Genome-Wide Association Mapping of Prostrate/Erect Growth Habit in Winter Durum Wheat
Abstract
1. Introduction
2. Results
2.1. Phenotypic Variation for the Trait
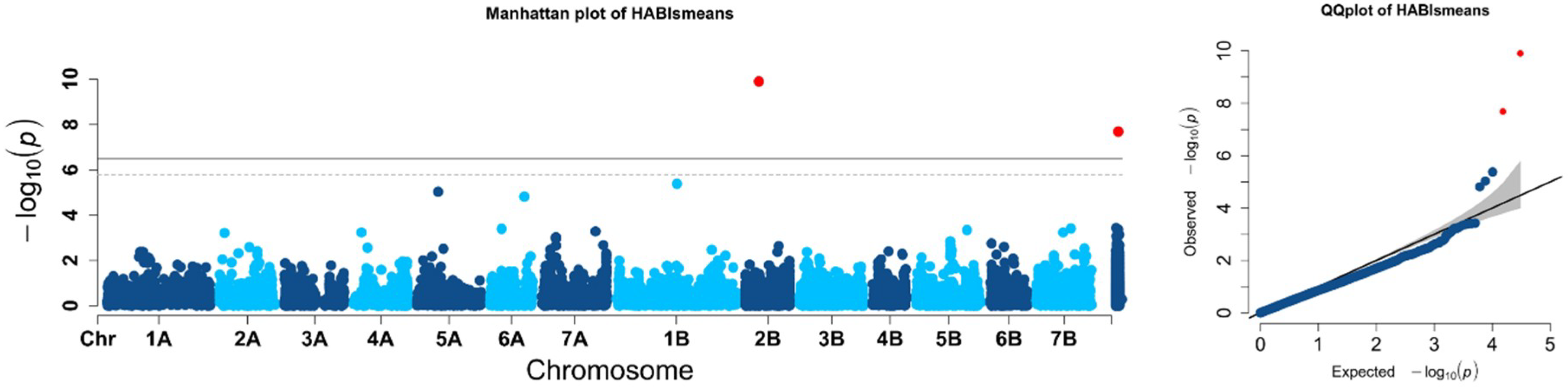
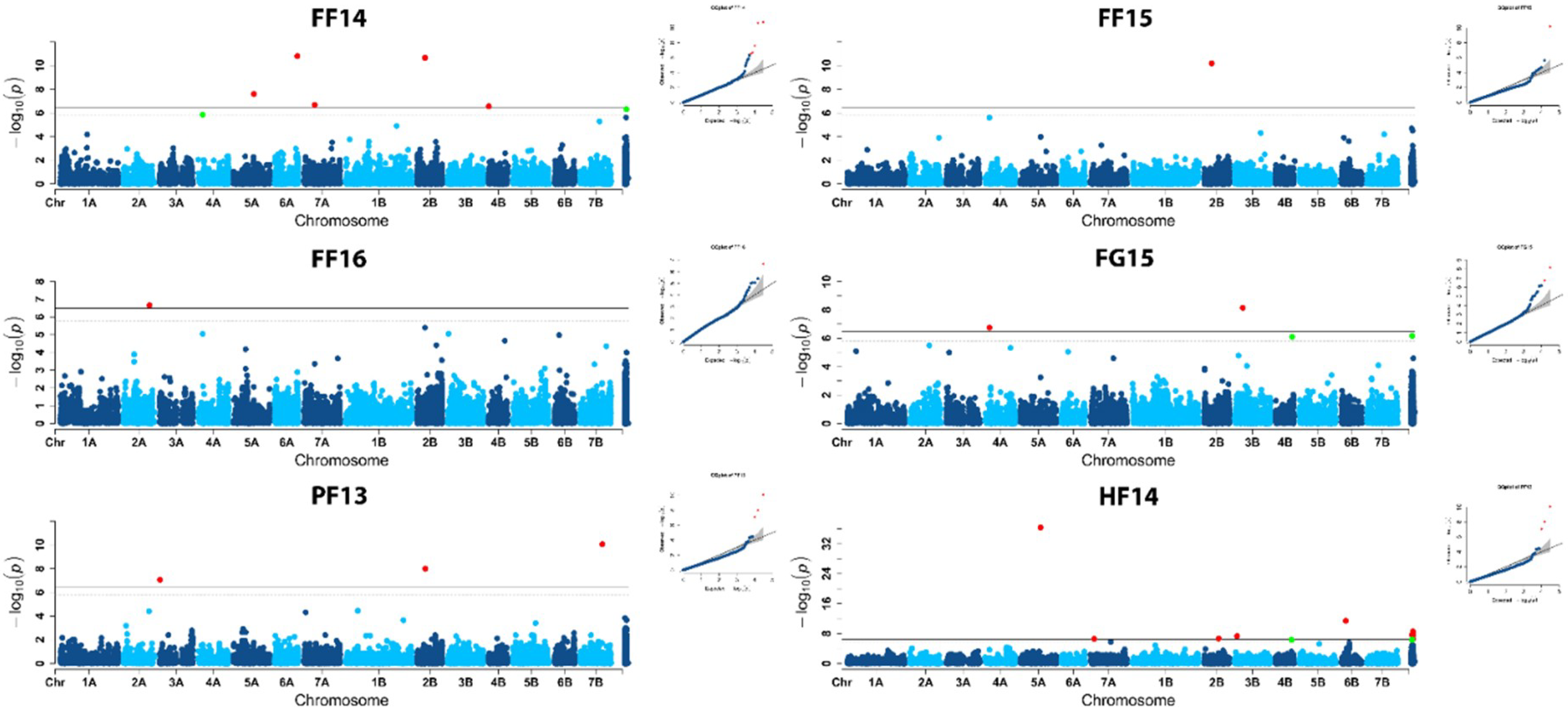
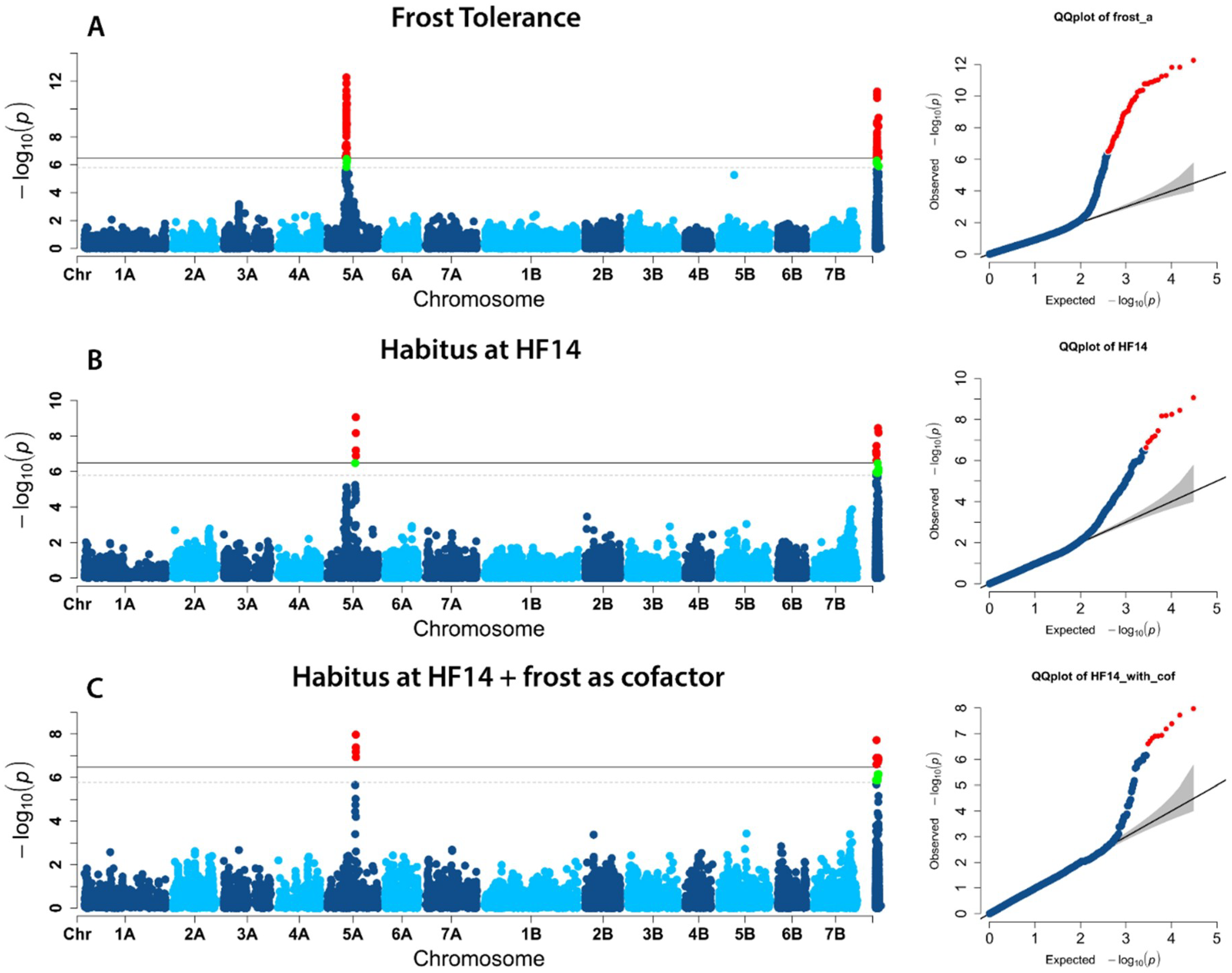
2.2. Population Structure and Association Mapping
2.3. Candidate genes
2.4. Search for Orthologs of the Rice Genes PROG1, LAZY1, TAC1 and SD1
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Field and Greenhouse Trials and Phenotyping
4.3. Statistical Analyses
4.4. Population Structure and GWA Analyses
4.5. Identification of Candidate Genes
4.6. Search for Orthologous of the Rice Genes PROG1, TAC1, LAZY1, SD1 in Wheat
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sakamoto, T.; Matsuoka, M. Generating high-yielding varieties by genetic manipulation of plant architecture. Curr. Opin. Biotechnol. 2004, 15, 144–147. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. Green revolution genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef]
- Donald, C.M. The breeding of crop ideotypes. Euphytica 1968, 17, 385–403. [Google Scholar] [CrossRef]
- Intergovernmental Panel on Climate Change. Climate Change 2014 Mitigation of Climate Change; Cambridge University Press: Cambridge, UK, 2015; ISBN 9781107415416. [Google Scholar]
- De Vita, P.; Taranto, F. Durum wheat (Triticum turgidum ssp. durum) breeding to meet the challenge of climate change. In Advances in Plant Breeding Strategies: Cereals; Al-Khayri, J.M., Jain, S., Johnson, M.D.V., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 5, pp. 471–524. [Google Scholar]
- De Vita, P.; Colecchia, S.A.; Pecorella, I.; Saia, S. Reduced inter-row distance improves yield and competition against weeds in a semi-dwarf durum wheat variety. Eur. J. Agron. 2017, 85, 69–77. [Google Scholar] [CrossRef]
- Hussien, A.; Tavakol, E.; Horner, D.S.; Muñoz-Amatriaín, M.; Muehlbauer, G.J.; Rossini, L. Genetics of tillering in rice and barley. Plant Genome 2014, 7. [Google Scholar] [CrossRef]
- Giunta, F.; De Vita, P.; Mastrangelo, A.M.; Sanna, G.; Motzo, R. Environmental and genetic variation for yield-related traits of durum wheat as affected by development. Front. Plant Sci. 2018, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Baum, M.; Grando, S.; Backes, G.; Jahoor, A.; Sabbagh, A.; Ceccarelli, S. QTLs for agronomic traits in the Mediterranean environment identified in recombinant inbred lines of the cross ‘Arta’ × H. spontaneum 41-1. Theor. Appl. Genet. 2003, 107, 1215–1225. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Kamiunten, H.; Ogawa, T.; Tsuzuki, E.; Terao, H.; Lin, D.; Matsuo, M. Mapping of QTLs for leaf developmental behavior in rice (Oryza sativa L.). Euphytica 2004, 138, 169–175. [Google Scholar] [CrossRef]
- Xu, Y.; McCouch, S.R.; Shen, Z. Transgressive segregation of tiller angle in rice caused by complementary gene action. Crop Sci. 1998, 38, 12–19. [Google Scholar] [CrossRef]
- Yu, C.Y.; Liu, Y.Q.; Jiang, L.; Wang, C.M.; Zhai, H.Q.; Wan, J.M. QTLs mapping and genetic analysis of tiller angle in rice (Oryza sativa L.). Yi Chuan Xue Bao 2005, 32, 948–954. [Google Scholar]
- Jin, J.; Huang, W.; Gao, J.P.; Yang, J.; Shi, M.; Zhu, M.Z.; Luo, D.; Lin, H.X. Genetic control of rice plant architecture under domestication. Nat. Genet. 2008, 40, 1365–1369. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, Y.; Qian, Q.; Fu, Z.; Wang, M.; Zeng, D.; Li, B.; Wang, X.; Li, J. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 2007, 17, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Li, X.; Liu, F.; Sun, X.; Li, C.; Zhu, Z.; Fu, Y.; Cai, H.; Wang, X.; Xie, D.; et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 2008, 40, 1360–1364. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Lin, Z.; Li, H.; Li, X.; Li, J.; Wang, Y.; Zhang, X.; Zhu, Z.; Zhai, W.; Wang, X.; et al. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 2007, 52, 891–898. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, X.; Moolhuijzen, P.; Bellgard, M.; Lance, R.; Appels, R.; Li, C. Towards isolation of the barley green revolution gene. In Proceedings of the 12th Australian Barley Technical Symposium, Hobart, Australia, 11–14 September 2005. [Google Scholar]
- Kuczyńska, A.; Mikołajczak, K.; Ćwiek, H. Pleiotropic effects of the sdw1 locus in barley populations representing different rounds of recombination. Electron. J. Biotechnol. 2014, 17, 217–223. [Google Scholar] [CrossRef]
- Talamè, V.; Sanguineti, M.; Chiapparino, E.; Bahri, H.; Ben Salem, A.; Forster, B.; Ellis, R.; Rhouma, S.; Zoumarou, W.; Waugh, R.; et al. Identification of Hordeum spontaneum QTL alleles improving field performance of barley grown under rainfed conditions. Ann. Appl. Biol. 2004, 144, 309–319. [Google Scholar] [CrossRef]
- Li, W.L.; Nelson, J.C.; Chu, C.Y.; Shi, L.H.; Huang, S.H.; Liu, D.J. Chromosomal locations and genetic relationships of tiller and spike characters in wheat. Euphytica 2002, 125, 357–366. [Google Scholar] [CrossRef]
- Liu, Y.; Lin, Y.; Gao, S.; Li, Z.; Ma, J.; Deng, M.; Chen, G.; Wei, Y.; Zheng, Y. A genome-wide association study of 23 agronomic traits in Chinese wheat landraces. Plant J. 2017, 91, 861–873. [Google Scholar] [CrossRef]
- Giraldo, P.; Royo, C.; González, M.; Carrillo, J.M.; Ruiz, M. Genetic diversity and association mapping for agromorphological and grain quality traits of a structured collection of durum wheat landraces including subsp. durum, turgidum and diccocon. PLoS ONE 2016, 11, e0166577. [Google Scholar] [CrossRef]
- Jia, Q.; Zhang, X.-Q.; Westcott, S.; Broughton, S.; Cakir, M.; Yang, J.; Lance, R.; Li, C. Expression level of a gibberellin 20-oxidase gene is associated with multiple agronomic and quality traits in barley. Theor. Appl. Genet. 2011, 122, 1451–1460. [Google Scholar] [CrossRef]
- Hashjin, J.M. Genotypic and Environmental Effects on Growth Habit in Wheat (Triticum aestivum L.): A Thesis Presented in Partial Fulfilment of the Requirements for the Degree of Master of Agricultural Science in Plant Science (Plant Breeding) at Massey University. Ph.D. Thesis, Massey University, Palmerston North, New Zealand, 1992. [Google Scholar]
- Reynolds, M.P.; Ortiz-Monasterio, J.I.; McNab, A. Application of Physiology in Wheat Breeding; CIMMYT: Mexico City, Mexico, 2001; ISBN 970-648-077-3. [Google Scholar]
- Roberts, D.W.A. Identification of loci on chromosome 5A of wheat involved in control of cold hardiness, vernalization, leaf length, rosette growth habit, and height of hardened plants. Genome 1990, 33, 247–259. [Google Scholar] [CrossRef]
- Nishio, K.; Nakagawa, M.; Watanabe, S.; Gocho, H. Studies on the growth habit of wheat varieties. 4. Relation between temperature effect and photoperiodism and its effect on the growth habit of wheat. Ikushugaku Zasshi 1962, 12, 101–107. [Google Scholar] [CrossRef]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genome J. 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Canè, M.A.; Maccaferri, M.; Nazemi, G.; Salvi, S.; Francia, R.; Colalongo, C.; Tuberosa, R. Association mapping for root architectural traits in durum wheat seedlings as related to agronomic performance. Mol. Breed. 2014, 34, 1629–1645. [Google Scholar] [CrossRef]
- Maccaferri, M.; El-Feki, W.; Nazemi, G.; Salvi, S.; Canè, M.A.; Colalongo, M.C.; Stefanelli, S.; Tuberosa, R. Prioritizing quantitative trait loci for root system architecture in tetraploid wheat. J. Exp. Bot. 2016, 67, 1161–1178. [Google Scholar] [CrossRef] [PubMed]
- Sabiel, S.A.I.; Huang, S.; Hu, X.; Ren, X.; Fu, C.; Peng, J.; Sun, D. SNP-based association analysis for seedling traits in durum wheat (Triticum turgidum L. durum (Desf.)). Breed. Sci. 2017, 67. [Google Scholar] [CrossRef]
- Laidò, G.; Mangini, G.; Taranto, F.; Gadaleta, A.; Blanco, A.; Cattivelli, L.; Marone, D.; Mastrangelo, A.M.; Papa, R.; De Vita, P. Genetic diversity and population structure of tetraploid wheats (Triticum turgidum L.) estimated by SSR, DArT and pedigree data. PLoS ONE 2013, 8, e67280. [Google Scholar]
- Marone, D.; Laidò, G.; Gadaleta, A.; Colasuonno, P.; Ficco, D.B.M.; Giancaspro, A.; Giove, S.; Panio, G.; Russo, M.A.; De Vita, P.; et al. A high-density consensus map of A and B wheat genomes. Theor. Appl. Genet. 2012, 125, 1619–1638. [Google Scholar] [CrossRef]
- Liu, W.; Maccaferri, M.; Rynearson, S.; Letta, T.; Zegeye, H.; Tuberosa, R.; Chen, X.; Pumphrey, M. Novel sources of stripe rust resistance identified by genome-wide association mapping in Ethiopian durum wheat (Triticum turgidum ssp. durum). Front. Plant Sci. 2017, 8, 774. [Google Scholar] [CrossRef]
- Laidò, G.; Panio, G.; Marone, D.; Russo, M.A.; Ficco, D.B.M.; Giovanniello, V.; Cattivelli, L.; Steffenson, B.; De Vita, P.; Mastrangelo, A.M. Identification of new resistance loci to African stem rust race TTKSK in tetraploid wheats based on linkage and genome-wide association mapping. Front. Plant Sci. 2015, 6, 1033. [Google Scholar]
- Aoun, M.; Breiland, M.; Kathryn Turner, M.; Loladze, A.; Chao, S.; Xu, S.S.; Ammar, K.; Anderson, J.A.; Kolmer, J.A.; Acevedo, M. Genome-wide association mapping of leaf rust response in a durum wheat worldwide germplasm collection. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Ghavami, F.; Elias, E.M.; Mamidi, S.; Ansari, O.; Sargolzaei, M.; Adhikari, T.; Mergoum, M.; Kianian, S.F.; De Koning, D.-J. Mixed model association mapping for Fusarium head blight resistance in Tunisian-derived durum wheat populations. Genetics 2011, 1, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Laidò, G.; Marone, D.; Russo, M.A.; Colecchia, S.A.; Mastrangelo, A.M.; De Vita, P.; Papa, R. Linkage disequilibrium and genome-wide association mapping in tetraploid wheat (Triticum turgidum L.). PLoS ONE 2014, 9, e95211. [Google Scholar]
- Mengistu, D.K.; Kidane, Y.G.; Catellani, M.; Frascaroli, E.; Fadda, C.; Pè, M.E.; Dell’Acqua, M. High-density molecular characterization and association mapping in Ethiopian durum wheat landraces reveals high diversity and potential for wheat breeding. Plant Biotechnol. J. 2016, 14, 1800–1812. [Google Scholar] [CrossRef]
- Soriano, J.M.; Malosetti, M.; Roselló, M.; Sorrells, M.E.; Royo, C. Dissecting the old Mediterranean durum wheat genetic architecture for phenology, biomass and yield formation by association mapping and QTL meta-analysis. PLoS ONE 2017, 12, e0178290. [Google Scholar] [CrossRef] [PubMed]
- Reimer, S.; Pozniak, C.J.; Clarke, F.R.; Clarke, J.M.; Somers, D.J.; Knox, R.E.; Singh, A.K. Association mapping of yellow pigment in an elite collection of durum wheat cultivars and breeding lines. Genome 2008, 51, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Sieber, A.-N.; Longin, C.F.H.; Würschum, T. Molecular characterization of winter durum wheat (Triticum durum) based on a genotyping-by-sequencing approach. Plant Genet. Resour. 2015, 15, 36–44. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Ricci, A.; Salvi, S.; Milner, S.G.; Noli, E.; Martelli, P.L.; Casadio, R.; Akhunov, E.; Scalabrin, S.; Vendramin, V.; et al. A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol. J. 2015, 13, 648–663. [Google Scholar] [CrossRef] [PubMed]
- Le Gouis, J.; Bordes, J.; Ravel, C.; Heumez, E.; Faure, S.; Praud, S.; Galic, N.; Remoué, C.; Balfourier, F.; Allard, V.; et al. Genome-wide association analysis to identify chromosomal regions determining components of earliness in wheat. Theor. Appl. Genet. 2012, 124, 597–611. [Google Scholar] [CrossRef]
- Lowe, I.; Jankuloski, L.; Chao, S.; Chen, X.; See, D.; Dubcovsky, J. Mapping and validation of QTL which confer partial resistance to broadly virulent post-2000 North American races of stripe rust in hexaploid wheat. Theor. Appl. Genet. 2011, 123, 143–157. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Singh, P.K.; Dreisigacker, S.; Singh, S.; Lillemo, M.; Duveiller, E. Dwarfing genes Rht-B1b and Rht-D1b are associated with both type I FHB susceptibility and low anther extrusion in two bread wheat populations. PLoS ONE 2016, 11, e0162499. [Google Scholar] [CrossRef]
- Didon, U.M.E. Variation between barley cultivars in early response to weed competition. J. Agron. Crop. Sci. 2002, 188, 176–184. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Bajaj, D.; Srivastava, R.; Daware, A.; Basu, U.; Tripathi, S.; Bharadwaj, C.; Tyagi, A.K.; Parida, S.K. Genetic dissection of plant growth habit in chickpea. Funct. Integr. Genom. 2017, 17, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Limin, A.E.; Fowler, D.B. Morphological and cytological characters associated with low-temperature tolerance in wheat (Triticum aestivum L. em Thell.). Can. J. Plant Sci. 2000, 80, 687–692. [Google Scholar] [CrossRef]
- Cheng, K.; Zhao, R.; Jiang, Z.; Bie, T.; Zang, S.; Zhang, B.; Wu, X.; Gao, D.; Wu, R. Quantitative trait loci responsible for sharp eyespot resistance in common wheat CI12633. Sci. Rep. 2017, 7, 11799. [Google Scholar]
- Jing, H.C.; Bayon, C.; Kanyuka, K.; Berry, S.; Wenzl, P.; Huttner, E.; Kilian, A.; Hammond-Kosack, K.E. DArT markers: Diversity analyses, genomes comparison, mapping and integration with SSR markers in Triticum monococcum. BMC Genom. 2009, 10, 458. [Google Scholar] [CrossRef]
- Rostoks, N.; Ramsay, L.; MacKenzie, K.; Cardle, L.; Bhat, P.R.; Roose, M.L.; Svensson, J.T.; Stein, N.; Varshney, R.K.; Marshall, D.F.; et al. Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. Proc. Natl. Acad. Sci. USA 2006, 103, 18656–18661. [Google Scholar] [CrossRef]
- Borghi, L.; Kang, J.; Ko, D.; Lee, Y.; Martinoia, E. The role of ABCG-type ABC transporters in phytohormone transport. Biochem. Soc. Trans. 2015, 43, 924–930. [Google Scholar] [CrossRef]
- Agarwal, P.; Kapoor, S.; Tyagi, A.K. Transcription factors regulating the progression of monocot and dicot seed development. BioEssays 2011, 33, 189–202. [Google Scholar] [CrossRef]
- Sharma, M.; Pandey, G.K. Expansion and function of repeat domain proteins during stress and development in plants. Front. Plant Sci. 2016, 6, 1218. [Google Scholar] [CrossRef] [PubMed]
- Kuczyńska, A.; Kosmala, A.; Surma, M.; Adamski, T. Identification of tillering node proteins differentially accumulated in barley recombinant inbred lines with different juvenile growth habits. Int. J. Mol. Sci. 2012, 13, 10410–10423. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, T.; Iino, M. Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and-independent gravity signaling pathways. Plant Cell Physiol. 2007, 48, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Li, C.; Shang, Y.; Zhu, J.; Hua, W.; Wang, J.; Yang, J.; Zhang, G. Molecular characterization and functional analysis of barley semi-dwarf mutant Riso no. 9265. BMC Genom. 2015, 16, 927. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.J.; Zhang, J.J.; Westcott, S.; Zhang, X.Q.; Bellgard, M.; Lance, R.; Li, C.D. GA-20 oxidase as a candidate for the semidwarf gene sdw1/denso in barley. Funct. Integr. Genom. 2009, 9, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, J.C.; Chang, T.; Konzak, C. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- UPOV. Durum Wheat. Guidelines for The Conduct of Tests for Distinctness, Uniformity and Stability; International Union for the Protection of New Varieties of Plants: Geneva, Switzerland, 2012. [Google Scholar]
- Schabenberger, O. Introducing the GLIMMIX procedure for generalized linear mixed models. In Proceedings of the Thirtieth Annual SAS Users Group International Conference, Philadelphia, PA, USA, 10–13 April 2005. [Google Scholar]
- Piepho, H.P.; Mohring, J.; Melchinger, A.E.; Buchse, A. BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 2008, 161, 209–228. [Google Scholar] [CrossRef]
- Piepho, H.-P.; Mohring, J. Computing heritability and selection response from unbalanced plant breeding trials. Genetics 2007, 177, 1881–1888. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; Von Holdt, B.M. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Zhang, Z.; Ersoz, E.; Lai, C.-Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Tian, F.; Pan, Y.; Buckler, E.S.; Zhang, Z. A super powerful method for genome wide association study. PLoS ONE 2014, 9, e107684. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Segura, V.; Vilhjálmsson, B.J.; Platt, A.; Korte, A.; Seren, Ü.; Long, Q.; Nordborg, M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 2012, 44, 825–830. [Google Scholar] [CrossRef]
- Miao, C.; Yang, J.; Schnable, J.C. Optimising the identification of causal variants across varying genetic architectures in crops. Plant Biotechnol. J. 2019, 17, 893–905. [Google Scholar] [CrossRef]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef]
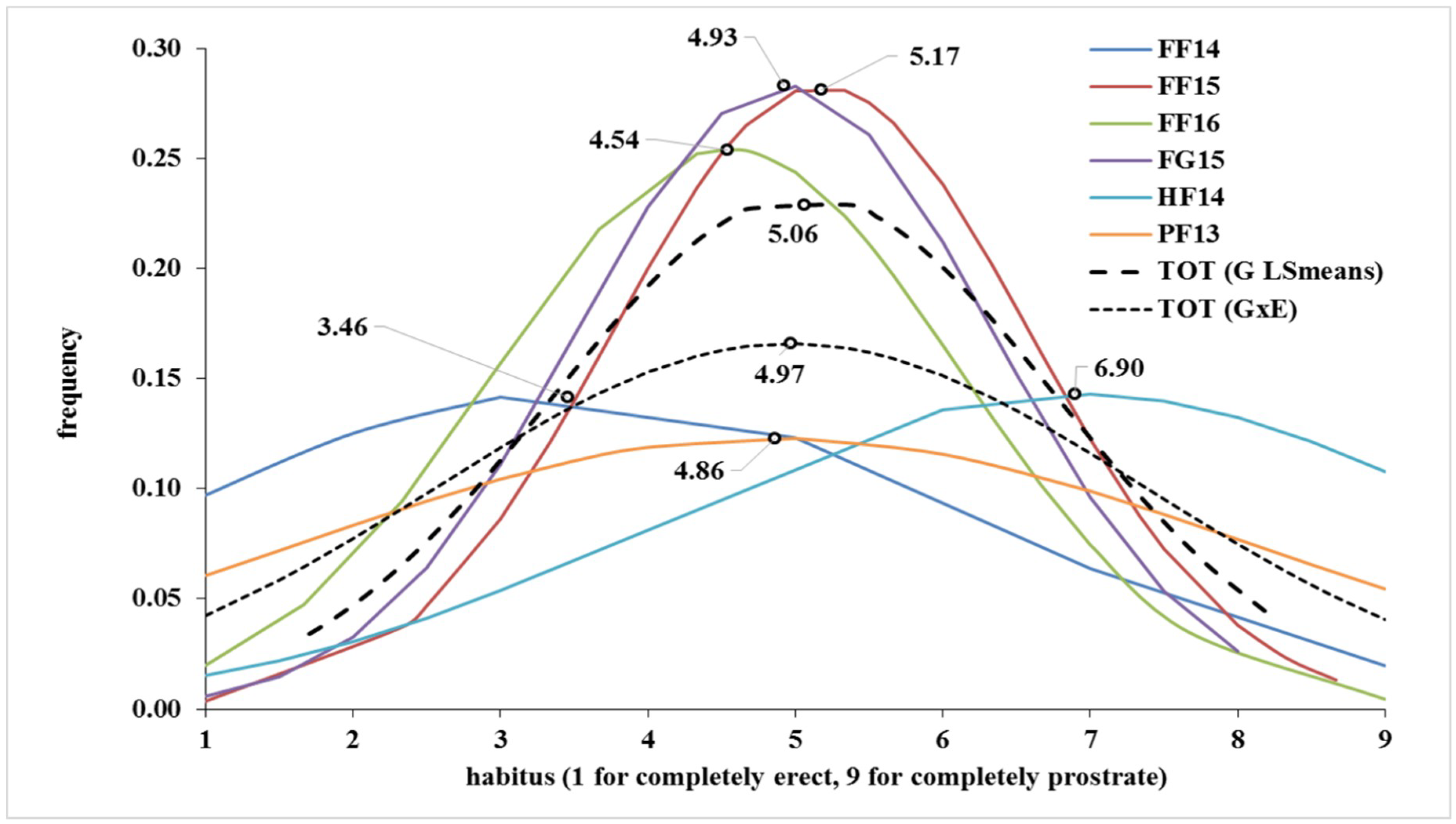

| FF14 | FF15 | FF16 | FG15 | HF14 | PF13 | TOT (G LSmeans) | TOT (G × E) | |
|---|---|---|---|---|---|---|---|---|
| Mean | 3.46 | 5.17 | 4.54 | 4.93 | 6.90 | 4.86 | 5.06 | 4.97 |
| Standard Error | 0.25 | 0.10 | 0.12 | 0.11 | 0.25 | 0.27 | 0.13 | 0.08 |
| Coefficient of Variation | 80.4 | 27.3 | 34.5 | 28.6 | 40.4 | 66.8 | 33.9 | 48.4 |
| σ2G | n.a. | 5.05 | 7.35 | 3.65 | 15.55 | 14.02 | n.a. | 30.29 |
| F | n.a. | 4.05 | 16.26 | 18.46 | 45.38 | 9.94 | n.a. | 10.69 |
| P | n.a. | <0001 | <0001 | <0001 | <0001 | <0001 | n.a. | <0001 |
| Kurtosis | −1.01 | −0.37 | −0.43 | −0.08 | −0.12 | −1.78 | −1.40 | −0.93 |
| Skewness | 0.61 | 0.06 | −0.46 | −0.21 | −1.28 | 0.08 | −0.28 | −0.06 |
| Min | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.7 | 1.0 |
| Max | 9.0 | 8.7 | 9.0 | 8.0 | 9.0 | 9.0 | 8.2 | 9.0 |
| Range | 8.0 | 7.7 | 8.0 | 7.0 | 8.0 | 8.0 | 6.5 | 8.0 |
| h2B with covariance structure (Mixed Model) | n.a. | 0.997 | 0.998 | 0.999 | 0.999 | 0.991 | n.a. | 0.910 |
| h2B with fixed effects (GLM) | n.a. | 0.941 | 0.903 | 0.999 | 0.978 | 0.793 | n.a. | 0.652 |
| Source of Variation | df | Habit MS | p | Cov |
|---|---|---|---|---|
| Environment (E) | 5 | 92.005 | <0001 | 0.315 |
| Blocks within Environment | 7 | 2.080 | 0.0048 | 0.010 |
| Genotype (G) | 182 | 30.287 | <0001 | 3.124 |
| G × E | 899 | 3.082 | <0001 | 1.191 |
| Error | 1229 | 0.708 | - | 0.708 |
| Marker | Environment | Chr | cM | p Value | PEV * | Literature |
|---|---|---|---|---|---|---|
| S1133336 | FF16 | 2A | 217.7 | 2.22 × 10−07 | 0.15 | |
| D1202558 | FF14 | 2B | 62.3 | 2.15 × 10−11 | 0.27 | Ppd-B1 |
| FF15 | 2B | 62.3 | 6.30 × 10−11 | 0.10 | Ppd-B1 | |
| LSmeans | 2B | 62.3 | 1.26 × 10−10 | 0.16 | Ppd-B1 | |
| D2294169 | PF13 | 2B | 65.1 | 9.91 × 10−09 | 0.12 | Ppd-B1 |
| D1137224 | HF14 | 2B | 120.3 | 2.07 × 10−07 | 0.05 | |
| D1271842 | PF13 | 3A | 2.7 | 8.54 × 10−08 | 0.11 | |
| D1266232 | HF14 | 3B | 23.9 | 4.73 × 10−08 | 0.04 | |
| S1049173 | FG15 | 3B | 71.8 | 7.11 × 10−09 | 0.07 | |
| D1665929 | FF14 | 4A | 37.2 | 1.39 × 10−06 | 0.24 | |
| FG15 | 4A | 37.2 | 1.80 × 10−07 | 0.06 | ||
| D1110414 | FF14 | 4B | 0 | 2.77 × 10−07 | 0.25 | |
| D1395268 | HF14 | 4B | 133.6 | 4.83 × 10−07 | 0.01 | Vrn-B2 |
| D1720107 | FG15 | 4B | 138.4 | 7.89 × 10−07 | 0.07 | Vrn-B2 |
| D2276320 | HF14 | 5A | 167.7 | 4.51 × 10−07 | 0.51 | Vrn-A1 |
| D1721703 | FF14 | 5A | 168.6 | 2.48 × 10−08 | 0.03 | Vrn-A1 |
| D1076422 | FF14 | 6A | 188.7 | 1.57 × 10−11 | 0.20 | |
| D2289020 | HF14 | 6B | 36.8 | 3.92 × 10−12 | 0.22 | |
| D2295851 | HF14 | 7A | 32.3 | 2.74 × 10−07 | 0.08 | |
| D1031337 | FF14 | 7A | 91.8 | 2.05 × 10−07 | 0.22 | Vrn-A3 |
| D1112046 | PF13 | 7B | 184.4 | 8.66 × 10−11 | 0.08 | |
| D4004513 | HF14 | unmapped | 5.56 × 10−07 | 0.18 | ||
| D1744736 | HF14 | unmapped | 2.28 × 10−08 | 0.19 | ||
| D3944539 | FG15 | unmapped | 6.76 × 10−07 | 0.04 | ||
| D3946194 | HF14 | unmapped | 3.54 × 10−07 | 0.18 | ||
| D2277949 | LSmeans | unmapped | 2.07 × 10−08 | 0.21 | ||
| D3935715 | HF14 | unmapped | 2.60 × 10−09 | 0.12 | ||
| D3533805 | HF14 | unmapped | 2.74 × 10−07 | 0.08 | ||
| S984195 | FF14 | unmapped | 4.83 × 10−07 | 0.03 | ||
| S1218298 | HF14 | unmapped | 2.50 × 10−08 | 0.10 |
| MTA | chr | CI (cM) | CI Start (bp) | CI End (bp) | CI (Mbp) | Number of Annotated Genes | Number of Related-Growth Habit Genes |
|---|---|---|---|---|---|---|---|
| S1133336 | 2A | 217.5–219.7 | 734724817 | 741838496 | 7.1 | 97 | 7 |
| D1202558 | 2B | 60.3–64.7 | 45229865 | 51452747 | 6.2 | 38 | 3 |
| D1137224 | 2B | 117.7–124.3 | 117320738 | 149930630 | 32.6 | 73 | 5 |
| D1271842 | 3A | 0.6–6.5 | 2640850 | 15387176 | 12.7 | 58 | 1 |
| D1266232 | 3B | 19.7–29.4 | 4996463 | 17173269 | 12.2 | 58 | 4 |
| S1049173 | 3B | 68.2–75.3 | 45389928 | 63515801 | 18.1 | 49 | 3 |
| D1665929 | 4A | 37.1–39.8 | 47676808 | 77735733 | 30 | 107 | 7 |
| D1110414 | 4B | 0–3 | 2180245 | 13531313 | 11.3 | 113 | 9 |
| D1395268 | 4B | 132.4–138 | 657599661 | 661590000 | 5.6 | 44 | 2 |
| D2276320 | 5A | 164.3–168.9 | 575035656 | 590239189 | 15.2 | 140 | 14 |
| D1076422 | 6A | 185.2–191.1 | 607923652 | 618547327 | 10.6 | 129 | 12 |
| D2289020 | 6B | 35.5–36.8 | 32844660 | 47681687 | 14.8 | 104 | 5 |
| D2295851 | 7A | 31.4–38 | 15519984 | 23593672 | 8.1 | 35 | 2 |
| D1031337 | 7A | 91.2–92.4 | 50882080 | 56636443 | 5.7 | 57 | 6 |
| D1112046 | 7B | 181.9–188.8 | 680402861 | 698138699 | 17.7 | 35 | 3 |
| MTA | chr | CI (cM) | CI Start (bp) | CI End (bp) | CI (Mbp) | Number of Annotated Genes | Number of Related-Growth Habit Genes |
|---|---|---|---|---|---|---|---|
| S1133336 | 2A | 217.5–219.7 | 739004624 | 746220682 | 7.2 | 202 | 10 |
| D1202558 | 2B | 60.3–64.7 | 44096854 | 49732398 | 5.6 | 148 | 7 |
| D1137224 | 2B | 117.7–124.3 | 109991884 | 143184509 | 33.2 | 358 | 20 |
| D1271842 | 3A | 0.6–6.5 | 963147 | 7004107 | 6 | 196 | 5 |
| D1266232 | 3B | 19.7–29.4 | 4675018 | 15213712 | 10.5 | 235 | 13 |
| S1049173 | 3B | 68.2–75.3 | 49798003 | 61284099 | 11.5 | 178 | 9 |
| D1665929 | 4A | 37.1–39.8 | 44642299 | 76324355 | 31.7 | 461 | 10 |
| D1110414 | 4B | 0–3 | 1729305 | 13558565 | 11.8 | 281 | 11 |
| D1395268 | 4B | 132.4–138 | 658918900 | 659539176 | 0.6 | 33 | 1 |
| D2276320 | 5A | 164.3–168.9 | 532806577 | 556694196 | 23.9 | 419 | 14 |
| D1076422 | 6A | 185.2–191.1 | 602368652 | 614383914 | 12 | 325 | 17 |
| D2289020 | 6B | 35.5–36.8 | 30090166 | 54319106 | 24.2 | 319 | 13 |
| D2295851 | 7A | 31.4–38 | 18289584 | 27148666 | 8.8 | 240 | 9 |
| D1031337 | 7A | 91.2–92.4 | 55718952 | 60630163 | 4.9 | 127 | 5 |
| D1112046 | 7B | 181.9–188.8 | 657142046 | 679568205 | 22.4 | 326 | 10 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marone, D.; Rodriguez, M.; Saia, S.; Papa, R.; Rau, D.; Pecorella, I.; Laidò, G.; Pecchioni, N.; Lafferty, J.; Rapp, M.; et al. Genome-Wide Association Mapping of Prostrate/Erect Growth Habit in Winter Durum Wheat. Int. J. Mol. Sci. 2020, 21, 394. https://doi.org/10.3390/ijms21020394
Marone D, Rodriguez M, Saia S, Papa R, Rau D, Pecorella I, Laidò G, Pecchioni N, Lafferty J, Rapp M, et al. Genome-Wide Association Mapping of Prostrate/Erect Growth Habit in Winter Durum Wheat. International Journal of Molecular Sciences. 2020; 21(2):394. https://doi.org/10.3390/ijms21020394
Chicago/Turabian StyleMarone, Daniela, Monica Rodriguez, Sergio Saia, Roberto Papa, Domenico Rau, Ivano Pecorella, Giovanni Laidò, Nicola Pecchioni, Julia Lafferty, Matthias Rapp, and et al. 2020. "Genome-Wide Association Mapping of Prostrate/Erect Growth Habit in Winter Durum Wheat" International Journal of Molecular Sciences 21, no. 2: 394. https://doi.org/10.3390/ijms21020394
APA StyleMarone, D., Rodriguez, M., Saia, S., Papa, R., Rau, D., Pecorella, I., Laidò, G., Pecchioni, N., Lafferty, J., Rapp, M., Longin, F. H., & De Vita, P. (2020). Genome-Wide Association Mapping of Prostrate/Erect Growth Habit in Winter Durum Wheat. International Journal of Molecular Sciences, 21(2), 394. https://doi.org/10.3390/ijms21020394









