kiss, R.M.; Priscu, J.C.; Pocock, T.; Gudynaite-Savitch, L.; Huner, N.P.A. Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiol. Mol. Biol. Rev. 2006, 70, 222–252. [Google Scholar] [CrossRef]
- Tehei, M.; Zaccai, G. Adaptation to extreme environments: Macromolecular dynamics in complex systems. Biochim. Biophys. Acta 2005, 1724, 404–410. [Google Scholar] [CrossRef]
- Sikanyika, M.; Aragao, D.; McDevitt, C.A.; Maher, M.J. The structure and activity of the glutathione reductase from Streptococcus pneumoniae. Acta Crystallogr. F Struct. Biol. 2019, 75, 54–61. [Google Scholar] [CrossRef]
- Loprasert, S.; Whangsuk, W.; Sallabhan, R.; Mongkolsuk, S. The unique glutathione reductase from Xanthomonas campestris: Gene expression and enzyme characterization. Biochem. Biophys. Res. Commun. 2005, 331, 1324–1330. [Google Scholar] [CrossRef]
- Ji, M.; Barnwell, C.V.; Grunden, A.M. Characterization of recombinant glutathione reductase from the psychrophilic Antarctic bacterium Colwellia psychrerythraea. Extremophiles 2015, 19, 863–874. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kamio, Y.; Higuchi, M. Cloning, nucleotide sequence, and disruption of Streptococcus mutans glutathione reductase gene (gor). Biosci. Biotechnol. Biochem. 1999, 63, 1056–1062. [Google Scholar] [CrossRef]
- Hodges, D.M.; Andrews, C.J.; Johnson, D.A.; Hamilton, R.I. Antioxidant enzyme responses to chilling stress in differentially sensitive inbred maize lines. J. Exp. Bot. 1997, 48, 1105–1113. [Google Scholar] [CrossRef]
- Wu, T.M.; Lin, W.R.; Kao, Y.T.; Hsu, Y.T.; Yeh, C.H.; Hong, C.Y.; Kao, C.H. Identification and characterization of a novel chloroplast/mitochondria co-localized glutathione reductase 3 involved in salt stress response in rice. Plant Mol. Biol. 2013, 83, 379–390. [Google Scholar] [CrossRef]
- Zhang, T.G.; Nie, T.T.; Sun, W.C.; Zhong, F.; Wang, J. Effects of diverse stresses on gene expression and enzyme activity of glutathione reductase in Brassica campestris. J. Appl. Ecol. 2018, 29, 213–222. [Google Scholar] [CrossRef]
- Mittl, P.R.E.; Schulz, G.E. Structure of glutathione reductase from Echerichia coli at 1.86 Å resolution: Comparison with the enzyme from human erythrocytes. Protein Sci. 1994, 3, 799–809. [Google Scholar] [CrossRef]
- Achary, V.M.M.; Reddy, C.S.; Pandey, P.; Islam, T.; Kaul, T.; Reddy, M.K. Glutathione reductase a unique enzyme: Molecular cloning, expression and biochemical characterization from the stress adapted C4 plant, Pennisetum glaucum (L.) R. Br. Mol. Biol. Rep. 2015, 42, 947–962. [Google Scholar] [CrossRef]
- Rouhier, N.; Couturier, J.; Jacquot, J.P. Genome-wide analysis of plant glutaredoxin systems. J. Exp. Bot. 2006, 57, 1685–1696. [Google Scholar] [CrossRef]
- Emnet, P.; Gaw, S.; Northcott, G.; Storey, B.; Graham, L. Personal care products and steroid hormones in the Antarctic coastal environment associated with two Antarctic research stations, McMurdo Station and Scott Base. Environ. Res. 2015, 136, 331–342. [Google Scholar] [CrossRef]
- Vorrath, M.E.; Muller, J.; Esper, O.; Mollenhauer, G.; Haas, C.; Schefuss, E.; Fahl, K. Highly branched isoprenoids for Southern Ocean sea ice reconstructions: A pilot study from the Western Antarctic Peninsula. Biogeosciences 2019, 16, 2961–2981. [Google Scholar] [CrossRef]
- Hou, Y.H.; Qiao, C.H.; Wang, Y.F.; Wang, Y.T.; Ren, X.L.; Wei, Q.F.; Wang, Q.F. Cold-adapted glutathione S-transferases from Antarctic psychrophilic bacterium Halomonas sp. ANT108: Heterologous expression, characterization, and oxidative resistance. Mar. Drugs 2019, 17, 147. [Google Scholar] [CrossRef]
- Wang, Y.F.; Hou, Y.H.; Wang, Y.T.; Lu, Z.B.; Song, C.; Xu, Y.F.; Wei, N.N.; Wang, Q.F. Cloning, expression and enzymatic characteristics of a 2-Cys peroxiredoxin from Antarctic sea-ice bacterium Psychrobacter sp. ANT206. Int. J. Biol. Macromol. 2019, 129, 1047–1055. [Google Scholar] [CrossRef]
- VuThi, H.; Sei-Heon, J.; Lee, C.W. Cloning and characterization of a thermostable glutathione reductase from a psychrophilic Arctic bacterium Sphingomonas sp. FEMS Microbiol. Lett. 2019, 366, fnz218. [Google Scholar] [CrossRef]
- Perry, A.C.; Ni Bhriain, N.; Brown, N.L.; Rouch, D.A. Molecular characterization of the gor gene encoding glutathione reductase from Pseudomonas aeruginosa: Determinants of substrate specificity among pyridine nucleotide-disulphide oxidoreductases. Mol. Microbiol. 1991, 5, 163–171. [Google Scholar] [CrossRef]
- Zheng, Y.Y.; Li, Y.J.; Liu, W.D.; Chen, C.C.; Ko, T.P.; He, M.; Xu, Z.X.; Liu, M.X.; Luo, H.Y.; Guo, R.T.; et al. Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase. J. Struct. Biol. 2016, 193, 206–211. [Google Scholar] [CrossRef]
- Li, F.L.; Shi, Y.; Zhang, J.X.; Gao, J.; Zhang, Y.W. Cloning, expression, characterization and homology modeling of a novel water-forming NADH oxidase from Streptococcus mutans ATCC 25175. Int. J. Biol. Macromol. 2018, 113, 1073–1079. [Google Scholar] [CrossRef]
- Gholampour-Faroji, N.; Farazmand, R.; Hemmat, J.; Haddad-Mashadrizeh, A. Modeling, stability and the activity assessment of glutathione reductase from Streptococcus Thermophilus; Insights from the in-silico simulation study. Comput. Biol. Chem. 2019, 83, 107121. [Google Scholar] [CrossRef]
- Patel, M.P.; Marcinkeviciene, J.; Blanchard, J.S. Enterococcus faecalis glutathione reductase: Purification, characterization and expression under normal and hyperbaric O-2 conditions. FEMS Microbiol. Lett. 1998, 166, 155–163. [Google Scholar] [CrossRef]
- Scrutton, N.S.; Berry, A.; Perham, R.N. Purification and characterization of glutathione-reductase encoded by a cloned and over-expressed gene in Escherichia-coli. Biochem. J. 1987, 245, 875–880. [Google Scholar] [CrossRef]
- Libreros-Minotta, C.A.; Pardo, J.P.; Mendoza-Hernández, G.; Rebdon, J.L. Purification and characterization of glutathione reductase from Rhodospirillum rubrum. Arch. Biochem. Biophys. 1992, 298, 247–253. [Google Scholar] [CrossRef]
- Ding, Y.; Miao, J.L.; Wang, Q.F.; Zheng, Z.; Li, G.Y.; Jian, J.C.; Wu, Z.H. Purification and characterization of a psychrophilic glutathione reductase from Antarctic ice microalgae Chlamydomonas sp. strain ICE-L. Polar Biol. 2007, 31, 23–30. [Google Scholar] [CrossRef]
- Arias, D.G.; Marquez, V.E.; Beccaria, A.J.; Guerrero, S.A.; Iglesias, A.A. Purification and characterization of a glutathione reductase from Phaeodactylum tricornutum. Protist 2010, 161, 91–101. [Google Scholar] [CrossRef]
- Mata, A.M.; Pinto, M.C.; Lopezbarea, J. Redox interconversion of Escherichia-coli glutathione-reductase—A study with permeabilized and intact-cells. Mol. Cell Biochem. 1985, 68, 121–130. [Google Scholar] [CrossRef]
- Mata, A.M.; Pinto, M.C.; Lopez-Barea, J. Purification by affinity chromatography of glutathione reductase (EC 1.6.4.2) from Escherichia coli and characterization of such enzyme. Z. Nat. C 1984, 39, 908–915. [Google Scholar] [CrossRef]
- Chung, Y.C.; Hurlbert, R.E. Purification and properties of the glutathione reductase of Chromatium vinosum. J. Bacteriol. 1975, 123, 203–211. [Google Scholar] [CrossRef]
- Arora, K.; Ahmad, R.; Srivastava, A.K. Purification and characterization of glutathione reductase (E.C. 1.8.1.7) from bovine filarial worms Setaria cervi. J. Parasit. Dis. 2013, 37, 94–104. [Google Scholar] [CrossRef][Green Version]
- Bowman, J.P.; McCammon, S.A.; Brown, M.V.; Nichols, D.S.; McMeekin, T.A. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 1997, 63, 3068–3078. [Google Scholar] [CrossRef]
- Lonhienne, T.; Gerday, C.; Feller, G. Psychrophilic enzymes: Revisiting the thermodynamic parameters of activation may explain local flexibility. Biophys. Acta 2000, 1543, 1–10. [Google Scholar] [CrossRef]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Microbiol. 2003, 1, 200–208. [Google Scholar] [CrossRef]
- Vanoni, M.A.; Wong, K.K.; Ballou, D.P.; Blanchard, J.S. Glutathione Reductase: Comparison of steady-state and rapid reaction primary kinetic isotope effects exhibited by the yeast, spinach, and Escherichia coli. Biochemistry 1990, 29, 5790–5796. [Google Scholar] [CrossRef]
- Struvay, C.; Feller, G. Optimization to low temperature activity in psychrophilic enzymes. Int. J. Mol. Sci. 2012, 13, 11643–11665. [Google Scholar] [CrossRef]
- Xue, D.S.; Zeng, X.H.; Gong, C.J.; Lin, D.Q.; Yao, S.J. A cold adapt and ethanol tolerant endoglucanase from a marine Bacillus subtilis. Chin. J. Chem. Eng. 2018, 26, 2601–2606. [Google Scholar] [CrossRef]
- Zhou, J.P.; Liu, Y.; Shen, J.D.; Zhang, R.; Tang, X.H.; Li, J.J.; Wang, Y.Y.; Huang, Z.X. Kinetic and thermodynamic characterization of a novel low-temperature-active xylanase from Arthrobacter sp. GN16 isolated from the feces of Grus nigricollis. Bioengineered 2015, 6, 111–114. [Google Scholar] [CrossRef][Green Version]
- Hausladen, A.; Alscher, R.G. Cold-hardiness-specific glutathione-reductase isozymes in Red Spruce—Thermal-dependence of kinetic-parameters and possible regulatory mechanisms. Plant Physiol. 1994, 105, 215–223. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.F.; Hou, Y.H.; Wang, Y.T.; Zheng, L.; Wang, Q.F. A novel cold-adapted nitroreductase from Psychrobacter sp. ANT206: Heterologous expression, characterization and nitrobenzene reduction capacity. Enzyme Microb. Technol. 2019, 131, 109434. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]





| PsGR | EcGR | Expected Effect on PsGR | |
|---|---|---|---|
| Electrostatic interactions | 782 | 836 | Protein stability |
| Salt bridges | 23 | 32 | |
| Hydrogen bonds | 697 | 741 | |
| Aromatic interactions | 32 | 40 | |
| Cation-Pi interactions | 30 | 23 | |
| Hydrophobic interactions | 750 | 792 | Thermolability |
| G (Gly) | 10.4% | 9.4% | Flexibility |
| P (Pro) | 3.3% | 4.5% | |
| R (Arg) | 3.3% | 3.8% | |
| Arg/(Arg+Lys) | 0.34 | 0.42 | |
| Gly/Pro | 3.13 | 2.09 |
| Pose Mode | Docking Scores Based on kcal/mol Via the GR-NADPH Interactions | Docking Scores Based on kcal/mol Via the GR-GSSG Interactions | ||
|---|---|---|---|---|
| PsGR | EcGR | PsGR | EcGR | |
| 1 | −6.9 | −5.2 | −5.0 | −4.8 |
| 2 | −6.6 | −5.0 | −5.0 | −4.7 |
| 3 | −6.5 | −5.0 | −5.0 | −4.5 |
| 4 | −6.3 | −4.9 | −4.9 | −4.5 |
| 5 | −6.1 | −4.9 | −4.8 | −4.3 |
| 6 | −6.0 | −4.9 | −4.7 | −4.3 |
| 7 | −5.8 | −4.8 | −4.7 | −4.3 |
| 8 | −5.8 | −4.8 | −4.7 | −4.2 |
| 9 | −5.6 | −4.8 | −4.7 | −4.2 |
| Source | MW (kDa) | Optimal Temperature (°C) | Optimal pH | Half-Life of Activity (min) | Activator | Km(μM) | kcat (1/s) | Ref | |
|---|---|---|---|---|---|---|---|---|---|
| GSSG | NADPH | ||||||||
| E. coli SG5 | 49 | — | — | — | — | 70 | 25 | — | [24] |
| E. faecalis | 49 | — | — | — | — | 80 | 9 | 145 | [23] |
| E. coli S33 | 55 | — | 7.5 | — | — | 66 | 16 | — | [29] |
| X. campestris | 50 | — | — | — | — | — | 52.6 | 37.5 | [5] |
| S. sp. | 50 | 60 | 7.5 | 60 (70 °C) | — | 178 | — | 160 | [18] |
| C. psychrerythraea | 48.7 | — | — | 40 (50 °C) | — | — | 10.9 | 700 | [6] |
| R. rubrum | 54.4 | — | — | — | — | 90 | — | [25] | |
| S. pneumoniae | 100 | — | — | — | — | 231.2 | 23.2 | — | [4] |
| C. vinosum | 52 | — | 7.0 | — | Na+, NH4+ | 7000 | — | — | [30] |
| P. sp. ANT206 | 53.5 | 25 | 7.5 | 30 (50 °C) | Na+, Mg2+ | 68.96 | 16.95 | 95.78 | This study |
| Reagent | Concentration | Relative Activity (%) | Reagent | Concentration | Relative Activity (%) |
|---|---|---|---|---|---|
| None | — | 100 ± 0.0 | — | — | — |
| KCl | 0.25 mM | 90.3 ± 2.5 | KCl | 1 mM | 81.0 ± 2.0 |
| CoCl2 | 0.25 mM | 27.3 ± 3.0 | CoCl2 | 1 mM | 0.0 ± 0.0 |
| MgCl2 | 0.25 mM | 109.1 ± 2.6 | MgCl2 | 1 mM | 70.3 ± 2.2 |
| CaCl2 | 0.25 mM | 90.9 ± 3.1 | CaCl2 | 1 mM | 78.1 ± 2.5 |
| ZnCl2 | 0.25 mM | 33.6 ± 2.5 | ZnCl2 | 1 mM | 13.4 ± 2.0 |
| FeCl2 | 0.25 mM | 44.5 ± 2.2 | FeCl2 | 1 mM | 5.4 ± 2.7 |
| CuCl2 | 0.25 mM | 20.3 ± 2.6 | CuCl2 | 1 mM | 0.0 ± 0.0 |
| HgCl2 | 0.25 mM | 10.9 ± 2.2 | HgCl2 | 1 mM | 0.0 ± 0.0 |
| CrCl2 | 0.25 mM | 47.3 ± 2.9 | CrCl2 | 1 mM | 15.8 ± 1.9 |
| CdCl2 | 0.25 mM | 19.1 ± 2.5 | CdCl2 | 1 mM | 2.3 ± 3.0 |
| Pb(NO3)2 | 0.25 mM | 0.0 ± 0.0 | Pb(NO3)2 | 1 mM | 0.0 ± 0.0 |
| BaCl2 | 0.25 mM | 37.3 ± 2.7 | BaCl2 | 1 mM | 8.9 ± 1.6 |
| EDTA | 0.25 mM | 28.2 ± 2.7 | EDTA | 1 mM | 10.2 ± 2.2 |
| Substrate | Vm (mmol/min/mg) | Km (μM) | Kcat (1/s) |
|---|---|---|---|
| GSSG | 172.41 | 68.96 | 95.78 |
| NADPH | 169.49 | 16.95 | 94.16 |
| Temperature (°C) | △H (KJ/mol) | △S (J/mol K) | △G (KJ/mol) | Kcat (1/s) |
|---|---|---|---|---|
| 0 | 38.48 | −79.05 | 60.07 | 18.90 |
| 5 | 38.44 | −78.07 | 60.15 | 29.87 |
| 10 | 38.39 | −77.44 | 60.32 | 44.80 |
| 15 | 38.35 | −78.21 | 60.89 | 56.12 |
| 20 | 38.31 | −79.78 | 61.70 | 63.13 |
| 25 | 38.27 | −78.93 | 61.80 | 94.16 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wang, Q.; Hou, Y. A New Cold-Adapted and Salt-Tolerant Glutathione Reductase from Antarctic Psychrophilic Bacterium Psychrobacter sp. and Its Resistance to Oxidation. Int. J. Mol. Sci. 2020, 21, 420. https://doi.org/10.3390/ijms21020420
Wang Y, Wang Q, Hou Y. A New Cold-Adapted and Salt-Tolerant Glutathione Reductase from Antarctic Psychrophilic Bacterium Psychrobacter sp. and Its Resistance to Oxidation. International Journal of Molecular Sciences. 2020; 21(2):420. https://doi.org/10.3390/ijms21020420
Chicago/Turabian StyleWang, Yatong, Quanfu Wang, and Yanhua Hou. 2020. "A New Cold-Adapted and Salt-Tolerant Glutathione Reductase from Antarctic Psychrophilic Bacterium Psychrobacter sp. and Its Resistance to Oxidation" International Journal of Molecular Sciences 21, no. 2: 420. https://doi.org/10.3390/ijms21020420
APA StyleWang, Y., Wang, Q., & Hou, Y. (2020). A New Cold-Adapted and Salt-Tolerant Glutathione Reductase from Antarctic Psychrophilic Bacterium Psychrobacter sp. and Its Resistance to Oxidation. International Journal of Molecular Sciences, 21(2), 420. https://doi.org/10.3390/ijms21020420




